Abstract
Introduction
The propagation of mechanochemical signals from the extracellular matrix to the cell nucleus has emerged as a central feature in regulating cellular differentiation and de-differentiation. This process of outside-in signaling and the associated mechanotransduction pathways have been well described in numerous developmental and pathological contexts. However, it remains less clear how mechanotransduction influences the activity of chromatin modifying enzymes that direct gene expression programs.
Objectives
The primary objective of this study was to explore how matrix mechanics and geometric confinement influence histone deacetylase (HDAC) activity in fibroblast culture.
Methods
Polyacrylamide hydrogels were formed and functionalized with fibronectin patterns using soft lithography. Primary mouse embryonic fibroblasts (MEFs) were cultured on the islands until confluent, fixed, and immunolabeled for microscopy.
Results
After 24 h MEFs cultured on soft hydrogels (0.5 kPa) show increased expression of class I HDACs relative to MEFs cultured on stiff hydrogels (100 kPa). A member of the class II family, HDAC4 shows a similar trend; however, there is a pronounced cytoplasmic localization on soft hydrogels suggesting a role in outside-in cytoplasmic signaling. Pharmacological inhibitor studies suggest that the opposing activities of extracellular related kinase 1/2 (ERK) and protein phosphatase 2a (PP2a) influence the localization of HDAC4. MEFs cultured on the soft hydrogels show enhanced expression of markers associated with a fibroblast–myofibroblast transition (Paxillin, αSMA).
Conclusions
We show that the phosphorylation state and cellular localization of HDAC4 is influenced by matrix mechanics, with evidence for a role in mechanotransduction and the regulation of gene expression associated with fibroblast–myofibroblast transitions. This work establishes a link between outside-in signaling and epigenetic regulation, which will assist efforts aimed at controlling gene regulation in engineered extracellular matrices.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The mechanical properties of the cell and tissue microenvironment have been shown to influence cellular functions including proliferation,34,42,46,55 migration,7 and differentiation.1,15,41,43,48,53,59 Cells sense the extracellular matrix (ECM) through cell surface receptors, and the formation of intracellular plaques called focal adhesions.25,60 Recruitment of signaling proteins, including paxilin, vinculin, and talin amongst others, serves to stabilize adhesion to facilitate cellular traction, and the subsequent propagation of force through the cellular cytoskeleton. The cytoskeleton then responds to mechanical stimuli such as changes in substrate stiffness by rearranging actin microfilaments, microtubules, and intermediate filaments47,60 which in turn influences nuclear shape and structure,40,54 chromatin condensation,23,29 and gene expression.16,26,28,45 The nucleus is generally the stiffest point within a eukaryotic cell12 and plays an essential role in mechanical strain transduction from the extracellular environment.22 The nucleus senses these changes in substrate stiffness via several proteins including the LINC (linker of nucleoskeleton and cytoskeleton) complex which is a molecular linker of the nucleus and the cytoskeleton5 and Nesprin-1, a protein which links the nucleus to the actomyosin cytoskeleton.11,30 These proteins act in a mechanosensory network with activities that convey mechanical information outside of the cell to biochemical activities within the cell. Cells respond to changes in substrate stiffness by altering nuclear morphology which then influences gene expression by controlling gene accessibility to transcription factors. On softer substrates, the nucleus is more rounded as opposed to a more flattened morphology on stiffer substrates,38 partially mediated by Nesprin-1 and the actomyosin network.11 Other proteins are also involved in mechanosensor networks, such as YAP (Yes-associated protein) and TAZ (PDZ-binding motif) which respond to differences in substrate rigidity by shuttling between the nucleus and the cytoplasm to guide the activity of transcription factors.39 YAP/TAZ sensing is dependent on changes in the actin cytoskeletal tension and requires Rho activity. On stiffer matrices, YAP/TAZ has been shown to localize to the nucleus, thus influencing the regulation of gene expression.14 While the mechanism between nuclear morphology and stiffness have been studied, direct relationships between mechanochemical signals, and the epigenetic modulators that establish the chromatin state have yet to be elucidated.
Histones play a major role in gene expression by influencing chromatin structure and by controlling the accessibility of regulatory factors to genes along the DNA.20 The affinity between DNA and histones can be regulated via a variety of reversible posttranslational modifications of the amino-acid residues within histone tails. One key modification is acetylation of lysine residues which neutralize positive charge and decrease affinity for the negative charged DNA backbone, thus relaxing the chromatin structure. In turn, deacetylation is important for transcriptional repression by allowing chromatin compaction and impeding binding of transcription factors. The regulation of histone acetylation and deacetylation effectively sets the chromatin state for gene expression regulation50 and is controlled by two primary families of enzymes: histone acetyltransferases (HATs) and histone deacetylases (HDACs). In general, HDAC activity is associated with gene silencing and transcriptional repression, though there is some evidence in promoting gene activation.62 Mammalian HDACs are further categorized into four families, class I, IIa, IIb, and IV, based on differences in structure, localization, function, and expression patterns. Class I HDACs, consisting of HDAC1, 2, 3, and 8, have high enzymatic activity and are ubiquitously expressed in mammalian cells. Class II HDACs, consisting of HDAC4, 5, 7, and 9, have differential expression patterns dependent on tissue type, and in some cases will shuttle between the nucleus and cytoplasm.63 The presence of a nuclear localization signal (NLS) located at the N-terminus, and a nuclear export sequence (NES) present at the C-terminus, serves as binding sites for chaperone protein 14-3-3, which facilitates translocation from the cytoplasm to the nucleus.64
Of the Class II HDACs, HDAC4 in particular has been implicated in controlling gene expression for a variety of cellular functions63 including chondrocyte hypertrophy during skeletogenesis,57 neuronal homeostasis,35,66 myoblast differentiation,35 and fibroblast–myofibroblast transition.17,19 When phosphorylated by protein kinases such as calcium/calmodulin-dependent protein kinase (CaMK),6 extracellular signal-regulated kinases 1 and 2 (ERK1/2),67 and glycogen synthase kinase 3 (GSK3),9 phospho-HDAC4 binds to chaperone protein 14-3-3 and is shuttled from the nucleus to the cytoplasm. In contrast, when dephosphorylated by protein phosphatases such as protein phosphatase 2A (PP2A), HDAC4 is shuttled from the cytoplasm to the nucleus.44
In this paper, we explore how the properties of the extracellular matrix influence the expression and localization of HDACs. Using model protein-conjugated hydrogels we explore nuclear morphological characteristics, acetylation state, and the expression of HDAC enzymes. We reveal a role for HDAC4 in mechanotransduction, where phosphostate—and subsequent cellular localization—are dynamically regulated in response to matrix mechanics.
