Abstract
The NUP98::NSD1 fusion gene is associated with extremely poor prognosis in patients with acute myeloid leukemia (AML). NUP98::NSD1 induces self-renewal and blocks differentiation of hematopoietic stem cells, leading to development of leukemia. Despite its association with poor prognosis, targeted therapy for NUP98::NSD1-positive AML is lacking, as the details of NUP98::NSD1 function are unknown. Here, we generated 32D cells (a murine interleukin-3 (IL-3)-dependent myeloid progenitor cell line) expressing mouse Nup98::Nsd1 to explore the function of NUP98::NSD1 in AML, including comprehensive gene expression analysis. We identified two properties of Nup98::Nsd1 + 32D cells in vitro. First, Nup98::Nsd1 promoted blocking of AML cell differentiation, consistent with a previous report. Second, Nup98::Nsd1 increased dependence on IL-3 for cell proliferation, due to overexpression of the alpha subunit of the IL-3 receptor (IL3-RA, also known as CD123). Consistent with our in vitro data, IL3-RA was also upregulated in samples from patients with NUP98::NSD1-positive AML. These results highlight CD123 as a potential new therapeutic target in NUP98::NSD1-positive AML.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Fusions of Nucleoporin 98 (NUP98) with various partner genes are associated with the development of hematological malignancies [1]. Nuclear receptor-binding SET domain protein 1 (NSD1) is a NUP98 fusion partner, and the NUP98::NSD1 fusion gene is formed as a result of the t(5;11)(q35;p15.5) chromosomal translocation [2]. The NUP98::NSD1 protein interacts with KMT2A to drive the HOXA and MEIS1 oncogenes, leading primarily to the development of acute myeloid leukemia (AML) [3, 4]. NUP98::NSD1 is most often detected in pediatric AML, occurring at a frequency of approximately 5%. NUP98::NSD1-positive AML is commonly accompanied by hyperleukocytosis at diagnosis and its most notable feature is its association with extremely poor patient prognosis; induction failure occurs in around 70% of patients with NUP98::NSD1-positive AML and most experience early relapse, even after hematopoietic stem cell transplantation [5, 6]. In addition, approximately 70–80% of patients with NUP98::NSD1-positive AML harbor a FLT3 internal tandem duplication, and prognosis is even worse in these cases [7]. There is no targeted therapy for NUP98::NSD1-positive AML at present, as the details of NUP98::NSD1 function are unclear. Here, we conducted functional analyses of NUP98::NSD1 using murine cell lines, with the aim of identifying a novel therapeutic approach for patients with NUP98::NSD1-positive AML.
Materials and methods
Cell lines and cell culture
The interleukin-3 (IL-3)-dependent myeloid progenitor cell line, 32D, was purchased from Riken BioResource Research Center (Ibaraki, Japan). The lymphoblastic leukemia cell line, WEHI-3, which secretes IL-3 was kindly provided by Dr. M. Suzuki (Tohoku University, Sendai, Japan). Plat-E retroviral packaging cells were kindly provided by Dr. T. Kitamura (The Institute of Medical Science, Tokyo University, Tokyo, Japan) [8].
WEHI-3 cells were cultured in RPMI1640 medium (Nacalai Tesque, Kyoto, Japan), supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, Waltham, MA, USA), 100 U/ml penicillin, and 10 mg/ml streptomycin (standard supplements). Further, 32D cells were also cultured in RPMI1640 medium with standard supplements plus 20% filtered supernatant of WEHI-3 cell culture medium [9]. Plat-E cells were cultured in DMEM (Nacalai Tesque) with standard supplements plus 1 μg/ml puromycin and 10 μg/ml blasticidin. All cells were cultured at 37 ℃ in a 5% CO2 humidified atmosphere.
