Abstract
Introduction
Bacterial surface display is a valuable biotechnology technique for presenting proteins and molecules on the outer surface of bacterial cells. However, it has limitations, including potential toxicity to host bacteria and variability in display efficiency. To address these issues, we investigated the removal of abundant non-essential outer membrane proteins (OMPs) in E. coli as a new strategy to improve the surface display of recombinant proteins.
Methods
We targeted OmpA, a highly prevalent OMP in E. coli, using the lambda red method. We successfully knocked out ompA in two E. coli strains, K-12 MG1655 and E. coli BL-21, which have broad research and therapeutic applications. We then combined ompA knockout strains and two OMPs with three therapeutic proteins including an anti-toxin enzyme (ClbS), interleukin 18 (IL-18) for activating cytotoxic T cells and an anti- CTLA4 nanobody (αCTLA4) for immune checkpoint blockade.
Results
A total of six different display constructs were tested for their display levels by flow cytometry, showing that the ompA knockout strains increased the percentage as well as the levels of display in bacteria compared to those of isogenic wild-type strains.
Conclusions
By removing non-essential, highly abundant surface proteins, we develop an efficient platform for displaying enzymes and antibodies, with potential industrial and therapeutic applications. Additionally, the enhanced therapeutic efficacy opens possibilities for live bacteria-based therapeutics, expanding the technology’s relevance in the field.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The bacterial surface display is a cutting-edge technique in biotechnology and molecular biology that involves the presentation of proteins or peptides on the surface of bacterial cells, of which Escherichia coli represents the primary technology platform [1]. To date, the bacterial surface display has garnered significant attention due to its wide range of applications in areas such as vaccine development [2], screening of peptide libraries [3], and catalysis [4]. Advantages of bacterial surface display have included ease of genetic manipulation, cost-effectiveness compared to mammalian cell surface display, and rapid screening of large peptide or protein libraries, which is invaluable in drug development and protein engineering [5,6,7,8,9].
The bacterial surface display has a relatively long history. The concept of bacterial surface display likely traces its roots back to the 1980s when scientists began to understand protein targeting and export mechanisms in bacteria [10]. This period saw significant advancements in genetic engineering and recombinant DNA technology, setting the stage for more complex applications like protein display. The first instances of the bacterial surface display were developed in the late 1980s to early 1990s. Researchers started to fuse foreign proteins or peptides to bacterial surface proteins. One of the early systems used was the fusion of proteins to the outer membrane protein (OMP) LamB of E. coli [11]. This pioneering work demonstrated that foreign proteins could be displayed on the surface of bacteria while retaining their functionality. In the late 1990s, the concept began to be applied in diverse fields such as vaccine development, where surface-displayed antigens could directly stimulate immune responses, and in bio-catalysis, where enzymes displayed on bacterial surfaces were used for industrial processes [12]. In the 2000s, with advancements in genomic and proteomic technologies, bacterial surface display systems became more sophisticated. This period saw the integration of high throughput screening methods, allowing for the rapid analysis and optimization of displayed proteins [13]. Recent developments have focused on enhancing the stability and expression levels of displayed proteins and expanding the range of applications. The technique is now a crucial tool in areas like antibody engineering, biosensor development, and environmental applications. Throughout its history, bacterial surface display has evolved from a novel concept to a widely used technology, demonstrating the power of combining molecular biology with innovative biotechnological approaches. The future of this field holds promise for further groundbreaking applications and improvements.
While bacterial surface display offers numerous advantages in terms of efficiency, cost, and versatility, it also faces challenges related to suboptimal display, improper protein folding, size limitations, and post-translational modifications. Specifically, proteins displayed on bacterial surfaces may not be present at high levels due to possible physical constraints, potentially affecting their functionality. Additionally, there is a limit to the size of the proteins or peptides that can be effectively displayed on the bacterial surface although different OMPs differ in their capacity to display proteins of various sizes [14,15,16]. Moreover, compared to the yeast surface display, bacteria lack the sophisticated post-translational modification systems found in eukaryotic cells, which can be crucial for the functionality of antibodies [17]. To address the limitations in surface levels and functionality of displayed proteins, ongoing efforts have been largely centered around identifying different OMPs as anchor protein scaffolds in a trial-and-error process [18,19,20,21]. This is because different recombinant target proteins have preference for the choice of anchor protein, and it is important to select an anchor protein that ensures stable and efficient display of the target protein on the bacterial surface. Previously, four genes encoding OMPs (OmpA, OmpC, OmpF, and LamB) were deleted in a series of single, double, triple, or quadruple knock-outs [22]. While these knockout strains were used to improve production and purification of recombinant OMPs, it provides an alternative approach to improving the bacterial surface display. Inspired by this work, the current study aims to alleviate constraints on the surface of E. coli by knocking out a nonessential yet highly abundant OMP. This strategy could potentially serve as a universal method to enhance bacterial surface display. Given that E. coli possesses one of the most well-documented proteomes, we conducted a literature survey to identify OMPs with characterized copy numbers and abundances. It is estimated that there are approximately 105 copies of MulI, 105 copies of OmpA, 104 copies of OmpC, and 104 copies of OmpF per bacterium [23, 24]. Notably, we selected OmpA as the candidate OMP for its extensive characterization in the literature. Our goal is to demonstrate whether knocking out OmpA enhances the surface display of heterologous recombinant proteins in E. coli.
Materials and Methods
Chemicals and Reagent
All chemicals and bacterial culture medium were purchased from Fisher Scientific International Inc. (Cambridge, MA, USA) unless otherwise noted, and were of the highest purity or analytical grade commercially available. Competent cells were purchased from New England Biolabs (Ipswich, MA, USA). PurePlasmid Miniprep Kits were purchased from CWBIO (Jiangsu Cowin Biotech Co., Ltd).