Materials and Methods
Materials
Unless otherwise noted, all materials were purchased from Sigma-Aldrich Inc. Tissue culture plastic ware was purchased from VWR. Glass coverslips were purchased from Fisher Scientific. Cell culture media and reagents were purchased from Gibco. Mouse anti-HDAC1 (5356), mouse anti-HDAC2 (5113), mouse anti-HDAC3 (3949), rabbit anti-acetylated lysine (9441), rabbit anti-beta-actin (8457s), anti-rabbit IgG HRP-linked antibody (7074 s), and anti-mouse IgG HRP-linked antibody (7076P2) antibodies were purchased from Cell Signaling. Rabbit anti-HDAC4 (sc-11418) antibody is purchased from Santa Cruz Biotechnology. Rabbit anti-PP2A (ab32141), mouse anti-αSMA (ab7817), and rabbit anti-paxillin (ab32084) antibody is purchased from Abcam. Mouse anti-vinculin (V9131) and rabbit anti-pHDAC4 (SAB4504422) was purchased from Sigma-Aldrich Inc. Secondary antibodies Goat 488-anti-rabbit (ab150077) is purchased from Abcam and Goat 647-anti-mouse (A21236) along with Hoechst 33342 is purchased from Invitrogen. FR180204 (ERK inhibitor) and SP600125 (JNK inhibitor) is purchased from Calbiochem.
Cell Culture
Mouse embryonic fibroblasts (MEFs) were a generous donation from Dr. Quanxi Li in the Department of Comparative Biosciences at the University of Illinois at Urbana-Champaign. MEFs were isolated from uteri of 13.5 day pregnant CD-1 mice following an approved protocol by the Illinois Institutional Animal Use and Care Committee. MEFs were cultured in high glucose (5 g/mL) DMEM with 15% fetal bovine serum (FBS, Invitrogen) and 1% penicillin/streptomycin. Media was changed every 3–4 days and passaged at ~80% confluency with 0.5% Trypsin:EDTA. MEFs were seeded on patterned polyacrylamide hydrogels at ~200,000 cells/cm2 before fixation. For inhibition studies, FR180204 (ERK inhibitor) or SP600125 (JNK inhibitor) was supplemented to the media at 6 µM at initial seeding and each media change.
Immunocytochemistry
MEFs on surfaces were fixed with 4% paraformaldehyde (Alfa Aesar) for 20 min at room temperature. 0.1% Triton X-100 in PBS was added for 30 min to permeabilize cells which were then blocked with 1% bovine serum albumin (BSA) for 15 min. Cells were then labeled with primary antibody in 1% BSA in PBS in 4 °C overnight with mouse anti-HDAC1(1:200 dilution), mouse anti-HDAC2 (1:200 dilution), mouse anti-HDAC3 (1:200 dilution), rabbit anti-HDAC4 (1:50 dilution), rabbit anti-pHDAC4 (1:50 dilution), rabbit anti-PP2A (1:200 dilution), mouse anti-αSMA (1:200 dilution), rabbit anti-AcK (1:200 dilution), mouse anti-vinculin (1:200 dilution) or rabbit anti-paxillin (1:200 dilution). Secondary antibody labeling was performed with Goat 488-anti-rabbit (1:200 dilution) and Goat 647-anti-mouse (1:200 dilution) along with Hoechst 33342 (1:3000 dilution) for 20 min in a humid chamber (37 °C). Immunofluorescence microscopy was conducted with an INCell Analyzer 2000 (GE) or Leica Microsystems DMi8 confocal.
Western Blot
For determination of protein levels, MEFs were lysed with RIPA buffer with protease inhibitor (Santa Cruz) and protein concentration determined with Pierce BCA Protein Assay Kit (Thermo Fisher). 20 µg of protein was then mixed with 4 × Laemmli sample loading buffer (BioRad), boiled for 5 min, and separated by 10% Tris-Tricine SDS-PAGE (Bio-Rad) at 175 V for 40 min. Proteins were then electrophoretically transferred onto a nitrocellulose membrane at 100 V for 45 min. The membrane was blocked with 5% BSA in TBST for 30 min and incubated in primary antibody (1:1000) for 1 h in room temperature and detected with horseradish peroxidase-conjugated anti-mouse or anti-rabbit antibody (1:10,000) followed by Clarity Western ECL Substrate detection system (Bio-Rad).
