Abstract
The tumor-suppressing role of miR-455-3p has been reported in lung cancer, but the working mechanism remains to be fully elucidated. This study aims to explore the possible mechanism of miR-455-3p in regulating epithelial–mesenchymal transition (EMT) progression and angiogenesis in non-small cell lung cancer (NSCLC) cells.
The expressions of miR-455-3p, HSF1, GLS1, and EMT-related proteins (E-cadherin, N-cadherin, vimentin, and Snail-1) in both NSCLC tissues and cell lines were determined by RT-qPCR and western blot. After cell transfection, cell proliferation and angiogenesis ability on NSCLC cells were assessed by MTT and tube formation assay. The binding of miR-455-3p with HSF1 was measured by luciferase reporter gene assay, while the interaction between HSF1 and GLS1 was determined by co-immunoprecipitation assay (Co-IP).
HSF1 was highly expressed in NSCLC tissues and cells. Inhibition of HSF1 expression or overexpression of miR-455-3p in NSCLC cells can suppress cell proliferation, angiogenesis ability, and EMT progression. miR-455-3p was found to negatively regulate HSF1 expression. Co-transfection of miR-455-3p overexpression and HSF1 inhibition in NSCLC cells showed that miR-455-3p can partially counteract the effect of HSF1 in NSCLC cells. HSF1 can interact with GLS1 and elevate the expression of GLS1. GLS1 can partially abolish the suppressive effect of miR-455-3p in NSCLC cells.
miR-455-3p can bind HSF1 to suppress the GLS1 in NSCLC cells, therefore suppressing EMT progression and angiogenesis of NSCLC cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Improvements in the treatment of non-small cell lung cancer (NSCLC) have been achieved in the last decades, including higher efficiency on early diagnosis and better understanding on the tumor biological function (Duma et al. 2019). Despite that, the overall survival rate for NSCLC patients remains far from satisfactory, making it more urgent to probe for new drugs and therapy strategies (Herbst et al. 2018). The molecular diagnosis and targeted therapy for NSCLC are being explored, and better understanding on the implication of gene alternation in disease progression may facilitate this process (Jonna and Subramaniam, 2019).
Heat shock factor 1 (HSF1) was upregulated in many solid tumors (Wan et al. 2018), and the HSF1 stress pathway is strongly suggested as a therapeutic target for various cancers (Workman 2020). Previous study showed that as an anti-apoptotic protein, BCL-2-interacting cell death suppressor (BIS) was subjected to the regulation of HSF1 upon various stresses in A549 NSCLC cells (Yun et al. 2018). However, how HSF1 was regulated in NSCLC cells remains to be determined.
To explore the upstream target gene of HSF1 in NSCLC cells, five miRs (miR-3187-3p, miR-217, miR-6905-3p, miR-455-3p, and miR-652-3p) with binding sites with HSF1 were selected in Starbase, among which miR-455-3p was selected for further exploration. miR-455 is a tumor suppressor in NSCLC cells and suggested as a potential therapeutic target (Li et al. 2016a, b). Moreover, not much evidence supported the interaction of miR-455-3p with HSF1 in NSCLC cells, except for a study focused on liver fibrosis suggesting that miR-455-3p can suppress the activation of hepatic stellate cells through regulating HSF1 (Wei et al. 2019). This study attached its importance on the regulation of miR-455-3p/HSF1 axis on epithelial–mesenchymal transition (EMT) progression and angiogenesis of NSCLC cells.
Glutaminase 1 (GLS1) is a potential promising therapeutic target of pulmonary fibrosis (Cui et al. 2019) and is associated with tumor metabolism (Cao et al. 2019). In colorectal cancer cells, HSF1 stimulated GLS1-dependent mTOR activation to promote colorectal carcinogenesis (Li et al. 2018). In this study, we applied co-immunoprecipitation assay (Co-IP) to validate the interaction of HSF1 with GLS1. Therefore, this study mainly focused on exploring the possible effect of miR-455-3p in biological activities of NSCLC cells. The results demonstrated the regulation of miR-455-3p on HSF1/GLS1 axis and highlighted the potential of miR-455-3p as a therapeutic target for NSCLC treatment.