Strains and Plasmids
Genetic knockout strains were constructed in E. coli K-12 MG1655 and BL21 (DE3), with the deletion of the ompA gene achieved via the Lambda Red system method [25]. Plasmids utilized for construct testing, generated through Gibson assembly, underwent prior validation through Sanger sequencing before the next steps. A list of plasmids used in this manuscript is shown in Table 1.
Generation of a Kanamycin Cassette with Homologous Arms Specific to ompA
DNA fragments consisting of a Kanamycin resistance cassette flanked by flippase recognition target (FRT), 25 bp priming sites, and 50 bp homologous arms were amplified from the pKD4 plasmid via a high-fidelity 2× Super Pfx MasterMix (CWBIO: CW2965M). The primer sequences are provided in Table 2. After the successful replacement of ompA by the Kanamycin cassette, the removal of the antibiotic resistance genes was achieved through the expression of the flippase recombination enzyme (FLP) recombinase from pCP20. The PCR reaction systems were as follows: 98 °C for 3 min, followed by 30 cycles of 98 °C for 30 s, 53 °C for 30 s, and 72 °C for 1 min. The resulting PCR reactions were digested with 0.5 μl DpnI per 30 μl PCR reaction for 1 h at 37 °C. The PCR products were purified using DNA cleanup buffers (Qiagen: 19066) and Zymo-Spin columns (ZYMO: C1003-50).
Recombineering
Bacteria harboring pTKRED were cultured in 1 ml SOC with 50 μg/ml kanamycin and 50 μg/ml Spectinomycin, allowing overnight growth at 220 rpm and 30 °C. The resulting culture was then diluted 1/100 into a baffled 125 ml flask containing 20 ml SOB supplemented with 50 μg/ml Spectinomycin and 2 mM IPTG. Continuous growth at 220 rpm and 30 °C was sustained until the OD600nm reached 0.4–0.8. After chilling the bacterial cells on ice for 30 min, they were centrifuged at 4000 rpm for 5 min at 4 °C. The supernatant was discarded, and the cells were resuspended in 10 ml sterile ice-cold 10% v/v glycerol using vortex mixing. Bacteria were centrifuged for 5 min at 4000 rpm and 4 °C. The cells were washed three times in total, and then they were concentrated 100 times by resuspension in 200 μl ice-cold 10% glycerol. Fifty microliters of electrocompetent cells were mixed with 100–500 ng of DNA and incubated on ice for at least 1 min, with the option to extend the duration up to 30 min. The mixture was transferred to a 1 mm electroporation cuvette and placed in the Shock Pod. 150 μl to 200 μl of SOC was added to the cuvette immediately after electroporation (1.8 kV, 25 µF, 200 Ω using a Bio-Rad GenePulser Xcell Electroporation System). The mixture was transferred to 1.5 ml microcentrifuge tubes and shaken at 220 rpm, 37 °C for 2 h. Once the recovery phase was completed, bacteria were spun down at 12,000 rpm for 1 min. The supernatant was discarded, and the pellet was resuspended in the residual medium and spread onto LB agar plates containing Kanamycin. The plates were incubated overnight at 30 °C in an air incubator. The next day, positive colonies that were randomly selected from the LB agar plates were verified for integration by colony PCR, and the resulting PCR fragments were then sequenced by Sanger sequencing (Genewiz).
Removal of pTKRED and the Kanamycin Cassette
Selected clones, confirmed to harbor the desired fragment, were inoculated into 1 ml of LB and cultured overnight in a shaker at 42 °C. The following day, samples were plated on both 100 μg/ml Spectinomycin LB agar plate and 50 μg/ml Kanamycin LB agar plate to validate the elimination of pTKRED. Spectinomycin-sensitive but Kanamycin-resistant colonies were electroporated with pCP20 and plated on Carbenicillin plates for overnight growth at 30 °C. Individual colonies were inoculated into 1 ml nonselective LB at 220 rpm and 42 °C for 6–16 h to eliminate the Kanamycin resistance cassette and cure pCP20. One microliter of bacterial culture was tri-streaked on LB agar plates and grown overnight at 37 °C. 10–20 colonies were randomly selected and streaked successively on nonselective LB plates, Carbenicillin-containing LB plates, and Kanamycin-containing LB plates. Colonies that lost resistance to Carbenicillin and Kanamycin were chosen for further analysis.
Protein Identification by Liquid Chromatography-Tandem Mass Spectrometry
For in-gel digestion, denatured bacterial lysates were separated by SDS-PAGE and stained with Coomassie blue stain. The protein samples were processed and analyzed at the Mass Spectrometry Facility of the Department of Pathology at the University of Michigan. Gel slice was destained with 30% methanol for 4 h. Upon reduction (10 mM DTT) and alkylation (65 mM 2-Chloroacetamide) of the cysteines, proteins were digested overnight with 500 ng of sequencing grade, modified trypsin (Promega) at 37 °C. Peptides were extracted by incubating the gel with 150 μl of 50% acetonitrile/0.1% TFA for 30 min at room temperature. A second extraction with 150 μl of 100% acetonitrile plus 0.1% TFA was also performed. Both extracts were combined and dried in a vacufuge (Eppendorf). For mass spectrometry, resulting peptides were dissolved in 9 μl of 0.1% formic acid/2% acetonitrile solution. Two microliters of the resulting peptide solution were resolved on a nano-capillary reverse phase column (Acclaim PepMap C18, 2 micron, 50 cm, Thermo Scientific) using a 0.1% formic acid/acetonitrile gradient at 300 nl/min over a period of 90 min (2–25% acetonitrile in 35 min; 25–50% acetonitrile in 20 min followed by a 90% acetonitrile wash for 5 min and a further 30 min re-equilibration with 2% acetonitrile). Eluent was directly introduced into Q Exactive HF mass spectrometer (Thermo Scientific, San Jose CA) using an EasySpray source. MS1 scans were acquired at 60 K resolution (AGC target = 3x106; max IT = 50 ms). Data-dependent collision induced dissociation MS/MS spectra were acquired on 20 most abundant ions following each MS1 scan (NCE ~ 28%; AGC target 1x105; max IT 45 ms). For database search, proteins were identified by searching the data against E coli K12 protein database (4362 entries; uniprotkb_proteome_E. coli K12_UP000000625) using Proteome Discoverer (v3.0, Thermo Scientific). Search parameters included MS1 mass tolerance of 10 ppm and fragment tolerance of 0.2 Da; two missed cleavages were allowed; carbamidomethylation of cysteine (+ 57.012 Da) was considered fixed modification and oxidation of methionine (+ 15.994 Da), deamidation of asparagine and glutamine (+ 0.984 Da), were considered as variable modifications. False discovery rate (FDR) was determined using Percolator and proteins/peptides with an FDR of ≤ 1% were retained for further analysis.