Image Analysis and Statistical Analysis
Immunofluorescent images were analyzed using ImageJ (NIH). Average intensity of over 20 cells per pattern was measured for at least 15 patterns per condition and background intensity was subtracted. For HDAC4 and pHDAC4, the channel staining of nuclei was used to create a selection overlay for the HDAC4 and pHDAC4 channel in order to locate nuclear boundary. Western blot images were analyzed using ImageJ (NIH). Background was subtracted from the average intensity of each band and data was normalized to β-actin control.
One-way ANOVA was then employed for comparing two groups or multiple groups for statistical analysis and values of p < 0.05 were considered statistically significant. Error bars represent standard error of the mean (SEM).
Gel Preparation and Micropatterning
0.5 kPa and 100 kPA polyacryamide hydrogels gels were fabricated as previously described.56 Briefly, a mixture of 5% polyacrylamide and 0.15% bis-acylamide were created for each desired stiffness which was then conjugated with 0.1% Ammonium Persulfate (APS) and 0.1% Tetramethylenediamine (TEMED) and pipetted onto a hydrophobically treated glass slide. An amino-silanized glass coverslip was then placed on top of the mixture to create a sandwich. After polymerization, gels were lifted off and immersed in 55% hydrazine hydrate (Fisher) for 2 h to convert amide groups to reactive hydrazide groups and washed in 5% glacial acetic acid for 1 h then stored in DI water for later use.
Gel Patterning
Polydimethysiloxane (PDMS, Polysciences, Inc) was polymerized on top of SU-8 patterned silion masters fabricated via conventional photolithography to create PDMS stamps of 0.001 cm2 circles. 25 µg/ml fibronectin was incubated with Sodium Periodate for 20 min to oxidize sugar groups into aldehydes and pooled on top of the patterned PDMS stamps for 30 min. Stamps were then dried under air and applied to the surface of hydrogels for 30 s to form desired patterns.
Results and Discussion
Matrix Mechanics Influence the Expression of Histone Deacetylase Enzymes
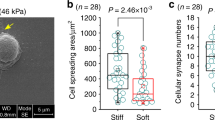
Gene expression in cells is regulated through complex networks of activity with chromatin modifying enzymes. One particular class of enzymes that influences the accessibility of DNA are the histone deacetylases (HDACs). To explore the influence of substrate stiffness on HDAC activity, we used the well-established material polyacrylamide that can be formulated to span all physiologically relevant moduli.13,65 Cells cultured on matrix coated polyacrylamide will freely migrate and self-organize in a heterogeneous fashion. In contrast, cells in tissue are spatially organized in well-defined architectures that are related to functional activity. In order to control the spatial coordination of cell populations on hydrogel substrates, we used an approach based on soft lithography to micropattern islands of covalently conjugated matrix proteins across our polyacrylamide substrates.2–4,31–33 PDMS stamps fabricated through photolithography were ‘inked’ with oxidized fibronectin and used to pattern 100,000 µm2 circles across polyacrylamide hydrogels pre-treated with hydrazine hydrate to enable covalent hydrazone linkages (Fig. 1a). Primary mouse embryonic fibroblasts (MEFs) were seeded onto the patterned substrates at high cell density and allowed to become confluent. To explore how stiffness may influence nuclear signaling, we first explored differences in nuclear area of cells adherent to our hydrogels. Both cell and nuclear area were significantly higher on the stiff (100 kPa) hydrogels compared to those cultured on soft (0.5 kPa) hydrogels (Fig. 1b). This confirms previous studies which have shown that cells cultured on softer substrates have smaller nuclear areas due to transmittance of forces via the cytoskeleton38 and differences in cell spreading.37 To ask whether differences in nuclear size were also related to changes in the chromatin state, we fixed and immunostained our cultures with an antibody against global acetylated lysine residues (Ac-K). Figure 1b shows on average there is higher Ac-K in cells cultured on the stiff hydrogels as opposed to those cultured on soft hydrogels. Higher histone acetylation may correspond to decondensed chromatin in a more open form with more active gene transcriptions21 and changes in histone modifications such as acetylation have been linked to changes in nuclear size and shape.10,27,36,49 Micropattern and mechanical strain has also been shown to modulate nuclear shape and HDAC activity36 and HDAC1 and HDAC2 has been shown to bind to nuclear actin.52 Thus, we immunostained our cultures for the class I HDACs that are known to be involved in a wide variety of cellular processes. HDAC 1 shows similar expression in cells on soft and stiff hydrogels, while HDAC 2 and 3 are more highly expressed in MEFs cultured on the soft hydrogels (Fig. 1c). Western blot analysis reveals increased HDAC1 and HDAC 2 expression on soft hydrogels; however, only the difference in HDAC 1 is statistically significant (Fig. S1). These results are consistent with the pattern in Ac-K where we would expect high HDAC activity would correspond with low Ac-K levels. Correlation plots of nuclear area and HDAC2 reveals no significant correlation (Fig. S2).
Changes in substrate stiffness leads to changes in the chromatin state for fibroblasts in culture. (a) Scheme depicting our polyacrylamide hydrogel patterning approach. (i) microcontact printing of oxidized protein; (ii) cell seeding (b) Cell nuclear area and global lysine acetylation (Ac-K) is lower in fibroblasts cultured on softer substrates. N = 12, N = 3 respectively. (c) Changes in class I HDAC expression as a function of stiffness. N = 3. Error bars represent ± SEM **p value < 0.0001; *p value < 0.05 scale bar = 90 µm.