Materials and methods
Collection of clinical samples
The tumor tissues and adjacent normal margin tissues (ANM tissues) were collected from 30 NSCLC patients admitted in our hospital between October 2018 and December 2020. The included subjects received neither radio-/chemotherapy nor clinical treatment before diagnosis. The collected tissues were stored in liquid nitrogen within 5 min and then preserved in a refrigerator at − 80 °C. The experimental design of this study was approved by the ethical committee of Hainan Affiliated Hospital of Hainan Medical University. The clinical tissues were collected with the informed consent signed by each included patient. The study was performed strictly based on the Declaration of Helsinki.
Cell culture and transfection
Human NSCLC cell lines (A549, H1299), human normal pulmonary epithelial cells (BEAS-2B), and human umbilical vein endothelial cells (HUVECs) were purchased from ATCC (NY, USA). Cells were cultured in RPMI 1640 culture medium (A1049101, gbico, NY, USA) which contains 10% fetal bovine serum (FBS, 16,140,071, gbico, NY, USA) in an incubator at 37 °C with 5% CO2.
The plasmids overexpressing HSF1 (OE-HSF1) or inhibiting HSF1 (sh-HSF1), the plasmids overexpressing GLS1 (OE-GLS1) or inhibiting GLS1 (sh-GLS1), miR-455-3p mimic (miR-455-3p mimic), plasmid inhibiting miR-455-3p (miR-455-3p inhibitor), and corresponding negative controls (OE-NC, sh-NC, mimic-NC, and inhibitor-NC) were purchased from Genechem (Shanghai Genechem Co., Ltd., Shanghai, China). Cells were seeded into 60-nm culture disks at the density of 3.0 × 105 cells/disk for 24 h before cell transfection. The transfection vector (3 μg) was supplemented with Lipofectamine 2000 regent (11,668,019, Invitrogen, California, USA), Opti-MEM I Reduced Serum Medium (31,985,062, gibco, NY, USA), and 8 ng/mL polybrene (TR-1003, Sigma-Aldrich, St. Louis, MO, USA) for cell incubation for 48 h.
MTT
Cells were harvested and adjusted into 1 × 104 cells/mL before inoculation into a 96-well plate for incubation at 37 ℃ with 5% CO2 for, respectively, 24, 48, and 72 h before 20 μL 5 mg/mL MTT was added into the cells for further incubation at a constant temperature incubator at 37 °C for 4 h. After that, the excessive liquid was removed, and 200 μL DMSO was added for fully mixing. The optical density (OD) for cells in each well was measured at a microplate (Bio-Rad Laboratories) with the wave length of 490 nm.
Dual luciferase reporter gene assay
Starbase (http://starbase.sysu.edu.cn/) predicted the binding sites of HSF1 with miR-455-3p. The miRNA target button in the toolbar was chosen, and the human in genome was chosen, after which miR-455-3p was input in the microRNA search box and HSF1 was inserted in the target gene search box for submission to obtain the binding sequence of miR-455-3p and HSF1. The mutant and wide sequences (MT-HSF1 and WT-HSF1) were designed and synthesized accordingly and then inserted into the luciferase reporter vector (pGL3-Basic), followed by co-transfection with miR-455-3p mimic (30 nM) or negative control (mimic-NC) into NSCLC cells. The activities of firefly luciferase and Renilla luciferase were measured, and the relative luciferase activity was calculated using Renilla luciferase activity as the internal control.
Tube formation assay
Matrigel (50 μL, BD Bioscience, USA) was coated into a 96-well plate and incubated with for 0.5 h at 37 °C. The HUVECs and NSCLC cells were centrifuged at 1000 rpm for 5 min, from which the supernatant of tumor cells was used to re-suspend HUVECs. HUVECs (4 × 104 cell/well) were inoculated into the Matrigel for incubation of 6 h at 37 °C with 5% CO2. The tube formation was observed under an inverted microscope (CKX40, Olympus, NY, USA).