Bacterial Growth Measurement
To generate growth curves, bacteria were inoculated in 1 ml of LB medium. For the analysis of four different strains, each fresh overnight bacterial suspension was diluted 1:10. The OD600nm was measured using NanoDrop Microvolume Spectrophotometers. Based on these measurements, the bacterial suspensions were further diluted to an OD600nm of 0.01 in 50 ml of pre-warmed LB medium within 125 ml flasks. The flasks were incubated at 37 °C with shaking at 220 rpm. Samples (1 ml) were taken from each flask at 0, 1, 2, 3, 4, 5, 6, 7, and 8 h, and OD600nm was recorded. The growth values from three biological replicates were averaged to create the growth curves. For monitoring the four different strains under protein production load, each fresh overnight bacterial suspension was split into two groups: one diluted 1:10 in 50 ml of pre-warmed LB medium with 0.2% glucose, and the other diluted 1:10 in 50 ml of pre-warmed LB medium with 10 mM Rhamnose, each in 125 ml flasks. These flasks were incubated at 25 °C with shaking at 220 rpm. Samples (1 ml) were collected at 0, 3, 6, 9, 11, 13, and 24 h, and OD600nm was recorded. The values from three biological replicates were averaged to plot the growth curves for each treatment.
Detection of SYTOX Green Intensity
The fresh overnight bacterial suspension of K-12 MG1655, ΔompA::FRT_K-12 MG1655, BL21 (DE3), and ΔompA::FRT_BL21 (DE3) underwent a single wash with FACS buffer and was subsequently incubated with 1 µM SYTOX Green (Thermo Fisher Scientific: S7020). An equivalent number of bacteria from each strain was subject to boiling at 100 °C for 10 min to serve as the SYTOX Green positive control. The SYTOX Green’s fluorescence signal was detected by Attune Flow Cytometer (Thermo Fisher Scientific), and the data were subsequently analyzed using FlowJo.
Preparation of Chemically Competent Cells and Transformation
Chemically competent cells were made by the Zymo Research Mix & Go E. coli Transformation Kit & Buffer Set (ZYMO: T3001). Briefly, 40 μl of overnight culture was inoculated into 2 ml ZymoMedium™. Shaking the culture at 220 rpm, 37 °C achieved an OD600nm of 0.4–0.6. Upon reaching the desired OD, the culture was incubated on ice for 10 min. The bacteria were then directly centrifuged in 2.0 ml tubes at 8000 rpm for 5 min in the cold room. After centrifugation, the supernatant was poured off, and cells were gently resuspended in 200 μl ice-cold 1× Wash Buffer, followed by re-pelleting. The supernatant was poured off again, and cells were gently resuspended in 200 μl ice-cold 1× Competent Buffer. Aliquots of 25 μl of the cell suspension were prepared in sterile tubes, making the cells ready for transformation or suitable for storage at − 80 °C. For plasmid transformation, 25 μl of competent cells were mixed with 2.5 μl of DNA plasmid and gently mixed for a few seconds. The transformation was incubated on ice for 10 min, followed by a 42 °C heat shock for 30 s. The cells were then put back on ice for up to 2 min. Cells were allowed to recover by adding 100 μl of SOC to the transformation mixture and shaking at 220 rpm at 37 °C for 1 h. Recovered cells were spun down for 1 min at 12,000 rpm, the supernatant was drained off, and the remaining cells were spread on pre-warmed plates and incubated at 37 °C.
Bacteria Surface Display Staining
All relevant constructs were inoculated into 1 ml of LB medium supplemented with 50 μg/ml Kanamycin. The cultures were allowed to grow overnight at 37 °C. Subsequently, the overnight bacterial suspension was diluted 1/50 in a pre-warmed LB medium containing 50 μg/ml Kanamycin and incubated in a shaker for 2 h. Afterward, rhamnose was added into the culture to a final concentration of 10 mM for induction at room temperature for 24 h. Fifty microliters of the induced bacterial culture with an OD600nm of approximately 1.5 were transferred to 0.5 ml FACS buffer (2% FBS, 0.05% w/v sodium azide, 2 mM Ethylenediaminetetraacetic acid dissolved in 1× PBS) pre-aliquoted in 1.5 ml tubes. The bacterial cultures were centrifuged at 8000 rpm for 2 min at room temperature. After discarding the supernatant, the bacterial incubated with 1 μg/ml Alexa Fluor® 647 anti-DYKDDDDK Tag Antibody (BioLegend: 637316) in FACS buffer and incubated on ice for 30–60 min. One milliliter of FACS buffer was added directly to the antibody–bacteria mixture, and then it was centrifuged at 8000 rpm for 2 min. After carefully pouring off the supernatant, the bacterial pellets were resuspended in 1 ml of 1× PBS through vortex mixing. The bacteria were kept on ice before the flow cytometry analysis.