Histone Deacetylase 4 Expression and Localization is Mechanosensitive
A class 2 deacetylase, HDAC4, has been implicated in regulating gene expression associated with a variety of cellular functions including fibroblast–myofibroblast differentiation.17,19,63 Immunofluorescence imaging of HDAC4 in our MEF cultures demonstrates a similar trend to our observations with the class 1 HDACs—higher expression on soft substrates compared to stiff substrates after 4 h of culture (Fig. S3) and at 24 h of culture (Fig. 2b). However, while HDAC1, 2, and 3 only show nuclear localization, HDAC4 will localize to both the nucleus and cytoplasm.63 In our patterned cultures after reaching confluence (24 h), we see a striking trend in localization on account of substrate stiffness, where there is significantly higher HDAC4 expression in the cytoplasm on soft gels (Fig. 2). HDAC4 localization and expression level was confirmed by confocal microscopy and western blot analysis (Figs. S4, S7c). HDAC4 is shuttled back and forth from the nucleus to the cytoplasm on account of phosphorylation state which is regulated by the opposing activities of kinase and phosphatase enzymes.18 To investigate the role of kinase and phosphatase activity in regulating HDAC4 localization, we selected inhibitors of several extracellular-related mitogen activated protein kinases (MAPK): SP600125 an inhibitor of c-Jun N-terminal Kinase (JNK), and FR180204, an inhibitor of Extracellular Related Kinase (ERK). Supplementation of our MEF cultures with the JNK inhibitor decreased total HDAC expression but did not demonstrate significant changes associated with HDAC localization (Fig. S6). However, treatment with the ERK inhibitor abrogated both the localization and stiffness-dependent differences in expression (Fig. 2). Previous studies have demonstrated how ERK activity can direct the translocation of HDAC4.67 Taken together, this suggests that ERK may play a role in regulating the phosphorylation state of HDAC4 in response to matrix stiffness.
HDAC4 localization is influenced by substrate stiffness. (a) Immunofluorescence images of HDAC4 with and without ERK inhibition in fibroblasts cultured on soft (0.5 kPa) and stiff (100 kPa) polyacrylamide hydrogels. (b) Quantitation of fluorescence intensity. N = 3. Error bars represent ± SEM. **p value <0.01; scale bar = 90 µm.
Next we investigated a phosphatase that has been previously associated with HDAC4 in regulating translocation from the cytoplasm to the nucleus, protein phosphatase 2a (PP2a).44 PP2a has been shown to dephosphorylate HDAC4 at multiple 14-3-3 binding sites, including S246, S467, S632, to regulate nuclear/cytoplasmic shuttling of HDAC4.44,61 Initially there is significantly higher PP2a and phosphorylated HDAC4 at S632 (pHDAC4) on soft gels compared to stiff gels (Fig. 3a). After 24 h we see the same trend in higher PP2a expression; however the results are not statistically significant. Interestingly, after 24 h there is a statistically significant increase in pHDAC4 on the stiff gels relative to the soft gels, which corresponds to the opposite trend in PP2a. This result suggest that PP2a activity may regulate the phosphostate and subsequent localization of HDAC4, which is consistent with previous reports of this phosphatase regulating nuclear/cytoplasmic shuttling (Figs. 3b, S7). To follow the dynamics of HDAC4 activity we performed a timecourse study and observe enhanced total HDAC4 levels in fibroblasts cultured on soft hydrogels over 5 days (Fig. 3c). On average we see higher expression of PP2a on soft hydrogels relative to stiff hydrogels over time. However, we do see a reproducible increase in nuclear PP2a over the first 2 days on stiff hydrogels; we suspect this is related to another bioactivity that is not associated with HDAC4 localization.
The phosphostate and nuclear localization of HDAC4 on soft hydrogels is guided by phosphatase PP2a. (a) Immunofluorescence images and quantitation of pHDAC4 and PP2a 4 h after fibroblast seeding. N = 3; Error bars represent ± SEM. *p value <0.001; scale bar = 90 µm. (b) Immunofluorescence images and quantitation of pHDAC4 and PP2a 24 h after fibroblast seeding. N = 3; Error bars represent ± SEM. *p value < 0.001; scale bar = 90 µm. (c) Timecourse analysis of nuclear HDAC4 and PP2a. N = 3; Error bars represent ± SEM. *0.5 kPa cytoplasmic HDAC4 is higher than nuclear, p < 0.05. §0.5 kPa cytoplasmic HDAC4 is higher than 100 kPa cytoplasmic HDAC4, p < 0.05. ¥0.5 kPa nuclear HDAC4 is significantly higher than 100 kPa nuclear HDAC4, p < 0.05. #0.5 kPa is significantly higher than 100 kPa, p value <0.05.