Co-IP
The proteins obtained from cell lysis were incubated with anti-GLS1 antibody (ab156876, Abcam, Cambridge, UK) and 20 µL protein A/G agarose (Pierce) at 4 °C for overnight. The sediments were washed with lysis buffer for 4 times and then suspended in 5 × SDS-PAGE loading buffer. After boiling for 5 min, the proteins were analyzed by western blot.
Quantitative reverse transcription polymerase chain reaction (RT-qPCR)
Cells or tissues were dissolved into 1 mL Trizol (Thermo Fisher Scientific, MA, USA) for extraction of total RNA. Then M-MLV reverse transcriptase and random primers were used to obtain the cDNA. The PCR reaction system was prepared using Premix Ex Taq™II kit (Takara, Dalian, China), and ABI7500 quantitative PCR instrument (Applied Biosystems, Shanghai, China) was used for RT-PCR. GAPDH was used as the internal control. Data were analyzed using 2−ΔΔCt method (Burja et al. 2019): ΔΔCt = [Ct(target gene)-Ct(internal control)]experimental group-[Ct(target gene)-Ct(internal control)]control group. The primer sequences are listed in Table 1.
Western blot
The tissues and cells were lysed to measure the concentration of total protein using the BCA protein kit (23,227, Thermo Fisher, USA). Then the proteins were diluted with 5 × loading buffer and electrophoresed in 12% separating gel for 90 min before being blocked in PBS blocking solution for 1 h, which contains 5% (w/v) skim milk powder. Proteins were incubated with primary antibodies of HSF1 (1:500, ab52757, Abcam, Cambridge, UK), GLS1 (1:500, ab156876, Abcam, Cambridge, UK), N-cadherin (1:1000, ab76057, Abcam, Cambridge, UK), anti-E-cadherin (1:1000, ab194982, Abcam, Cambridge, UK), anti-Snail 1 (1:1000, ab53519, Abcam, Cambridge, UK), or anti-vimentin (1:1000, ab92547, Abcam, Cambridge, UK), or GAPDH (1:500, ab8245, Abcam, Cambridge, UK) for overnight at 4 °C. After PBS washing, the proteins were then incubated with secondary antibody (1:500, ab150077, Abcam, Cambridge, UK) at room temperature for 1 h. The expressions of target protein were photographed using BioSpectrum Imaging System (UVP, USA).
Statistical analysis
All experiments were conducted for three times, unless otherwise indicated. Data analysis was achieved using SPSS 18.0 (IBM Corp., Armonk, NY, USA) and GraphPad Prism 6.0 (GraphPad Software Inc.). Measurement data were expressed as mean ± standard deviation. Comparison between two groups was analyzed using t test, while those among groups were conducted using one-way analysis of variance and post hoc Turkey’s tests. P value of less than 0.05 was considered to having statistical significance.
Results
Upregulated expression of HSF1 in lung cancer cells
Data in TCGA showed that HSF1 was highly expressed in squamous cell lung carcinoma and lung adenocarcinoma (Fig. 1A). RT-qPCR and western blot showed that compared with that in ANM tissues, the expression of HSF1 was higher in NSCLC tissues (Fig. 1B–C) (P < 0.01). Further measurement of HSF1 in lung cancer cells showed that HSF1 was highly expressed in A549 and H1299 cells when compared with BEAS-2B cell (Fig. 1D–E) (P < 0.01). Taken together, HSF1 expression may associate with the progression of NSCLC.