SDS-PAGE Analysis
Samples were mixed with Laemmli SDS-sample buffer (Boston BioProducts, Inc.: BP-111R) and heated to 100 °C for 10 min. The prepared samples were then electrophoresed on 10% polyacrylamide gels. The protein bands were visualized using the Coomassie BB R-250 Staining Solutions Kit (Fisher Scientific: NC0228390). Quantitative analysis of the bands was performed using ImageJ software.
Statistical Analysis
Statistical significance was evaluated using a Student’s unpaired t-test using GraphPad PRISM (San Diego, CA, USA). P values < 0.05 were considered statistically significant. The statistical significance is indicated in all figures according to the following scale: *p < 0.05, **p < 0.01, ***p < 0.001. All graphs are expressed as the means ± SEM.
Plasmids and Primers
Results
Genetic Knockout of ompA from the Genome of E. coli
Prior studies by others uncovered the presence of > 100 integral and lipid-linked membrane proteins on the E. coli cell envelope [24]. Of note, one constitutive OMP, named OmpA, is estimated to be present at 105 copies per cell in E. coli [27]. We speculate that the presence of OmpA may result in steric hindrance and limit the surface display of target proteins or peptides in E. coli. Moreover, enforced expression of recombinant proteins on crowded extracellular surfaces of E. coli can potentially affect the membrane integrity, which may explain the fact that high levels of bacterial surface display can often interfere with bacterial growth [5]. On the contrary, if we can remove some constitutively expressed proteins such as OmpA without affecting the growth and viability of E. coli, it could potentially reduce the steric hindrance and divert the resources required for protein synthesis and membrane translocation from making native membrane proteins to the expression and display of chimeric OMPs. While OmpA has been implicated in bacterial stress responses [28], virulence [29] and biofilm formation [30] for pathogenic E. coli strains to interact with host environments [31], OmpA may become a nonessential component for E. coli when the bacteria are repurposed for laboratory and industry applications (Fig. 1).
The overall design. A A traditional approach by evaluating different OMPs to enhance bacterial surface display. B A new strategy in this work involves knocking out OmpA, a nonessential OMP for E. coli. The hypothesis is that the removal of OmpA from the outer membrane may generate free space, enabling the improved display of protein cargoes on the surface of E. coli. This figure was created using BioRender
To test our hypothesis, we chose the E. coli strains K-12 MG1655 and BL21 (DE3) due to their well-established roles and characteristics in the field of biotechnology and microbial therapeutics [32,33,34]. We proposed to use the lambda red recombineering method to knock out the ompA gene in the E. coli genome (Fig. 2A) [25]. Specifically, a Kanamycin cassette flanked by short flippase (FLP) recognition target (FRT) sites and 50 bp DNA fragments homologous to the ompA gene at 5′ and 3′ ends were amplified by polymerase chain reaction (PCR) from the pKD4 plasmid. The linear PCR product was subsequently electroporated into E. coli strains K-12 MG1655 and BL21 (DE3) containing pTKRED, a plasmid encoding the lambda red proteins (Fig. S1). After colony PCR verification, the correct clones were transformed with pCP20, a plasmid expressing FLP to remove the Kanamycin resistance gene. The successful removal of the Kanamycin resistance gene was confirmed by colony PCR and Sanger sequencing (Figs. 2B, C, S2). In addition, we confirmed the loss of OmpA protein in ΔompA strains by resolving and staining the total proteins in SDS-PAGE. Of note, a protein band with an estimated molecular weight of 35 kDa was absent in ΔompA strains but present in the wild type (Fig. S3A). To verify if the protein band corresponds to OmpA, we excised the band present in the wild-type K-12 MG1655 but absent at the same position in the ΔompA K-12 MG1655 (Fig. S3A). As a negative control, we also sliced the gel band from the same molecular weight range in the ΔompA K-12 MG1655 strain. Both gel slices were submitted for protein identification via mass spectrometry, which confirmed the absence of the OmpA protein in the ΔompA strain (Fig. 2D).
Genetic knockout of ompA from the genome of E. coli. A Illustration of the genetic knockout of ompA from the E. coli genome. The process involves using a linear template containing a Kanamycin resistance cassette flanked by FRT recognition sites, which is amplified from pKD4. The target gene is then replaced with this linear template using Lambda red recombination. After recombination, the antibiotic fragment is removed through the expression of FLP recombinase. B Sanger sequencing analysis of the final recombinant is presented. The highlighted portion illustrates the sequencing of the FRT fragment. C The deletion of ompA and the final recombinant is validated through colony PCR using specific primers (S272, S245 and S273). Sequencing analysis of the final recombinant is also conducted to confirm the deletion. In the upper gel image, primers S272 located upstream of KanR, and S245 in the midsection of KanR are used. Six colonies were randomly picked and exhibited the PCR fragment of 649 bp. In the bottom gel image, primers S272 and S273, located downstream of the FRT site of KanR, are utilized. Four colonies were randomly picked after deletion, resulting in a PCR product of 479 bp. D Validation of OmpA protein knockout in the ΔompA strain by mass spectrometry. Heat maps of protein levels for the top 50 most abundant genes detected by mass spectrometry in K-12 MG1655 wild-type and ΔompA strains. The color in each tile represents the scaled abundance value
Knocking Out OmpA Does Not Affect the Growth of E. coli
The ompA gene in E. coli encodes the outer membrane protein A (OmpA), which plays several important roles in the bacterium’s biology including but not limited to stress response, host interaction, cell division, and structure integrity [35]. Using the lambda red recombineering, however, we successfully knocked out ompA, which confirmed that the ompA gene is a nonessential gene under the experimental settings. To further confirm if the ompA knockout may affect the physiology of bacteria, we compared the growth rates between the wild-type and ΔompA strains in both K12 MG1655 and BL21 (DE3) backgrounds. Following overnight culture to the stationary phase, bacteria were diluted to an OD600nm of 0.01, OD600nm measurements were conducted at 0, 1, 2, 3, 4, 5, 6, 7, and 8 h. As shown in Fig. 3A and B, there was no difference in growth rates between the wild-type and ΔompA strains in K12 MG1655 and BL21 (DE3), underscoring the fact that OmpA is a nonessential protein under laboratory growth conditions.