Myofibroblast Differentiation is Enhanced Within Microtissues on Soft Matrices
A key bioactivity where gene expression has been shown to be influenced by HDAC4 is the fibroblast–myofibroblast transition involved in wound healing17,19 where HDAC4 has been shown to be required by TFGβ1 induced myofibroblastic differentiation and silencing of HDAC4 blocks TFGβ1 stimulated αSMA expression. However, the relationship between HDAC4 localization and fibroblast–myofibroblast differentiation, and a potential role for matrix mechanics, remains to be explored. To investigate whether our MEFs are undergoing myofibroblast differentiation due to differences in HDAC4 localization on our engineered substrates, we immunostained our cultures for proteins involved in fibroblast adhesion to fibronectin. After 24 h in culture we see a trend of higher paxillin staining for MEFs cultured on soft hydrogels relative to stiff hydrogels (Figs. 4, S8). Paxillin is a focal adhesion protein which has been shown to have a regulatory role in pulmonary arterial smooth muscle cells58 and is an early marker for cardiac fibroblast to myofibroblast differentiation.8,51 Vinculin, another focal adhesion protein indicated as an early marker for cardiac fibroblast to myofibroblast differentiation,8 showed a trend of increased expression as substrate stiffness increased. When our cultures are stained for smooth muscle actin (αSMA), the actin isoform employed by myofibroblasts, we see increased expression in cells cultured on soft hydrogels, however these differences were not statistically significant. Western blot analysis corroborates the immunofluorescence results (Fig. 4c, S8). Taken together, these results suggest that matrix mechanics may influence the fibroblast to myofibroblast transition in cultured MEFs. This is interesting considering previous research with collagen coated hydrogels which indicated increased myofibroblast differentiation on stiffer environments due to TFGβ activation,24 though it is worth noting that our system has a wider range of stiffness than those previously utilized (0.5–100 kPa vs. 0.5–20 kPa). Our results suggest that HDAC4 localization plays a role in fibroblast to myofibroblast differentiation in response to stiffness. While we did not explore TGFβ signaling in our system, it is reasonable to expect that matrix mechanics will influence the secretory profile from MEFs. For instance, previous research has shown that matrix mechanics can influence secretory profiles of mesenchymal stem cells.4 Future research will involve profiling the secretome to discern relationships between matrix mechanics and cytokine secretion during fibroblast–myofibroblast transitions on micropatterned hydrogels.
Fibroblast-myofibroblast transition is influenced by matrix mechanics. (a) Immunofluorescence images of fibroblasts cultured on soft and stiff hydrogels stained for focal adhesion proteins and markers of smooth muscle. (b) Quantitation of immunofluorescence images. N = 3; Error bars represent ± SEM. (c) Western blot of focal adhesion proteins and αSMA in fibroblasts. N = 4, Error bars represent ± SEM.
Conclusions
Morphogenetic processes in vivo are coordinated by interplay between the microenvironment and intracellular pathways that regulate changes in gene expression. In this paper we report the discovery of a relationship between matrix stiffness, mechanotransduction, the regulation of HDAC localization, and fibroblast–myofibroblast differentiation. HDAC4 expression increases when fibroblasts are cultured on soft hydrogels (0.5 kPa) relative to stiff hydrogels (100 kPa), with preferential localization to the cytoplasm through phosphorylation. The opposing activities of HDAC4 and PP2a influences the phosphostate of HDAC4 which in turn determines the enzymes’ localization. We propose that fibroblasts regulate this balance in response to extracellular mechanical cues to influence HDAC4 localization, and the acetylation state during fibroblast–myofibroblast transition. This result provides an example of how class II histone deacetylase enzymes may be involved in mechanosensing as direct players in regulating cellular epigenetics and gene expression associated with morphogenesis. Since myofibroblast differentiation is involved in several physiological processes, including angiogenesis and wound healing, we believe this work will be of broad fundamental interest, and help guide the design of materials for regenerative engineering.
References
Abdeen, A. A., J. Lee, N. A. Bharadwaj, R. H. Ewoldt, and K. A. Kilian. Temporal modulation of stem cell activity using magnetoactive hydrogels. Adv. Healthc. Mater. 5(19):2536–2544, 2016.
Abdeen, A. A., J. Lee, Y. Li, and K. A. Kilian. Cytoskeletal priming of mesenchymal stem cells to a medicinal phenotype. Regen. Eng. Transl. Med. 3:5–14, 2017.
Abdeen, A. A., J. Lee, S. H. Mo, and K. A. Kilian. Spatially defined stem cell-laden hydrogel islands for directing endothelial tubulogenesis. J. Mater. Chem. B 3(40):7896–7898, 2015.
Abdeen, A. A., J. B. Weiss, J. Lee, and K. A. Kilian. Matrix composition and mechanics direct proangiogenic signaling from mesenchymal stem cells. Tissue Eng. Part A 20(19–20):2737–2745, 2014.
Alam, S. G., Q. Zhang, N. Prasad, Y. Li, S. Chamala, R. Kuchibhotla, B. Kc, V. Aggarwal, S. Shrestha, A. L. Jones, S. E. Levy, K. J. Roux, J. A. Nickerson, and T. P. Lele. The mammalian LINC complex regulates genome transcriptional responses to substrate rigidity. Sci. Rep. 6:38063, 2016.
Backs, J., K. Song, S. Bezprozvannaya, S. Chang, and E. N. Olson. CaM kinase II selectively signals to histone deacetylase 4 during cardiomyocyte hypertrophy. J. Clin. Invest. 116(7):1853–1864, 2006.
Banerjee, S., and M. C. Marchetti. Controlling cell-matrix traction forces by extracellular geometry. New J. Phys. 15(3):35015, 2013.
Baum, J., and H. S. Duffy. Fibroblasts and myofibroblasts: what are we talking about? J. Cardiovasc. Pharmacol. 57(4):376–379, 2011.
Cernotta, N., A. Clocchiatti, C. Florean, and C. Brancolini. Ubiquitin-dependent degradation of HDAC4, a new regulator of random cell motility. Mol. Biol. Cell 22(2):278–289, 2011.
Chambliss, A. B., P.-H. Wu, W.-C. Chen, S. X. Sun, and D. Wirtz. Simultaneously defining cell phenotypes, cell cycle, and chromatin modifications at single-cell resolution. FASEB J. 27(7):2667–2676, 2013.