Retroviral construct and transduction of Nup98::Nsd1
The representative human NUP98::NSD1 structure comprises amino acids (aa) 1–518 of NUP98 fused to aa 1166–2596 of NSD1 (Fig. 1a). Human aa 518 of NUP98 (GenBank Accession number: U41815.1) is equivalent to murine aa 535 of Nup98 (NM_001287164.1), and aa 1–518 and 1–535 of human and murine NUP98, respectively, exhibit approximately 98% identity with one another. Similarly, aa 1166 of human NSD1 (AF322907.1) is equivalent to aa 1167 of murine Nsd1 (AF064553.1), with aa 1166–2596 and 1167–2588, respectively, of human and mouse NSD1 sharing approximately 85% identity (supplementary Fig. 1). Therefore, codons 1–1605 and 4598–8866 of Nup98 and Nsd1, respectively, encoding aa 1–535 and 1167–2588 of the respective murine proteins, were cloned into the pCR™Blunt II-TOPO® vector (Thermo Fisher Scientific). Then the FLAG-tagged full-length of Nup98::Nsd1 coding sequence was cloned into the pMSCVpuro retroviral vector (Addgene, Watertown, MA, USA), using an In-Fusion® HD Cloning Kit (TAKARA Bio USA, Mountain View, CA, USA), according to the manufacturer’s instructions.
Nup98::Nsd1 construct and confirmation of its expression. a Schematic diagram of a human NUP98::NSD1 construct. Amino acids (aa) 1–518 of wild-type NUP98 are fused to aa 1166–2596 of wild-type NSD1. The murine Nup98::Nsd1 construct was cloned into the pMSCVpuro retroviral vector. FG, phenylalanine-glycine repeat domain; PHD, plant homeodomain; PWWP, proline-tryptophan-tryptophan-proline sequence motif domain; SET, Su(Var)3–9, enhancer-of-zeste, trithorax domain; SAC, SET domain-associated cysteine-rich domain; CH, cysteine-histidine rich domain; NID, nuclear receptor interaction domain; aa, amino acid. b Western blotting with anti-FLAG antibody revealed expression of Nup98::Nsd1 (about 220 kDa). Nup98::Nsd1 extracted from Plat-E retroviral packaging cells was used for western blotting. Representative images from three independent experiments are shown
Retroviral supernatants produced by Plat-E cells were used to establish 32D cells expressing Nup98::Nsd1 (Nup98::Nsd1+ 32D cells) and mock-transduced 32D cells, as described previously [10]. Nup98::Nsd1 expression was confirmed by western blotting.
Cell proliferation and cell cycle analyses
Cells were seeded in 12-well culture plates at 5 × 104 cells per well, with or without IL-3. Cell numbers and viability were measured using a Countess™ II automated cell counter (Thermo Fisher Scientific) every 12 or 24 h.
For cell cycle analysis, cells were seeded in 6-well culture plates at 1 × 106 cells per well, with or without IL-3. After 24 h, cells were stained with propidium iodide solution (Sigma-Aldrich, St. Louis, MO, USA) and DNA content was analyzed using a BD FACSCalibur™ flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA).
Flow cytometric analysis and Annexin V assay
CD11b (MAC-1) and CD123 expression levels were analyzed using fluorescein isothiocyanate (FITC)-conjugated anti-mouse MAC-1 antibody or phycoerythrin-conjugated anti-mouse CD123 antibody (BioLegend®, San Diego, CA, USA) on a BD Accuri™ C6 Plus flow cytometer (BD Biosciences).
To detect apoptotic cells, Annexin V assays were conducted using a TACS™ Annexin V-FITC Apoptosis Detection Kit (R&D Systems, Minneapolis, MN, USA), according to the manufacturer’s instructions. Data were analyzed using FlowJo™ (BD Biosciences) and evaluated by calculating mean fluorescence intensity (MFI) values.
Western blotting analysis
Western blotting was performed as described previously [11]. Primary antibodies were as follows: anti-β-actin antibody at 1:10,000 dilution, anti-FLAG-M2 antibody at 1:1,000 dilution (Sigma-Aldrich), anti-phospho-Jak2 (Y1007/1008), and anti-Jak2 (D2E12) antibody at 1:400 dilution, anti-phospho-Stat5 (Tyr694) (D47E7) and anti-Stat5 antibody at 1:1,000 dilution (Cell Signaling Technology, Beverly, MA, USA). Secondary antibodies were as follows: anti-mouse IgG, HRP-linked and anti-rabbit IgG, HRP-linked antibody (Cell Signaling Technology).