Knocking out OmpA does not affect the growth rates or the membrane integrity of E. coli without protein induction. Bacterial growth was monitored for A wild-type and ΔompA strains in K-12 MG1655 and B wild-type and ΔompA strains in BL21 (DE3) in LB broth, with optical density OD600nm measurements taken at 0, 1, 2, 3, 4, 5, 6, 7, and 8 h post inoculation. Data represent mean ± SEM of three independent experiments. Flow cytometric analysis of SYTOX staining for C wild-type and ΔompA strains in K-12 MG1655, and D wild-type and ΔompA strains in BL21 (DE3). Live bacteria are SYTOX-negative while dead bacteria are SYTOX-positive. As a control, each strain is subject to boiling at 100 °C for 10 min. A representative graph is presented, based on three independent experiments. E, F Quantification of membrane integrity in both K-12 MG1655 and BL21 (DE3) expressed as the percentage of SYTOX-negative cells. Data = mean ± SEM, from three independent experiments
Knocking Out OmpA Does Not Affect the Membrane Integrity of E. coli
While we did not observe changes in growth rates resulting from the ompA knockout, it remains possible that OmpA may play a role in the outer membrane since there is an estimated copy number of 105 OmpA protein per cell. We employed SYTOX Green to test if knocking out OmpA can compromise the membrane integrity of E. coli. SYTOX Green is a high-affinity nucleic acid stain that penetrates cells with compromised membranes, making it a useful tool for assessing membrane integrity. In healthy cells with intact membranes, SYTOX Green cannot penetrate, and thus the cells will not fluoresce. However, in cells with damaged or leaky membranes, SYTOX Green enters and binds to DNA, resulting in a bright green fluorescence. We labeled the wild-type or ΔompA strains (K-12 MG1655 and BL21(DE3)) from the exponentially growing phase with SYTOX Green and analyzed the percentage of SYTOX Green-positive and -negative populations by flow cytometry. Interestingly, there was almost no detectable population for the SYTOX Green-positive bacteria in ΔompA strains compared to that of the wild-type strains (Fig. 3C–F). As SYTOX Green is also a cell viability dye, we concluded that knocking out ompA in K-12 MG1655 and BL21 (DE3) does not compromise the membrane intactness in E. coli or affect bacterial viability under the laboratory culture settings.
Knocking Out OmpA Does Not Change the E. coli Growth Rates Compared to Wild-Type Strains Under the Protein Induction Conditions
While knocking out OmpA does not interfere with bacterial proliferation or membrane integrity, we next examined the impact on bacterial growth in both wild-type and ΔompA strains when induced for protein surface display. We focused on the OmpA-mIL18 construct due to its recently reported therapeutic applications [36, 37]. It was found that induction of protein production in both wild-type and ΔompA strains resulted in a dramatic reduction in bacterial proliferation. This aligns with our observations and those of others that protein induction generally imposes a significant metabolic burden on bacteria. Moreover, overexpression of OMPs can further induce membrane stress. However, there was no statistically significant difference in growth between the wild-type and ΔompA strains under induction conditions in both K-12 and BL21 (DE3) backgrounds (Fig. 4). In contrast, when protein induction was suppressed by the presence of 0.2% glucose, both wild-type and ΔompA strains carrying the same construct exhibited faster growth rates compared to those under rhamnose induction (Fig. 4).
Knocking out OmpA does not change the E. coli growth rates compared to wild-type strains under the protein induction conditions. Bacterial growth was monitored for A wild-type and ΔompA strains in K-12 MG1655 and B wild-type and ΔompA strains in BL21 (DE3) in LB broth. Cultures were treated separately with 0.2% glucose to suppress OmpA-mIL18 expression (control) and with 10 mM Rhamnose to induce OmpA-mIL18 expression. Optical density (OD600nm) measurements were taken at 0, 3, 6, 9, 11, 13, and 24 h. Data represent the mean ± SEM of three independent experiments. ns refers to no statistical significance
OmpA Knockout Markedly Improves the Surface Display of Three Representative Recombinant Proteins
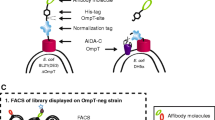
Having established that ΔompA strains (K-12 MG1655 and BL21(DE3)) exhibit normal proliferation and membrane integrity, we next explore the use of ΔompA strains for bacterial surface display. We first chose an OmpA derivative as an anchor protein since it represents one of the most popular OMPs for bacterial surface display. Specifically, the OmpA variant consists of a signal peptide, the first nine residues of Braun’s lipoprotein (referred to as Lpp), and five of the eight membrane-spanning segments of the OmpA protein (amino acid residues 46–159). The OmpA variant is responsible for targeting proteins of interest to the outer membrane via genetic fusion at the C-terminus of OmpA [38]. In addition to OmpA, we also extended our work to another anchor protein named YiaT. YiaT is a putative OMP in E. coli although its function has not been fully well characterized. By predicting the outer membrane topology, a previous study by others determined that the fifth extracellular loop (at the 232th amino acid) of YiaT is an ideal target for the C-terminal fusion with a protein of interest [39]. In our unpublished works, we have explored the use of OmpA to display a bacterial enzyme named ClbS, that is known to antagonize the DNA damaging effects by colibactin [40, 41], as well as a recombinant murine cytokine IL-18, which is known to promote the cytotoxicity of CD8 T cells and natural killer cells for cancer immunotherapy [36, 37]. Additionally, we also identified YiaT that can display a camelid-derived nanobody against Cytotoxic T-lymphocyte associated protein 4 (CTLA4), which is a protein receptor that functions as an immune checkpoint and downregulates immune responses [42]. Of note, these three different cargo proteins have distinct biophysical and biochemical properties as summarized in Supplemental Table 1, which allow us to test the versatility of our approach. By comparing three proteins from different species (bacteria, mouse, and camelid) along with two different anchor proteins (OmpA and YiaT), we aim to demonstrate the versatility of the ΔompA strains to enhance the bacterial surface display (Fig. 5A).