Chancellor, T. J., J. Lee, C. K. Thodeti, and T. Lele. Actomyosin tension exerted on the nucleus through nesprin-1 connections influences endothelial cell adhesion, migration, and cyclic strain-induced reorientation. Biophys. J. 99(1):115–123, 2010.
Dahl, K. N., A. J. S. Ribeiro, and J. Lammerding. Nuclear shape, mechanics, and mechanotransduction. Circ. Res. 102(11):1307–1318, 2008.
Discher, D. E., D. J. Mooney, and P. W. Zandstra. Growth factors, matrices, and forces combine and control stem cells. Science 324(5935):1673–1677, 2009.
Dupont, S., L. Morsut, M. Aragona, E. Enzo, S. Giulitti, M. Cordenonsi, F. Zanconato, J. Le Digabel, M. Forcato, S. Bicciato, N. Elvassore, and S. Piccolo. Role of YAP/TAZ in mechanotransduction. Nature 474(7350):179–183, 2011.
Evans, N. D., C. Minelli, E. Gentleman, V. LaPointe, S. N. Patankar, M. Kallivretaki, X. Chen, C. J. Roberts, and M. M. Stevens. Substrate stiffness affects early differentiation events in embryonic stem cells. Eur. Cells Mater. 18:1–13, 2009.
Farge, E. Mechanical induction of Twist in the Drosophila foregut/stomodeal primordium. Curr. Biol. 13(16):1365–1377, 2003.
Glenisson, W., V. Castronovo, and D. Waltregny. Histone deacetylase 4 is required for TGF beta 1-induced myofibroblastic differentiation. Biochim. Biophys. Acta 1773(10):1572–1582, 2007.
Grozinger, C. M., and S. L. Schreiber. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3- 3-dependent cellular localization. Proc. Natl. Acad. Sci. USA. 97(14):7835–7840, 2000.
Guo, W., B. Shan, R. C. Klingsberg, X. Qin, and J. A. Lasky. Abrogation of TGF-beta1-induced fibroblast-myofibroblast differentiation by histone deacetylase inhibition. Am. J. Physiol. Lung Cell. Mol. Physiol. 297(5):L864–L870, 2009.
Haberland, M., R. L. Montgomery, and E. N. Olson. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat. Rev. Genet. 10(1):32–42, 2009.
Hebbes, T. R., A. W. Thorne, and C. Crane-Robinson. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 7(5):1395–1402, 1988.
Heo, S.-J., T. P. Driscoll, S. D. Thorpe, N. L. Nerurkar, B. M. Baker, M. T. Yang, C. S. Chen, D. A. Lee, and R. L. Mauck. Differentiation alters stem cell nuclear architecture, mechanics, and mechano-sensitivity. Elife, 5:e18207, 2016
Heo, S.-J., S. D. Thorpe, T. P. Driscoll, R. L. Duncan, D. A. Lee, and R. L. Mauck. Biophysical regulation of chromatin architecture instills a mechanical memory in mesenchymal stem cells. Sci. Rep. 5(1):16895, 2015.
Huang, X., N. Yang, V. F. Fiore, T. H. Barker, Y. Sun, S. W. Morris, Q. Ding, V. J. Thannickal, and Y. Zhou. Matrix stiffness-induced myofibroblast differentiation is mediated by intrinsic mechanotransduction. Am. J. Respir. Cell Mol. Biol. 47(3):340–348, 2012.
Ingber, D. E. Cellular mechanotransduction: putting all the pieces together again. FASEB J. 20(7):27, 2006.
Jagielska, A., A. L. Lowe, E. Makhija, L. Wroblewska, J. Guck, R. J. M. Franklin, G. V. Shivashankar, and K. J. Van Vliet. Mechanical strain promotes oligodendrocyte differentiation by global changes of gene expression. Front. Cell. Neurosci. 11:93, 2017.
Jain, N., K. V. Iyer, A. Kumar, and G. V. Shivashankar. Cell geometric constraints induce modular gene-expression patterns via redistribution of HDAC3 regulated by actomyosin contractility. Proc. Natl. Acad. Sci. USA 110(28):11349–11354, 2013.
Karamichos, D., R. A. Brown, and V. Mudera. Collagen stiffness regulates cellular contraction and matrix remodeling gene expression. J. Biomed. Mater. Res. A 83(3):887–894, 2007.
Kocgozlu, L., P. Lavalle, G. Koenig, B. Senger, Y. Haikel, P. Schaaf, J.-C. Voegel, H. Tenenbaum, and D. Vautier. Selective and uncoupled role of substrate elasticity in the regulation of replication and transcription in epithelial cells. J. Cell Sci. 123(Pt 1):29–39, 2010.
Kumar, A., and G. V. Shivashankar. Dynamic interaction between actin and nesprin2 maintain the cell nucleus in a prestressed state. Methods Appl. Fluoresc. 4(4):44008, 2016.
Lee, J., A. A. Abdeen, T. H. Huang, and K. A. Kilian. Controlling cell geometry on substrates of variable stiffness can tune the degree of osteogenesis in human mesenchymal stem cells. J. Mech. Behav. Biomed. Mater. 38:209–218, 2014.
Lee, J., A. A. Abdeen, A. S. Kim, and K. A. Kilian. Influence of biophysical parameters on maintaining the mesenchymal stem cell phenotype. ACS Biomater. Sci. Eng. 1(4):218–226, 2015.