HSF1 was found to be highly expressed in NSCLC tissues and cells. Note: (A) HSF1 expression in NSCLC tissues as shown in TCGA; (B–E) RT-qPCR (B, D) and western blot (C, E) were used to measure the mRNA and protein expressions of HSF1. Compared with ANM group or BEAS-2B group, **P < 0.01. HSF1, 85 kDa. NSCLC, non-small cell lung cancer; ANM, adjacent normal margin; BEAS-2B, a kind of human normal pulmonary epithelial cells; RT-qPCR, quantitative reverse transcription polymerase chain reaction
HSF1 promotes EMT and angiogenesis in NSCLC cells
OE-HSF1, OE-NC, sh-HSF1, or sh-NC was transfected into A549 and H1299 cells to explore the alternation of HSF1 on biological functions of NSCLC cells. RT-qPCR and western blot consistently displayed increased expression of HSF1 in OE-HSF1 group (Fig. 2A) vs OE-NC group (P < 0.01) and decreased expression of HSF1 in sh-HSF1 group (Fig. 2B) vs sh-NC group (P < 0.01), suggesting the overexpression or inhibition on HSF1 expression in NSCLC cells was successfully achieved. The cell viability of cells was determined by MTT assay. Compared with OE-NC group, the cell proliferation abilities of cells in OE-HSF1 were enhanced, while reversed expression pattern was found in sh-HSF1 group when compared with sh-NC group (Fig. 2C) (P < 0.01). The detection on the expressions of EMT-related proteins showed that compared with OE-NC group, OE-HSF1 group had suppressed expression of E-cadherin while elevated expressions of N-cadherin, vimentin, and Snail-1 (Fig. 2D–E) (P < 0.01). Different expression patterns were found in sh-HSF1 group when compared with sh-NC group (Fig. 2D–E) (P < 0.01). The supernatant in each group was collected to co-incubate with HUVECs to verify the possible effect of HSF1 on cell angiogenesis. The angiogenesis ability of HUVECs in OE-HSF1 group was enhanced, while that in sh-HSF group was inhibited when, respectively, compared with OE-NC group or sh-NC group (Fig. 2F) (P < 0.01). Collectively, the overexpression of HSF1 can promote the EMT progression and angiogenesis of NSCLC cells.
Alternation of HSF1 affects the EMT progression and angiogenesis of NSCLC cells. Note: the overexpression or inhibition of HSF1 was achieved in NSCLC cells. RT-qPCR (A) and western blot (B) were applied to detect the expression of HSF1 after cell transfection. MTT assay was applied to determine the proliferation ability of NSCLC cells (C). RT-qPCR (D) and western blot (E) measured the expressions of EMT-related proteins. The supernatant of NSCLC cells was collected to incubate with HUVECs to measure the angiogenesis ability (F). N = 3, *P < 0.05, **P < 0.01. HSF1, 85 kDa; E-cadherin, 97 kDa; N-cadherin, 100 kDa; vimentin, 54 kDa; Snail-1, 29 kDa. EMT, epithelial–mesenchymal transition; NSCLC, non-small cell lung cancer; RT-qPCR, quantitative reverse transcription polymerase chain reaction; HUVECs, human umbilical vein endothelial cells
miR-455-3p targets and negatively regulates HSF1 expression
To explore the upstream miR of HSF1 in NSCLC, five miRs with binding sites with HSF1 were selected in Starbase, whose expressions were detected in NSCLC cells. Among the five miRs, miR-455-3p was downregulated in both NSCLC tissues and cell lines (Fig. 3A) (P < 0.01). The binding sites of miR-455-3p with HSF1 are shown in Fig. 3B. The WT-HSF1 or MT-HSF1 was co-transfected with miR-455-3p mimic or mimic-NC. Detection on luciferase activity showed that co-transfection of WT-HSF1 with miR-455-3p mimic can suppress the luciferase activity (Fig. 3C) (P < 0.01), while co-transfection of MT-HSF1 with miR-455-3p mimic showed no significant effect on luciferase activity, suggesting that miR-455-3p can bind with HSF1.