OmpA knockout markedly improves the surface display of three representative recombinant proteins. A Experimental outline and schematic for engineering K-12 MG1655 and BL21 (DE3) strains to display heterologous proteins on the bacterial surface. Each protein is fused with an OMP (OmpA or YiaT232) along with a FLAG epitope driven by a rhamnose-inducible promoter. In total, six constructs (OmpA-αCTLA4, OmpA-mIL18, OmpA-ClbS, YiaT232-αCTLA4, YiaT232-mIL18, and YiaT232-ClbS) were induced with 10 mM l-rhamnose for 24 h at room temperature before staining for the FLAG epitope and flow cytometry. The percentage of wild-type and ΔompA E. coli displaying B OmpA or C YiaT fusion constructs are quantified, and representative flow cytometry histograms are shown to highlight the increase of surface display in ΔompA over the wild-type strains for both K-12 MG1655 and BL21 (DE3). Mean fluorescence intensity (MFI) of FLAG signals is quantified to compare ΔompA to wild-type strains in D K-12 MG1655 and E BL21 (DE3). Data = mean ± SEM, from three independent experiments, *p < 0.05, **p < 0.01, ***p < 0.001, unpaired t test
Therefore, we sought to comprehensively quantify and compare the display levels of six different constructs (OmpA-ClbS, YiaT232-ClbS, OmpA-mIL18, YiaT232-mIL18, OmpA-αCTLA4 and YiaT232-αCTLA4) between the wild-type and ΔompA strains in both K12 MG1655 and BL21 (DE3) backgrounds. Of note, all six constructs were fused with a FLAG epitope to facilitate the detection of proteins on the bacterial surface by flow cytometry. Following induction with rhamnose at room temperature for 24 h, bacteria were harvested, washed and directly stained with anti-FLAG antibody conjugated with a fluorescent dye. It was found that the isogenic ΔompA strains in both K-12 MG1655 and BL21 (DE3) strains markedly increased the percentage of bacterial population displaying ClbS, mIL18 or αCTLA4, compared to their wild-type strains (Fig. 5B, C). Moreover, by quantifying the mean fluorescence intensity, we also detected increased levels of display in the ΔompA strains over those of the wild-type background (Fig. 5D, E). Interestingly, when total cell lysates were analyzed by SDS-PAGE, the levels of total recombinant proteins (αCTLA4, mIL18 and ClbS), which combine both intracellular and extracellular fractions, were comparable between ΔompA and wild-type strains (Fig. S3B). Since fusion proteins are first translated in the cytoplasm followed by translocation through inner and outer membranes for surface display, knocking out OmpA protein itself may not change the protein translation efficiency and the total amount of fusion proteins. Instead, removal of native OmpA proteins from bacterial surface improved the extracellular fraction of recombinant proteins likely though increased surface availability. It is noteworthy that the OmpA-fused proteins displayed a more significant difference between WT and ΔompA strains compared to the YiaT232-fused proteins (Fig. 5D, E). In the literature, OmpA is a well-characterized protein in E. coli, whereas YiaT has been studied less extensively regarding its biophysical and biological functions [39]. Although the exact reasons are unclear, it is possible that different OMPs (e.g., OmpA versus YiaT) may be phase-separated into distinct domains on the outer membrane at the nanoscale according to a previous fundamental biophysical study [43]. Knocking out OmpA might favor OmpA chimeric fusion proteins occupying the same regions where native OmpA proteins are localized. Conversely, native YiaT and YiaT chimeric fusion proteins may occupy different regions on the outer membrane. Additionally, OmpA and YiaT might utilize slightly different cytoplasmic, inner, and outer membrane machineries to facilitate membrane translocation from the cytoplasm. Therefore, knocking out OmpA may free up more proteins such as chaperons to help with the translocation of the same protein cargo, such as OmpA fusion proteins.
Discussion
While a myriad of anchor proteins have been explored to optimize the bacterial surface display, this work presents a generic strategy by deleting OmpA, a constitutively expressed OMP in E. coli. Since OmpA has been estimated to present at a 105 copy number per bacterium, removing this protein could possibly reduce steric hindrance to accommodate chimeric OmpA fusion proteins on the outer membrane. Furthermore, knocking out OmpA may divert the resources required for protein synthesis and membrane translocation from making native membrane proteins to the expression and display of chimeric OMPs. Indeed, we demonstrated that knocking out OmpA markedly improve the display levels of three different proteins by two different anchor proteins. Furthermore, we found that deletion of ompA gene did not interfere with bacterial growth or induce membrane leakage at least under the laboratory growth conditions. However, our work does not rule out the possibilities that knocking out ompA could affect other physiological processes such as bacterial adhesion, protein secretion, biofilm formation, virulence, and stress responses [35]. Additionally, it is possible that knocking out OmpA may compromise the integrity of membrane organization and phase separation of OMPs at the nanoscale based on one previous study [43]. Nevertheless, we speculate that if the bacterial surface display is primarily employed for directed evolution by displaying libraries of peptides or protein variants, the ompA knockout strategy could potentially increase the diversity and size of displayed libraries for in vitro screening of optimal peptide or protein sequences. If the bacterial surface display is repurposed for delivery of protein therapeutics by engineering bacteria as living therapeutics in vivo, it warrants further characterization of how the deletion of ompA may contribute to the colonization or clearance of engineered bacteria in the host. For example, E. coli strains have been engineered to deliver immune cytokines or other proteins in tumors by harnessing their tumor tropism [36, 44]. Therefore, it is important to examine whether knocking out ompA can reduce the efficiency of the tumor-targeting potential of engineered E. coli.