Microarray analysis
Gene expression profiling of Nup98::Nsd1+ 32D and mock-transduced 32D cells was conducted using a GeneChip™ Mouse Genome 430 2.0 Array (Thermo Fisher Scientific). RNA was extracted from cells using an RNeasy Mini Kit (Qiagen, Venlo, Netherlands) and complementary RNA (cRNA) was synthesized and biotinylated using a GeneChip™ 3'IVT PLUS Reagent Kit (Thermo Fisher Scientific). cRNA was hybridized to the GeneChip™ Mouse Genome 430 2.0 Array and scanned using a GeneChip™ 3000 7G Scanner (Thermo Fisher Scientific). Data were normalized with Affymetrix Expression Console™ Software 1.4.1 (Thermo Fisher Scientific) and gene expression levels were compared between Nup98::Nsd1+ 32D and mock-transduced 32D cells using Transcriptome Analysis Console Software version 4.0 (Thermo Fisher Scientific). Pathway analysis was performed using Gene Set Enrichment Analysis (GSEA) version 4.1.0 (Broad Institute, Cambridge, MA, USA), and oncogenic signatures (C6) from GSEA Molecular Signature Database v7.4. Gene sets meeting criteria of nominal p value < 0.05 and FDR q value < 0.25 were defined as significantly enriched.
Reverse transcription polymerase chain reaction (RT-PCR) and quantitative real-time PCR (qPCR)
RT-PCR was performed to confirm the expression of Hoxa9 and qPCR for Meis1 mRNA in Nup98::Nsd1+ 32D cells. The primer pairs used were as follows: forward (5’-TCCACTCGGAAGAAGCGATG-3’) and reverse (5’-TTTCGGTGAGGTTGAGCAGC-3’) in Hoxa9, and forward (5’-GGGATAACAGCAGTGAGCAAGG-3’) and reverse (5’-TTTCGGTGAGGTTGAGCAGC-3’) in Meis1. Relative mRNA expression of target genes was determined by comparative cycle threshold using Gapdh as internal control.
Nup98::Nsd1+ cell colony formation assay
Colony formation assays were performed as described previously [12]. Lineage-negative (Lin-) bone marrow cells were harvested from C57BL/6 or BALB/c mice and transduced using retroviral vectors. Transduced Lin- cells (4 × 104) were plated in methylcellulose medium, supplemented with 0.05 μM β-mercaptoethanol, recombinant murine IL-3 (10 ng/ml), IL-6 (10 ng/ml), and SCF (100 ng/ml) (PeproTech, Cranbury, NJ, USA), and incubated at 37℃ in a 5% CO2 humidified atmosphere. Colony numbers and cell viability were measured every 7 days. Then 4 × 104 viable cells were re-plated in fresh methylcellulose medium. Nup98::Nsd1 expression was confirmed by reverse transcription PCR (RT-PCR). KMT2A::Aff1 transduced cells served as a positive control [13]. All animal experiments were approved by the Institutional Animal Care and Use Committee of Kyoto Prefectural University of Medicine.
Drug sensitivity test
Cytarabine, daunorubicin, and STK33 inhibitor, named as ML281, were purchased from Selleck Chemicals (Houston, TX, USA). Nup98::Nsd1+ or mock-transduced 32D cells were seeded in 12-well culture plates at 1 × 105 cells/ml and incubated with vehicle control, cytarabine (1, 10, 100 or 1,000 nM), daunorubicin (1, 10, 100 or 1,000 nM) or ML281 (0.01, 0.1, 1 or 10 μM). Every 24 h of culture, cells were lysed under hypotonic conditions and nuclei were counted with a particle analyzer (ERMA Inc, PARTICLE ANALYZER PA-2000). At 72 h, the concentration of each drug causing 50% growth inhibition (IC50) was determined by Image J (https://imagej.nih.gov/ij/).
Statistical analysis
Statistical analyses were performed using the unpaired Student’s t test or Mann–Whitney U test. p < 0.05 was considered statistically significant.
Results
Successful generation of Nup98::Nsd1+ 32D cells by retroviral transduction
We generated 32D cells expressing FLAG-tagged Nup98::Nsd1 by retroviral transduction and confirmed expression of Nup98::Nsd1 mRNA by RT-PCR (sFig. 2). Although we could not detect Nup98::Nsd1 in Nup98::Nsd1+ 32D cells by western blotting, we determined it in Plat-E cells expressing Nup98::Nsd1, suggesting that Nup98::Nsd1 was expressed in transduced 32D cells (Fig. 1b).