Lee, J., A. A. Abdeen, K. L. Wycislo, T. M. Fan, and K. A. Kilian. Interfacial geometry dictates cancer cell tumorigenicity. Nat Mater. 15(8):856–862, 2016.
Lee, C., A. Grodzinsky, and M. Spector. The effects of cross-linking of collagen-glycosaminoglycan scaffolds on compressive stiffness, chondrocyte-mediated contraction, proliferation and biosynthesis. Biomaterials 22(23):3145–3154, 2001.
Li, J., J. Chen, C. L. Ricupero, R. P. Hart, M. S. Schwartz, A. Kusnecov, and K. Herrup. Nuclear accumulation of HDAC4 in ATM deficiency promotes neurodegeneration in ataxia telangiectasia. Nat. Med. 18(5):783–790, 2012.
Li, Y., J. S. Chu, K. Kurpinski, X. Li, D. M. Bautista, L. Yang, K.-L. P. Sung, and S. Li. Biophysical regulation of histone acetylation in mesenchymal stem cells. Biophys. J. 100(8):1902–1909, 2011.
Li, Y., D. Lovett, Q. Zhang, S. Neelam, R. A. Kuchibhotla, R. Zhu, G. G. Gundersen, T. P. Lele, and R. B. Dickinson. Moving cell boundaries drive nuclear shaping during cell spreading. Biophys. J. 109(4):670–686, 2015.
Lovett, D. B., N. Shekhar, J. A. Nickerson, K. J. Roux, and T. P. Lele. Modulation of nuclear shape by substrate rigidity. Cell. Mol. Bioeng. 6(2):230–238, 2013.
Low, B. C., C. Q. Pan, G. V. Shivashankar, A. Bershadsky, M. Sudol, and M. Sheetz. YAP/TAZ as mechanosensors and mechanotransducers in regulating organ size and tumor growth. FEBS Lett. 588(16):2663–2670, 2014.
Maniotis, A. J., C. S. Chen, and D. E. Ingber. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc. Natl. Acad. Sci. 94(3):849–854, 1997.
Mao, A. S., J.-W. Shin, and D. J. Mooney. Effects of substrate stiffness and cell-cell contact on mesenchymal stem cell differentiation. Biomaterials 98:184–191, 2016.
Mih, J. D., A. Marinkovic, F. Liu, A. S. Sharif, and D. J. Tschumperlin. Matrix stiffness reverses the effect of actomyosin tension on cell proliferation. J. Cell Sci. 125(Pt 24):5974–5983, 2012.
Park, J. S., J. S. Chu, A. D. Tsou, R. Diop, Z. Tang, A. Wang, and S. Li. The effect of matrix stiffness on the differentiation of mesenchymal stem cells in response to TGF-β. Biomaterials 32:3921–3930, 2011.
Paroni, G., N. Cernotta, C. Dello Russo, P. Gallinari, M. Pallaoro, C. Foti, F. Talamo, L. Orsatti, C. Steinkuhler, and C. Brancolini. PP2A regulates HDAC4 nuclear import. Mol. Biol. Cell 19(2):655–667, 2007.
Provenzano, P. P., D. R. Inman, K. W. Eliceiri, and P. J. Keely. Matrix density-induced mechanoregulation of breast cell phenotype, signaling and gene expression through a FAK-ERK linkage. Oncogene 28(49):4326–4343, 2009.
Provenzano, P. P., and P. J. Keely. Mechanical signaling through the cytoskeleton regulates cell proliferation by coordinated focal adhesion and Rho GTPase signaling. J. Cell Sci. 124(Pt 8):1195–1205, 2011.
Ralphs, J. R., A. D. Waggett, and M. Benjamin. Actin stress fibres and cell-cell adhesion molecules in tendons: organisation in vivo and response to mechanical loading of tendon cells in vitro. Matrix Biol. 21(1):67–74, 2002.
Ren, K., T. Crouzier, C. Roy, and C. Picart. Polyelectrolyte multilayer films of controlled stiffness modulate myoblast cells differentiation. Adv. Funct. Mater. 18(9):1378–1389, 2008.
Rozwadowska, N., T. Kolanowski, E. Wiland, M. Siatkowski, P. Pawlak, A. Malcher, T. Mietkiewski, M. Olszewska, and M. Kurpisz. Characterisation of nuclear architectural alterations during in vitro differentiation of human stem cells of myogenic origin. PLoS ONE 8(9):e73231, 2013.
Ruthenburg, A. J., H. Li, D. J. Patel, and C. D. Allis. Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 8(12):983–994, 2007.
Santiago, J.-J., A. L. Dangerfield, S. G. Rattan, K. L. Bathe, R. H. Cunnington, J. E. Raizman, K. M. Bedosky, D. H. Freed, E. Kardami, and I. M. C. Dixon. Cardiac fibroblast to myofibroblast differentiation in vivo and in vitro: expression of focal adhesion components in neonatal and adult rat ventricular myofibroblasts. Dev. Dyn. 239(6):1573–1584, 2010.
Serebryannyy, L. A., C. M. Cruz, P. de Lanerolle, R. D. Mullins, and P. D. McCrea. A role for nuclear actin in HDAC 1 and 2 regulation. Sci. Rep. 6(1):28460, 2016.
Shih, Y.-R. V., K.-F. Tseng, H.-Y. Lai, C.-H. Lin, and O. K. Lee. Matrix stiffness regulation of integrin-mediated mechanotransduction during osteogenic differentiation of human mesenchymal stem cells. J. Bone Miner. Res. 26(4):730–738, 2011.