Notably, even under optimal conditions, at least 20% of the cells in this study do not exhibit detectable levels of the protein of interest. The efficiency of bacterial surface display, defined as the percentage of bacteria positive for staining, can vary depending on the specific fusion protein, cargo, or anchor proteins used. For instance, previous studies have reported a bimodal distribution of positive and negative populations in bacterial surface display [45]. Moreover, the presence of a population without any surface display has also been observed in yeast surface display platform [46, 47].
One interesting future direction is to develop a strain with minimal surface proteome, in a manner similar to the development of a genome-reduced strain that harbors the minimum number of genes required for replication [25,26,27]. Of note, dozens of OMPs have been identified through biochemical and genetic assays, which presents an opportunity to further modify the surface of E. coli by deleting all nonessential OMPs such that maximal free space is available to present recombinant proteins on the surface without affecting the viability of bacteria. Although this approach may appear a tedious task, it proposes an interesting fundamental question with regards to the minimal proteome of OMPs on the surface of E. coli necessary to support the growth of E. coli. Previously, four genes encoding abundant OMPs (OmpA, OmpC, OmpF, and LamB) were deleted in a series of single, double, triple, or quadruple knock-outs [22]. While these knockout strains were used to improve production and purification of recombinant OMPs, they provide an alternative approach to improving bacterial surface display by knocking out additional OMPs such as OmpC, OmpF, and LamB.
Data Availability
N/A.
References
Lee, S. Y., J. H. Choi, and Z. Xu. Microbial cell-surface display. Trends Biotechnol. 21:45–52, 2003.
Lee, J.-S., K.-S. Shin, J.-G. Pan, and C.-J. Kim. Surface-displayed viral antigens on Salmonella carrier vaccine. Nat. Biotechnol. 18:645–648, 2000.
Westerlund-Wikstrom, B., et al. Functional expression of adhesive peptides as fusions to Escherichia coli flagellin. Protein Eng. Des. Sel. 10:1319–1326, 1997.
Richins, R. D., I. Kaneva, A. Mulchandani, and W. Chen. Biodegradation of organophosphorus pesticides by surface-expressed organophosphorus hydrolase. Nat. Biotechnol. 15:984–987, 1997.
Salema, V., and L. Á. Fernández. Escherichia coli surface display for the selection of nanobodies. Microb. Biotechnol. 10:1468–1484, 2017.
Park, M. Surface display technology for biosensor applications: a review. Sensors. 20:2775, 2020.
Feng, X., M. Jin, W. Huang, W. Liu, and M. Xian. Whole-cell catalysis by surface display of fluorinase on Escherichia coli using N-terminal domain of ice nucleation protein. Microb. Cell Factories. 20:206, 2021.
Daugherty, P. S., G. Chen, M. J. Olsen, B. L. Iverson, and G. Georgiou. Antibody affinity maturation using bacterial surface display. Protein Eng. 11:825–832, 1998.
Daugherty, P. S., G. Chen, B. L. Iverson, and G. Georgiou. Quantitative analysis of the effect of the mutation frequency on the affinity maturation of single chain Fv antibodies. Proc. Natl Acad. Sci. U. S. A. 97:2029–2034, 2000.
Smith, G. P. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 228:1315–1317, 1985.
Charbit, A., J. C. Boulain, A. Ryter, and M. Hofnung. Probing the topology of a bacterial membrane protein by genetic insertion of a foreign epitope; expression at the cell surface. EMBO J. 5:3029–3037, 1986.
Strauss, A., and F. Götz. In vivo immobilization of enzymatically active polypeptides on the cell surface of Staphylococcus carnosus. Mol. Microbiol. 21:491–500, 1996.
Sergeeva, A., M. Kolonin, J. Molldrem, R. Pasqualini, and W. Arap. Display technologies: application for the discovery of drug and gene delivery agents. Adv. Drug Deliv. Rev. 58:1622–1654, 2006.
Daugherty, P. S., M. J. Olsen, B. L. Iverson, and G. Georgiou. Development of an optimized expression system for the screening of antibody libraries displayed on the Escherichia coli surface. Protein Eng. Des. Sel. 12:613–621, 1999.
Daugherty, P. S. Protein engineering with bacterial display. Curr. Opin. Struct. Biol. 17:474–480, 2007.
Rockberg, J., J. Löfblom, B. Hjelm, M. Uhlén, and S. Ståhl. Epitope mapping of antibodies using bacterial surface display. Nat. Methods. 5:1039–1045, 2008.
Teymennet-Ramírez, K. V., F. Martínez-Morales, and M. R. Trejo-Hernández. Yeast surface display system: strategies for improvement and biotechnological applications. Front. Bioeng. Biotechnol.9:794742, 2022.
Van Bloois, E., R. T. Winter, H. Kolmar, and M. W. Fraaije. Decorating microbes: surface display of proteins on Escherichia coli. Trends Biotechnol. 29:79–86, 2011.
Hernández-Rollán, C., et al. LyGo: a platform for rapid screening of lytic polysaccharide monooxygenase production. ACS Synth. Biol. 10:897–906, 2021.