Nup98::Nsd1 reduced MAC-1 expression levels in 32D cells
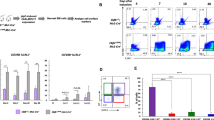
NUP98::NSD1 is reported to impair cell differentiation [1, 14]; thus, MAC-1 (CD11b, known as a myeloid differentiation marker) expression levels were compared between Nup98::Nsd1+ 32D and mock-transduced 32D cells by flow cytometry. A flow cytometry revealed that MAC-1 expression levels were lower in Nup98::Nsd1+ 32D cells, and MFI also showed a significant difference (MFI, 7475 vs. 10,601; p < 0.05) (Fig. 2a,b) suggesting that Nup98::Nsd1 perturbs the differentiation state of 32D cells, consistent with a previous report [15].
Flow cytometric analysis and growth curves of Nup98::Nsd1+ 32D cells. a A flow cytometry histogram and (b) MFI revealed Nup98::Nsd1 reduced macrophage-1 antigen (MAC-1) expression levels in 32D cells. The histograms are representative images from three independent experiments. MFI mean ± SD values from three independent experiments are shown. *p < 0.05 by two-tailed unpaired t test. c Nup98::Nsd1+ 32D cells did not proliferate in the absence of IL-3; however, in the presence of IL-3, they exhibited a growth advantage. **p < 0.01 by two-tailed unpaired t test. Mean ± SD values from three independent experiments are shown
Nup98::Nsd1 did not induce IL-3-independent growth, but led to a growth advantage in 32D cells exposed to IL-3
Some fusion genes expressed in leukemia cells can induce IL-3-independent growth of 32D cells [16, 17]. To examine whether Nup98::Nsd1+ 32D cells acquired IL-3 independence, we deprived Nup98::Nsd1+ 32D cells of IL-3. Nup98::Nsd1 did not confer IL-3-independent growth to 32D cells. By contrast, Nup98::Nsd1+ 32D cells exhibited growth advantage in the presence of IL-3 (Fig. 2c). Interestingly, in the absence of IL-3, death of Nup98::Nsd1+ 32D cells observed under a microscope appeared to be more rapid; however, no significant difference in cell numbers was detected.
Nup98::Nsd1 increased 32D cell dependence on IL-3
Nup98::Nsd1+ 32D cells showed reduced cell proliferation activity 24 h after IL-3 deprivation (Fig. 2c). Further, cell cycle analysis at this time point revealed that a sub-G1 peak was significantly detected in Nup98::Nsd1+ 32D cells (Fig. 3), indicating that Nup98::Nsd1 induced apoptosis of 32D cells in the absence of IL-3. Annexin V positivity was higher in Nup98::Nsd1+ 32D cells 48 h after IL-3 deprivation (38.6 vs. 22.2%; p < 0.01) (Fig. 4). We hypothesized that Nup98::Nsd1 altered the sensitivity of 32D cells to IL-3 and thus evaluated the expression levels of the alpha subunit of the IL-3 receptor (IL-3RA, also known as CD123). A flow cytometry revealed CD123 expression levels were higher in Nup98::Nsd1+ 32D cells, and MFI showed a significant difference (MFI, 910 vs. 673; p < 0.05) (Fig. 5), suggesting relevance of this molecule to the proliferation pattern observed in Nup98::Nsd1+ 32D cells. Though a previous study reported that overexpression of CD123 induced the phosphorylation of STAT5 [18], Nup98::Nsd1+ 32D cells did not show the phosphorylation of Jak2 and Stat5 by western blotting (sFig. 3).