Swift, J., I. L. Ivanovska, A. Buxboim, T. Harada, P. C. D. P. Dingal, J. Pinter, J. D. Pajerowski, K. R. Spinler, J.-W. Shin, M. Tewari, F. Rehfeldt, D. W. Speicher, and D. E. Discher. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science 341(6149):1240104, 2013.
Tan, P. S., and S. H. Teoh. Effect of stiffness of polycaprolactone (PCL) membrane on cell proliferation. Mater. Sci. Eng. C 27(2):304–308, 2007.
Tse, J. R., and A. J. Engler. Preparation of hydrogel substrates with tunable mechanical properties. Curr. Protoc. Cell Biol. 47:10.16:10.16.1–10.16.16, 2010
Vega, R. B., K. Matsuda, J. Oh, A. C. Barbosa, X. Yang, E. Meadows, J. McAnally, C. Pomajzl, J. M. Shelton, J. A. Richardson, G. Karsenty, and E. N. Olson. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell 119(4):555–566, 2004.
Veith, C., L. M. Marsh, M. Wygrecka, K. Rutschmann, W. Seeger, N. Weissmann, and G. Kwapiszewska. Paxillin regulates pulmonary arterial smooth muscle cell function in pulmonary hypertension. Am. J. Pathol. 181(5):1621–1633, 2012.
Wang, L.-S., J. Boulaire, P. P. Y. Chan, J. E. Chung, and M. Kurisawa. The role of stiffness of gelatin-hydroxyphenylpropionic acid hydrogels formed by enzyme-mediated crosslinking on the differentiation of human mesenchymal stem cell. Biomaterials 31(33):8608–8616, 2010.
Wang, N., J. P. Butler, and D. E. Ingber. Mechanotransduction across the cell surface and through the cytoskeleton. Science 260(5111):1124–1127, 1993.
Wang, A. H., M. J. Kruhlak, J. Wu, N. R. Bertos, M. Vezmar, B. I. Posner, D. P. Bazett-Jones, and X. J. Yang. Regulation of histone deacetylase 4 by binding of 14-3-3 proteins. Mol. Cell. Biol. 20(18):6904–6912, 2000.
Wang, A., S. K. Kurdistani, and M. Grunstein. Requirement of Hos2 histone deacetylase for gene activity in yeast. Science 298(5597):1412–1414, 2002.
Wang, Z., G. Qin, and T. C. Zhao. HDAC4: mechanism of regulation and biological functions. Epigenomics 6(1):139–150, 2014.
Wang, A. H., and X. J. Yang. Histone deacetylase 4 possesses intrinsic nuclear import and export signals. Mol. Cell. Biol. 21(17):5992–6005, 2001.
Yeung, T., P. C. Georges, L. A. Flanagan, B. Marg, M. Ortiz, M. Funaki, N. Zahir, W. Ming, V. Weaver, and P. A. Janmey. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil. Cytoskelet. 60(1):24–34, 2005.
Yuan, H., K. Denton, L. Liu, X. J. Li, S. Benashski, L. McCullough, and J. Li. Nuclear translocation of histone deacetylase 4 induces neuronal death in stroke. Neurobiol. Dis. 91:182–193, 2016.
Zhou, X., V. M. Richon, A. H. Wang, X.-J. Yang, R. A. Rifkind, and P. A. Marks. Histone deacetylase 4 associates with extracellular signal-regulated kinases 1 and 2, and its cellular localization is regulated by oncogenic Ras. Proc. Natl. Acad. Sci. 97(26):14329–14333, 2000.
Acknowledgments
The authors would like to thank Dr. Quanxi Li in the Department of Comparative Biosciences at the University of Illinois for the generous isolation and donation of MEFs and Dr. Karin Jensen in the Department of Bioengineering at the University of Illinois for guidance with western blots.
Conflict of interest
Yanfen Li, Claire B. Tang, and Kristopher A. Kilian declare that they have no conflicts of interest.
Funding
Funding was provided by the National Science Foundation Grant No. 1454616 CAR. Y.L. was supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE – 1144245.
Research Involved in Human and Animal Rights
No humans or animals were employed in this research by the authors.
Author information
Authors and Affiliations
Corresponding author
Additional information
Associate Editor Alyssa Panitch oversaw the review of this article.
Kristopher A. Kilian
received B.S. and M.S. degrees in Chemistry from the University of Washington in 1999 and 2003 respectively. He worked for Merck Research Labs in the Methods Development group from 2000 to 2004 before travelling to Sydney, Australia to do his PhD with Justin Gooding at the University of New South Wales. In 2007, he joined the laboratory of Milan Mrksich at the University of Chicago as a postdoctoral fellow to investigate new methods for directing the differentiation of stem cells. Kris joined the faculty of the University of Illinois at Urbana-Champaign in 2011 with appointments in the Departments of Materials Science and Engineering, and Bioengineering. Kris is a 2008 recipient of the NIH Ruth L. Kirchstein National Research Service Award, a 2014 Kavli Fellow of the 19th German-American Frontiers of Science, and a 2015 recipient of the National Science Foundation’s CAREER award. His research interests include the design and development of model extracellular matrices for cell engineering and fundamental studies in cell biology.

This article is part of the 2017 CMBE Young Innovators special issue.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, Y., Tang, C.B. & Kilian, K.A. Matrix Mechanics Influence Fibroblast–Myofibroblast Transition by Directing the Localization of Histone Deacetylase 4. Cel. Mol. Bioeng. 10, 405–415 (2017). https://doi.org/10.1007/s12195-017-0493-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12195-017-0493-8