Rutherford, N., and M. Mourez. Surface display of proteins by Gram-negative bacterial autotransporters. Microb. Cell Factories. 5:22, 2006.
Glass, D. S., and I. H. Riedel-Kruse. A synthetic bacterial cell–cell adhesion toolbox for programming multicellular morphologies and patterns. Cell. 174:649-658.e16, 2018.
Meuskens, I., M. Michalik, N. Chauhan, D. Linke, and J. C. Leo. A new strain collection for improved expression of outer membrane proteins. Front. Cell. Infect. Microbiol. 7:464, 2017.
Weiner, J. H., and L. Li. Proteome of the Escherichia coli envelope and technological challenges in membrane proteome analysis. Biochim. Biophys. Acta BBA Biomembr. 1778:1698–1713, 2008.
Molloy, M. P., et al. Proteomic analysis of the Escherichia coli outer membrane. Eur. J. Biochem. 267:2871–2881, 2000.
Datsenko, K. A., and B. L. Wanner. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. U.S.A. 97:6640–6645, 2000.
Kuhlman, T. E., and E. C. Cox. Site-specific chromosomal integration of large synthetic constructs. Nucleic Acids Res. 38:e92–e92, 2010.
Koebnik, R., K. P. Locher, and P. Van Gelder. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol. Microbiol. 37:239–253, 2000.
Alba, B. M., and C. A. Gross. Regulation of the Escherichia coli σE-dependent envelope stress response. Mol. Microbiol. 52:613–619, 2004.
Monteiro, R., et al. A secretome view of colonisation factors in Shiga toxin-encoding Escherichia coli (STEC): from enterohaemorrhagic E. coli (EHEC) to related enteropathotypes. FEMS Microbiol. Lett. 363:fnw179, 2016.
Chagnot, C., M. A. Zorgani, T. Astruc, and M. Desvaux. Proteinaceous determinants of surface colonization in bacteria: bacterial adhesion and biofilm formation from a protein secretion perspective. Front. Microbiol. 4:303, 2013.
Nie, D., et al. Outer membrane protein A (OmpA) as a potential therapeutic target for Acinetobacter baumannii infection. J. Biomed. Sci. 27:26, 2020.
Jeong, H., H. J. Kim, and S. J. Lee. Complete genome sequence of Escherichia coli strain BL21. Genome Announc. 3:e00134-e215, 2015.
Soupene, E., et al. Physiological studies of Escherichia coli strain MG1655: growth defects and apparent cross-regulation of gene expression. J. Bacteriol. 185:5611–5626, 2003.
Blattner, F. R., et al. The complete genome sequence of Escherichia coli K-12. Science. 277:1453–1462, 1997.
Wang, Y. The function of OmpA in Escherichia coli. Biochem. Biophys. Res. Commun. 292:396–401, 2002.
Yang, S., et al. Nonpathogenic E. coli engineered to surface display cytokines as a new platform for immunotherapy. Res. Square. 2024. https://doi.org/10.21203/rs.3.rs-4031911/v1.
Zhou, T., et al. IL-18BP is a secreted immune checkpoint and barrier to IL-18 immunotherapy. Nature. 583:609–614, 2020.
Francisco, J. A., and G. Georgiou. The expression of recombinant proteins on the external surface of Escherichia coli: biotechnological applications. Ann. N. Y. Acad. Sci. 745:372–382, 1994.
Han, M.-J., and S. H. Lee. An efficient bacterial surface display system based on a novel outer membrane anchoring element from the Escherichia coli protein YiaT. FEMS Microbiol. Lett. 362:1–7, 2015.
Bossuet-Greif, N., et al. Escherichia coli ClbS is a colibactin resistance protein. Mol. Microbiol. 99:897–908, 2016.
Tripathi, P., et al. ClbS is a cyclopropane hydrolase that confers colibactin resistance. J. Am. Chem. Soc. 139:17719–17722, 2017.
Gurbatri, C. R., et al. Engineered probiotics for local tumor delivery of checkpoint blockade nanobodies. Sci. Transl. Med. 12:eaax0876, 2020.
Benn, G., et al. Phase separation in the outer membrane of Escherichia coli. Proc. Natl Acad. Sci. U. S. A. 118:e2112237118, 2021.
Leventhal, D. S., et al. Immunotherapy with engineered bacteria by targeting the STING pathway for anti-tumor immunity. Nat. Commun. 11:2739, 2020.
Wendel, S., E. C. Fischer, V. Martínez, S. Seppälä, and M. H. H. Nørholm. A nanobody:GFP bacterial platform that enables functional enzyme display and easy quantification of display capacity. Microb. Cell Factories. 15:71, 2016.
Uchański, T., et al. An improved yeast surface display platform for the screening of nanobody immune libraries. Sci. Rep. 9:382, 2019.
Boder, E. T., and K. D. Wittrup. Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 15:553–557, 1997.
Acknowledgments
We would like to express our gratitude to the Proteomics Resource Facility at the University of Michigan to support our study. We also want to thank members of the Li lab for helpful discussions.
Funding
This work was supported by NIH 1DP2GM154019 (J.L.).
Author information
Authors and Affiliations
Contributions
JL conceived the ideas and supervised the experiments. RZ designed and conducted all the experiments. NY facilitated the protein analysis. ZW and SY prepared the plasmids and strains. All authors contributed to the discussion of the results and the writing and editing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declared that they have no conflict of interest.
Additional information
Associate Editor Rob Raphael oversaw the review of this article.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, R., Ye, N., Wang, Z. et al. A New Bacterial Chassis for Enhanced Surface Display of Recombinant Proteins. Cel. Mol. Bioeng. (2024). https://doi.org/10.1007/s12195-024-00819-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12195-024-00819-w