Cell cycle analysis of Nup98::Nsd1+ 32D cells. Cell cycle analysis (a, b) in the presence of IL-3 and (c, d) at 24 h after IL-3 deprivation. A sub-G1 peak was significantly detected in Nup98::Nsd1+ 32D cells following IL-3 deprivation. Representative images from three independent experiments are shown
Annexin V assay of (a) mock-transduced 32D cells and (b) Nup98::Nsd1+ 32D cells in the presence of IL-3, (c) mock-transduced 32D cells and (d) Nup98::Nsd1+ 32D cells 48 h after IL-3 deprivation. (e) Annexin V positivity was elevated in Nup98::Nsd1+ 32D cells. A dot plot representative of three independent experiments is shown. Mean ± SD values from three independent experiments are shown. **p < 0.01 by two-tailed unpaired t test
Flow cytometric analysis of Nup98::Nsd1+ 32D cells. a A flow cytometry histogram and (b) MFI revealed Nup98::Nsd1 enhanced CD123 expression levels in 32D cells. The histograms are representative images from three independent experiments. MFI mean ± SD values from three independent experiments are shown. *p < 0.05 by two-tailed unpaired t test
RNA sequencing analysis of pediatric NUP98::NSD1-positive AML samples revealed higher IL-3RA expression levels
Shiba et al. published RNA sequencing data from 140 samples from patients with pediatric AML [19]. Of the 140 patients, seven were NUP98::NSD1-positive and 133 were NUP98::NSD1-negative. Using this data, we compared IL-3RA expression levels between NUP98::NSD1-positive and -negative AML samples using the Mann–Whitney U test. Similar to our in vitro experiments, IL-3RA was overexpressed in clinical NUP98::NSD1-positive AML samples (Fig. 6).
IL-3RA expression levels in 140 pediatric AML samples determined by RNA sequencing. Comparison of IL-3RA expression levels between NUP98::NSD1-positive (n = 7) and -negative (n = 133) AML samples revealed overexpression of IL-3RA in NUP98::NSD1-positive samples. Data were analyzed using the Mann–Whitney U test. **p < 0.01 by two-tailed unpaired t test. FPKM, fragments per kilobase of exon per million reads mapped
Microarray analysis of Nup98::Nsd1+ 32D cells revealed upregulation of Il3rα
Gene expression levels of 45,037 genes were compared between Nup98::Nsd1+ and mock-transduced 32D cells. Among total genes, 1,400 were upregulated and 721 downregulated, based on a threshold of log twofold change > 2 or < -2 in Nup98::Nsd1+ 32D cells; for example, Psrc1, which is related to cell division, was the most highly elevated with log twofold change 93.27, while Il3rα was also upregulated with log twofold change 2.32 (Fig. 7a). These gene expression characteristics coincided with the specific proliferation pattern of Nup98::Nsd1+ 32D cells; that is, growth advantage in the presence of IL-3 and increased IL-3 dependence.
Gene expression profile of Nup98::Nsd1+ 32D cells. a Volcano plot showing genes with log twofold change > 2 (red; n = 1400 genes) and < -2 (green; n = 721 genes) and adjusted p < 0.01 of analysis of variance in Nup98::Nsd1+ 32D cells compared with mock-transduced 32D cells. Psrc1 and Il-3rα are highlighted. b GSEA analysis showed that genes differentially expressed in Nup98::Nsd1+ 32D cells were enriched for molecules regulated by STK33. NOM p value, Nominal p value; NES normalized enrichment score, FDR false discovery rate
GSEA analysis revealed that, compared with mock-transduced control 32D cells, Nup98::Nsd1+ 32D cells were enriched for genes regulated by STK33, which enhances cancer cell viability (Fig. 7b); however, Kmt2a, Hoxa9, and Meis1, which are overexpressed in NUP98::NSD1-positive leukemia cells, were not upregulated in our model. To confirm the data of microarray analysis, we performed qPCR to determine that the expression level of Meis1 was not upregulated in Nup98::Nsd1+ 32D cells (sFig. 4a). RT-PCR also revealed lack of expression of Hoxa9 in 32D cells irrespective of expression of Nup98::Nsd1 (sFig. 4b). This finding indicates that Nup98::Nsd1+ 32D cells did not exactly mimic the characteristics of NUP98::NSD1+ AML cells. We speculate that this may explain the observed differentiation state of 32D cells; that is, 32D cells were relatively well-differentiated for a leukemogenic model. Therefore, we next assessed the effect of Nup98::Nsd1 on colony formation using murine Lin- hematopoietic cells.