miR-455-3p negatively regulates HSF1 expression in NSCLC cells. Note: the upstream miRs of HSF1 in NSCLC were searched in Starbase, whose miR expressions were determined by RT-qPCR (A). The bind sites of miR-455-3p with HSF1 as predicted by Starbase (B). The binding of miR-455-3p with HSF1 was detected by luciferase reporter gene assay (C). The transfection efficiency of miR-455-3p in NSCLC cells was measured by RT-qPCR (D). The mRNA (E) and protein (F) expressions of HSF1 were determined by RT-qPCR and western blot. N = 3, compared with ANM group or BEAS-2B group, *P < 0.05, **P < 0.01. HSF1, 85 kDa. NSCLC, non-small cell lung cancer; RT-qPCR, quantitative reverse transcription polymerase chain reaction
To further explore the regulation of miR-455-3p on HSF1, the overexpression or inhibition of miR-455-3p expression was achieved in NSCLC cells, and then the expression of HSF1 was determined by RT-qPCR and western blot. Compared with mimic-NC group or inhibitor-NC group, the expression of miR-455-3p was elevated in miR-455-3p mimic group, while suppressed in miR-455-3p inhibitor group (Fig. 3D) (P < 0.01), suggesting miR-455-3p mimic and miR-455-3p inhibitor were successfully transfected into NSCLC cells. Moreover, compared with mimic-NC group, the expression of HSF1 was suppressed in miR-455-3p mimic group, while compared with inhibitor-NC group, the expression of HSF1 was elevated in miR-455-3p inhibitor group (Fig. 3E–F) (P < 0.01). Taken together, in NSCLC, miR-455-3p can target and negatively regulate the expression of HSF1.
miR-455-3p suppresses EMT and angiogenesis of NSCLC cells through regulating HSF1
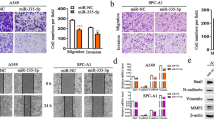
To determine whether miR-455-3p can regulate HSF1 expression to influence biological functions of NSCLC cells, miR-455-3p mimic, miR-455-3p inhibitor, OE-HSF1, or sh-HSF1 was transfected into NSCLC cells. The measurement on EMT-related proteins showed that in contrast to control group, the proliferation ability of cells was suppressed, and the expression of E-cadherin was increased, while the expressions of N-cadherin, vimentin, and Snail-1 were suppressed, and the angiogenesis was also inhibited in miR-455-3p mimic group (vs control group) and in miR-455-3p inhibitor + sh-HSF1 group (vs miR-455-3p inhibitor group). Different expression patterns were found in miR-455-3p mimic group and miR-455-3p mimic + OE-HSF1 group, when, respectively, compared with control group or miR-455-3p inhibitor group (Fig. 4A–F) (P < 0.01). Taken together, the overexpression of miR-455-3p can partially suppress the enhancive effect of HSF1 on EMT and angiogenesis in NSCLC cells.
miR-455-3p suppresses EMT and angiogenesis ability of NSCLC cells through HSF1. Note: the overexpression or inhibition of miR-455-3p and HSF1 was achieved in NSCLC cells. MTT assay was applied to measure the proliferation ability of cells (A). The mRNA (B) and protein (C) expressions of EMT-related proteins were determined by RT-qPCR and western blot. The angiogenesis of cells was measured in HUVECs by tube formation assay (D). N = 3, *P < 0.05, **P < 0.01. E-cadherin, 97 kDa; N-cadherin, 100 kDa; vimentin, 54 kDa; Snail-1, 29 kDa. NSCLC, non-small cell lung cancer; EMT, epithelial–mesenchymal transition; RT-qPCR, quantitative reverse transcription polymerase chain reaction
HSF1 binds GLS1 to enhance GLS1 expression
Evidence in colorectal cancer showed that HSF1 can enhance the expression of GLS1 (Li, et al. 2018). To verify such regulatory expression in NSCLC cells, we applied co-IP to verify the binding of HSF1 with GLS1. Western blot showed that HSF1 can specifically bind GLS1 in NSCLC cells (Fig. 5A). Then HSF1 overexpression and inhibition plasmid was transfected into NSCLC cells. RT-qPCR and western blot demonstrated that compared with OE-NC group, the expression of GLS1 was enhanced in OE-HSF1 group, while compared with sh-NC group, the expression of GLS1 in sh-HSF1 group was suppressed (Fig. 5B–C) (P < 0.01). Collectively, HSF can bind GLS1 and enhance the expression of GLS1 in NSCLC cells.