Nup98::Nsd1 did not immortalize murine Lin- bone marrow cells
First, we transduced Nup98::Nsd1 into Lin- bone marrow (BM) cells from C57BL/6 mice using a retrovirus vector. Transduced BM cells lost proliferation activity over time and did not form colonies (sFig. 5a). Expression of Nup98::Nsd1 mRNA in transduced BM cells was confirmed by RT-PCR (sFig. 5b). NUP98::NSD1 has been reported to confer mouse strain-dependent immortality on Lin- BM cells [20]; therefore, we also performed the same assay using BALB/c mice; however, BM cells from BALB/c mice were similarly not immortalized by Nup98::Nsd1 (sFig. 5c, 5d).
Sensitivity of Nup98::Nsd1 to chemotherapeutic agents
To evaluate whether Nup98::Nsd1 cause chemotherapeutic resistance or not, we performed drug sensitivity test for cytarabine and daunorubicin which were key drug for treatment of AML using 32D cells with or without Nup98::Nsd1 expression. In our analysis, cytarabine did not attenuate cell proliferation of Nup98::Nsd1+ 32D cells compared with mock-transduced 32D cells (sFig. 6a, 6b), but it was impossible to calculate IC50. On the other hand, daunorubicin did not visibly attenuate cell proliferation of Nup98::Nsd1+ 32D cells (sFig. 6c, 6d). IC50 in Nup98::Nsd1+ 32D cells was slightly higher than mock-transduced 32D cells, although not statistically significant (IC50, 20.3 vs. 11.2 nM; p = 0.15). Because GSEA analysis revealed enrichment of genes regulated by STK33 in Nup98::Nsd1+ 32D cells, we performed drug sensitivity test for ML281. However, our data showed that ML281 had no effect to suppress the proliferation of Nup98::Nsd1+ 32D cells (sFig. 6e, 6f).
Discussion
In our study, we generated Nup98::Nsd1-positive 32D cells to elucidate the function of the NUP98::NSD1 fusion gene. NUP98 is fused to various partner genes and the function of the resulting fusion molecules depends on the partner gene [14]. The pathogenic mechanism mediated by NUP98 rearrangement in AML is reported to involve interaction between NUP98 fusion proteins and KMT2A, leading to elevated expression of HOXA and MEIS1 oncogenes, resulting in poor prognosis for patients with AML [3]. Among NUP98 rearrangements in AML, NUP98::NSD1-positive AML is associated with particularly poor prognosis. NUP98 functions in nuclear transport, mitotic progression, and regulation of gene expression, while NSD1 is a transcriptional co-regulator and co-repressor. A previous study reported that NUP98::NSD1 promotes self-renewal and blocks differentiation of hematopoietic stem cells [21]. In terms of NSD1 function, NUP98::NSD1 is presumed to influence transcriptional regulation; however, the detailed relationship between its effects and poor patient prognosis is unclear.
Most previously reported NUP98::NSD1 transduction experiments in murine cells used human NUP98::NSD1 constructs [2,3,4]. Due to concern that a transcription factor derived from human genome sequence may not function properly in murine cells, despite the approximately 90% amino acid sequence identity between murine and human NUP98::NSD1, we used a murine Nup98::Nsd1 construct and cell lines in our functional experiments. In our Nup98::Nsd1+ 32D cells, Nup98::Nsd1 promoted blocking of 32D cell differentiation. Further, in the absence of IL-3, Nup98::Nsd1 restricted 32D cell growth and promoted apoptosis, while in the presence of IL-3, Nup98::Nsd1 provided 32D cells with a growth advantage. Given the function of NSD1, the NUP98::NSD1 fusion is unlikely to be directly relevant to cell growth. A recent report identified high expression of CDK6 downstream of NUP98::NSD1 in murine and human AML samples [22]. CDK6 is a cell cycle regulator essential for G1 phase progression, and aberrant CDK6 activation stimulates cell cycle progression. We consider that the proliferation of Nup98::Nsd1+ 32D cells likely reflects cell cycle progression meditated by Cdk6. In addition, our findings indicated that Il3-ra overexpression in Nup98::Nsd1+ 32D cells increased their dependence on IL-3 and influenced cell proliferation. IL3-RA, also referred to as CD123, is overexpressed in 45% of AML and its levels are correlated with cell cycle progression and resistance to apoptosis [18, 23]. Upregulation of IL3-RA was also confirmed in samples from patients with NUP98::NSD1-positive AML; therefore, we speculate that enhanced cell proliferation and chemoresistance due to IL3-RA overexpression likely contribute to the poor prognosis of