HSF1 enhances GLS1 expression in NSCLC cells. Note: co-IP was applied to measure the binding of HSF1 with GLS1 (A). The mRNA (B) and protein (C) expressions of GLS1 were determined by RT-qPCR and western blot. N = 3, **P < 0.01. GLS1, 73 kDa; H3, 15 kDa; HSF1, 85 kDa. NSCLC, non-small cell lung cancer; RT-qPCR, quantitative reverse transcription polymerase chain reaction; Co-IP, co-immunoprecipitation assay
miR-455-3p regulates EMT and angiogenesis of NSCLC cells through HSF1/GLS1 axis
miR-455-3p or GLS1 suppression was achieved in NSCLC cells to verify the regulatory role of miR-455-3p on GLS1 expression. RT-qPCR and western blot showed that compared with sh-NC group, the expression of GLS1 in sh-GLS1 group was suppressed, indicating GLS1 was successfully suppressed in NSCLC cells (Fig. 6A–B) (P < 0.01). Further measurement on EMT and angiogenesis showed that compared with miR-455-3p inhibitor group, the miR-455-3p inhibitor + sh-GLS11 group had suppressed proliferation and angiogenesis ability; inhibited expressions of N-cadherin, vimentin, and Snail-1; and increased E-cadherin expression (Fig. 6C–H) (P < 0.01). Those results suggested that GLS1 can partially counteract the suppressive effect of miR-455-3p on EMT progression and angiogenesis ability of NSCLC cells.
miR-455-3p regulates EMT and angiogenesis of NSCLC cells through HSF1/GLS1 axis. Note: NSCLC cells were transfected with sh-GLS1 and miR-455-3p inhibitor. The mRNA (A) and protein (B) expressions of GLS1 were determined by RT-qPCR and western blot. Cell proliferation ability was assessed by MTT (C). The mRNA (D) and protein (E) expressions of EMT-related proteins were determined by RT-qPCR and western blot. The angiogenesis of cells was measured in HUVECs by tube formation assay (F). N = 3, *P < 0.05, **P < 0.01. GLS1, 73 kDa; E-cadherin, 97 kDa; N-cadherin, 100 kDa; vimentin, 54 kDa; Snail-1, 29 kDa. NSCLC, non-small cell lung cancer; EMT, epithelial–mesenchymal transition; RT-qPCR, quantitative reverse transcription polymerase chain reaction; HUVECs, human umbilical vein endothelial cells
Discussion
Currently, lung cancer ranks the top for mortality associated with cancers, which make it an evitable appealing for new therapeutic approaches. In this study, we mainly focused on exploring the possible effect of HSF1 in the EMT and angiogenesis ability of NSCLC cells. The results showed that miR-455-3p can suppress HSF1/GLS1 axis and therefore hinder the EMT event and angiogenesis development of NSCLC cells.
We searched TCGA to identify the expression pattern of HSF1 in lung cancers, which showed that HSF1 was highly expressed in tumor tissues, which was consistent with the detection on lung cancer cells (A549 and H1299 cell lines), indicating the HSF1 may be implicated in the progression NSCLC and plays a tumor promotive role in NSCLC. Further measurement on phenotypes of NSCLC cells demonstrated the effect of HSF1 overexpression in promoting EMT and angiogenesis ability of NSCLC cells. Traditionally, HSF1 can be activated in response to several cellular stresses, including heat shock, which contributes to the expressions of HSF1-mediated heat shock proteins in order to ensure the proteome stability and proteostasis (Carpenter and Gokmen-Polar, 2019). HSF1 was also found to be highly expressed in tumor tissues and cells, supporting multiple facets of malignancy. The association of HSF1 with tumor aggressiveness has long been identified in previous studies, which identified several mechanisms for HSF1 regulation on cell proliferation, EMT, migration, and metastasis (Jiang et al. 2015). Therefore, HSF1 has long been exploited as a therapeutic target for various cancers, and evidence in previous studies identified that the activation or suppression of HSF1 expression was regulated by cell molecules or signals jointly (Wang et al. 2020). For instance, in colorectal cancer cells, miR455-3p suppression can induce m6A modification, which in turn regulates the translation of HSF1 mRNA (Song et al. 2020). But how HSF1 was regulated or how HSF1 regulates its downstream targets remains to be further determined.
Through data searching in Starbase, we identified five miRs may bind HSF1. After further detection in NSCLC tissues and cells, miR455-3p was finally selected for its downregulated expression pattern in NSCLC tissues and cells. Luciferase reporter gene assay supported the binding of miR455-3p with HSF1. Western blot and tube formation assay on EMT-related proteins and angiogenesis ability showed that miR-455-3p overexpression can slow down the EMT and angiogenesis progression, while co-transfection of miR-455-3p and HSF1 demonstrated that the tumor promotive role of HSF1 can be partially abolished by miR-455-3p overexpression. Taken together, miR-455-3p can suppress HSF1 to inhibit the EMT progression and angiogenesis ability in NSCLC cells. The regulation of miRs on HSF1 can be found in other diseases. For instance, in acute myeloid leukemia, miR-34b can target the expression of HSF1 to suppress the activation of downstream Wnt-β-catenin pathway, so as to inhibiting cell proliferation and enhancing cell apoptosis (Li et al. 2016a, b). In hepatocellular carcinoma, HSF1 was subjected to the regulation of miR-644a (Liang et al. 2018). Furthermore, we also noticed the regulation of HSF1 on GLS1 in colorectal cancer cells (Li, et al. 2018), which hinted the possibility of their interaction in NSCLC cells. The testing on metabolic dependency of NSCLC cell lines for glutamine supply showed that NSCLC cells are highly sensitive to the glutamine supply (Lee et al. 2016). Glutamine is one of the key nutrients responsible for various biological functions, including energy formation, cell signaling, and biosynthesis, and for supporting the biological hallmarks of malignancy (Hensley et al. 2013). Data in a previous study showed that inhibition on GLS1 can limit the tumor growth and metastatic progression of osteosarcoma cells (Ren et al. 2020). To this end, co-IP and western blot were applied to verify the bind of HSF1 with GLS1. Unsurprisingly, HSF1 can bind GLS1 and increase the expression level of GLS1 in NSCLC cells. Meanwhile, miR-455-3p was found to regulate the HSF1/GLS1 axis and therefore to hinder the progression of NSCLC.
Conclusions
In summary, this study supported the important role of miR-455-3p, HSF1, and GLS1 in EMT and angiogenesis ability of NSCLC cells. This study also showed that miR-455-3p can suppress the EMT progression and angiogenesis ability of NSCLC cells through regulating the HSF1/GLS1 axis. As miR-455-3p may not be the only one upstream target of HSF1 in lung cancer cells and this study only focused on miR-455-3p, more evidence is required, and more research are needed to better understand the implication of HSF1 in the progression of NSCLC cells.
Availability of data and materials
The datasets used or analyzed during the current study are available from the corresponding author on reasonable request.
Change history
16 February 2022
A Correction to this paper has been published: https://doi.org/10.1007/s12192-022-01259-z
References
Burja B et al (2019) Olive leaf extract attenuates inflammatory activation and DNA damage in human arterial endothelial cells. Front Cardiovasc Med 6(56.