patients with this type of AML. In addition, Psrc1, which was related to cell division, was also upregulated in Nup98::Nsd1+ 32D cells. This molecule may also influence cell growth and apoptosis resistance under IL-3 + condition, although the relationship of Psrc1 to CD123 is unknown. CD123 is preferentially overexpressed in leukemia cells compared to normal leukocytes; therefore, CD123 is a potential new therapeutic target in NUP98::NSD1-positive AML. Antibody–drug, antibody–drug conjugate, and chimeric antigen receptor T cell therapies have been developed targeting CD123 [24]. Tagraxofusp, a CD123-directed cytotoxin combining human IL-3 and truncated diphtheria toxin, was approved for treatment of blastic plasmacytoid dendritic cell neoplasm in the USA in 2018 and is now also in clinical trials to treat other hematological malignancies, including AML [25, 26]. Our data indicate that anti-CD123 therapy may be effective against NUP98::NSD1-positive AML; however, CD123 is also highly expressed in some organs, including lung, adipose tissue, retina, white matter, and breast; therefore, evaluation of the influence of such treatment on these healthy organs will be required, in addition to assessment of its anti-leukemia effects. Anti-CD123 drug treatment alone would be expected to generate insufficient anti-leukemia effects; therefore, investigation of whether CD123-targeted therapy combined with conventional chemotherapy that can improve the prognosis of patients with NUP98::NSD1-positive AML is also required. GSEA analysis revealed enrichment for genes regulated by STK33 in Nup98::Nsd1+ 32D cells. Association between STK33 and NUP98::NSD1 has not been reported currently; however, STK33 is reported to be overexpressed in patients with diffuse large B cell lymphoma and be associated with chemoresistance [27]. However, Nup98::Nsd1+ 32D cells did not show chemotherapeutic resistance and STK33 inhibitor did not suppress the proliferation of Nup98::Nsd1+ 32D cells, which is consistent with the previous study demonstrating that STK33 inhibitor did not block the growth of AML cell lines with KRAS activating mutation [28].
A limitation of this study was the gene expression profile of Nup98::Nsd1+ 32D cells, specifically that Kmt2a, Hoxa9, and Meis1 were not upregulated. NUP98::NSD1 is reported to drive the expression of these genes, resulting in leukemogenesis [3, 4]; however, microarray analysis of Nup98::Nsd1+ 32D cells did not show any change in their expression profiles. We speculate that this can be attributed to the differentiation state of 32D cells; that is, 32D cells are too differentiated to undergo leukemogenesis, especially in terms of non-expression of Hoxa9. In addition, we could not prove that Nup98:: Nsd1 conferred chemotherapeutic resistance on 32D cells, which also suggest that 32D cell was not adequate to analyze precise function of Nup98::Nsd1. We also assessed the influence of Nup98::Nsd1 on colony formation; however, Lin- hematopoietic cells did not acquire immortality in response to transduction of the fusion, possibly because of the cytokines and growth factors used in the colony formation assay. Although we supplied recombinant murine IL-3, IL-6, and SCF in the methylcellulose medium, further studies using modified combinations of cytokines and growth factors, for example, including FLT3 ligand, are required [19]. Our data indicate that cloning and transduction of the Nup98::Nsd1 construct alone were unable to confer immortality to Lin- hematopoietic cells. Hence, generation of chromosomal translocation using the CRISPR/Cas9 system or introduction of additional gene mutations, such as FLT3-ITD, may be required to produce an adequate murine leukemogenic model.
In summary, our Nup98::Nsd1-positive AML model cells showed high Il3-ra expression and a survival advantage in the presence of IL-3 in vitro. Since IL-3 is widely distributed in vivo, we speculate that it contributes to the rapid proliferation and acquisition of chemoresistance of NUP98::NSD1-positive AML cells in vivo. Therefore, CD123 inhibition is expected to restrain disease progression and ameliorate the chemoresistance of NUP98::NSD1-positive AML cells, and could represent a new therapeutic approach. For assessment of treatment effects, it will be necessary to generate faithful murine Nup98::Nsd1+ AML model cells in vitro, or use patient-derived xenograft models, where appropriate samples are available.