Cao J et al (2019) Expression of GLS1 in intrahepatic cholangiocarcinoma and its clinical significance. Mol Med Rep 20(2):1915–1924
Carpenter RL, Gokmen-Polar Y (2019) HSF1 as a cancer biomarker and therapeutic target. Curr Cancer Drug Targets 19(7):515–524
Cui H et al (2019) Inhibition of glutaminase 1 attenuates experimental pulmonary fibrosis. Am J Respir Cell Mol Biol 61(4):492–500
Duma N, Santana-Davila R, Molina JR (2019) Non-small cell lung cancer: epidemiology, screening, diagnosis, and treatment. Mayo Clin Proc 94(8):1623–1640
Hensley CT, Wasti AT, DeBerardinis RJ (2013) Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest 123(9):3678–3684
Herbst RS, Morgensztern D, Boshoff C (2018) The biology and management of non-small cell lung cancer. Nature 553(7689):446–454
Jiang S et al (2015) Multifaceted roles of HSF1 in cancer. Tumour Biol 36(7):4923–4931
Jonna S, Subramaniam DS (2019) Molecular diagnostics and targeted therapies in non-small cell lung cancer (NSCLC): an update. Discov Med 27(148):167–170
Lee JS, Kang JH, Lee SH, Lee CH, Son J, Kim SY (2016) Glutaminase 1 inhibition reduces thymidine synthesis in NSCLC. Biochem Biophys Res Commun 477(3):374–382
Li G, Song Y, Zhang Y, Wang H, Xie J (2016a) miR-34b targets HSF1 to suppress cell survival in acute myeloid leukemia. Oncol Res 24(2):109–116
Li J et al (2018) Heat shock factor 1 epigenetically stimulates glutaminase-1-dependent mTOR activation to promote colorectal carcinogenesis. Mol Ther 26(7):1828–1839
Li YJ, Ping C, Tang J, Zhang W (2016b) MicroRNA-455 suppresses non-small cell lung cancer through targeting ZEB1. Cell Biol Int 40(6):621–628
Liang W et al (2018) MicroRNA-644a promotes apoptosis of hepatocellular carcinoma cells by downregulating the expression of heat shock factor 1. Cell Commun Signal 16(1):30
Ren L et al (2020) Glutaminase-1 (GLS1) inhibition limits metastatic progression in osteosarcoma. Cancer Metab 8(4.
Song P et al (2020) Beta-catenin represses miR455-3p to stimulate m6A modification of HSF1 mRNA and promote its translation in colorectal cancer. Mol Cancer 19(1):129
Wan T, Shao J, Hu B, Liu G, Luo P, Zhou Y (2018) Prognostic role of HSF1 overexpression in solid tumors: a pooled analysis of 3,159 patients. Onco Targets Ther 11(383–393.
Wang G, Cao P, Fan Y (1874) Tan K (2020) Emerging roles of HSF1 in cancer: cellular and molecular episodes. Biochim Biophys Acta Rev Cancer 1:188390
Wei S et al (2019) miR-455–3p alleviates hepatic stellate cell activation and liver fibrosis by suppressing HSF1 expression. Mol Ther Nucleic Acids 16(758–769.
Workman P (2020) Reflections and outlook on targeting HSP90, HSP70 and HSF1 in cancer: a personal perspective. Adv Exp Med Biol 1243(163–179.
Yun HH, Baek JY, Seo G, Kim YS, Ko JH, Lee JH (2018) Effect of BIS depletion on HSF1-dependent transcriptional activation in A549 non-small cell lung cancer cells. Korean J Physiol Pharmacol 22(4):457–465
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original version of this article was revised: the affiliation has been updated
Rights and permissions
About this article
Cite this article
Meng, C., Liu, K., Cai, X. et al. Mechanism of miR-455–3 in suppressing epithelial–mesenchymal transition and angiogenesis of non-small cell lung cancer cells. Cell Stress and Chaperones 27, 107–117 (2022). https://doi.org/10.1007/s12192-022-01254-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12192-022-01254-4