Data availability
Original data presented in this manuscript are available upon request addressed to the corresponding author.
References
Gough SM, Slape CI, Aplan PD. NUP98 gene fusions and hematopoietic malignancies: common themes and new biologic insights. Blood. 2011;118(24):6247–57.
Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol. 2007;9(7):804–12.
Xu H, Valerio DG, Eisold ME, Sinha A, Koche RP, Hu W, et al. NUP98 fusion proteins interact with the NSL and MLL1 complexes to drive Leukemogenesis. Cancer Cell. 2016;30(6):863–78.
Heikamp EB, Henrich JA, Perner F, Wong EM, Hatton C, Wen Y, et al. The menin-MLL1 interaction is a molecular dependency in NUP98-rearranged AML. Blood. 2022;139(6):894–906.
Cerveira N, Correia C, Dória S, Bizarro S, Rocha P, Gomes P, et al. Frequency of NUP98–NSD1 fusion transcript in childhood acute myeloid leukaemia. Leukemia. 2003;17(11):2244–7.
Struski S, Lagarde S, Bories P, Puiseux C, Prade N, Cuccuini W, et al. NUP98 is rearranged in 3.8% of pediatric AML forming a clinical and molecular homogenous group with a poor prognosis. Leukemia. 2017;31(3):565–72.
Ostronoff F, Othus M, Gerbing RB, Loken MR, Raimondi SC, Hirsch BA, et al. NUP98/NSD1 and FLT3/ITD coexpression is more prevalent in younger AML patients and leads to induction failure: a COG and SWOG report. Blood. 2014;124(15):2400–7.
Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7(12):1063–6.
McNiece IK, Bradley TR, Kriegler AB, Hodgson GS. A growth factor produced by WEHI-3 cells for murine high proliferative potential GM-progenitor colony forming cells. Cell Biol Int Rep. 1982;6(3):243–51.
Fujiki A, Imamura T, Sakamoto K, Kawashima S, Yoshida H, Hirashima Y, et al. All-trans retinoic acid combined with 5-Aza-2’-deoxycytidine induces C/EBPalpha expression and growth inhibition in MLL-AF9-positive leukemic cells. Biochem Biophys Res Commun. 2012;428(2):216–23.
Yoshida H, Imamura T, Fujiki A, Hirashima Y, Miyachi M, Inukai T, et al. Post-transcriptional modulation of C/EBPalpha prompts monocytic differentiation and apoptosis in acute myelomonocytic leukaemia cells. Leuk Res. 2012;36(6):735–41.
Lavau C, Szilvassy SJ, Slany R, Cleary ML. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. EMBO J. 1997;16(14):4226–37.
Lin S, Luo RT, Ptasinska A, Kerry J, Assi SA, Wunderlich M, et al. Instructive role of MLL-fusion proteins revealed by a model of t(4;11) Pro-B acute lymphoblastic Leukemia. Cancer Cell. 2016;30(5):737–49.
Michmerhuizen NL, Klco JM, Mullighan CG. Mechanistic insights and potential therapeutic approaches for NUP98-rearranged hematologic malignancies. Blood. 2020;136(20):2275–89.
Deshpande AJ, Deshpande A, Sinha AU, Chen L, Chang J, Cihan A, et al. AF10 regulates progressive H3K79 methylation and HOX gene expression in diverse AML subtypes. Cancer Cell. 2014;26(6):896–908.
Mizuki M, Fenski R, Halfter H, Matsumura I, Schmidt R, Müller C, et al. Flt3 mutations from patients with acute myeloid leukemia induce transformation of 32D cells mediated by the Ras and STAT5 pathways. Blood. 2000;96(12):3907–14.
Peiris MN, Meyer AN, Nelson KN, Bisom-Rapp EW, Donoghue DJ. Oncogenic fusion protein BCR-FGFR1 requires the breakpoint cluster region-mediated oligomerization and chaperonin Hsp90 for activation. Haematologica. 2020;105(5):1262–73.
Testa U, Pelosi E, Frankel A. CD 123 is a membrane biomarker and a therapeutic target in hematologic malignancies. Biomark Res. 2014;2(1):4.
Shiba N, Yoshida K, Hara Y, Yamato G, Shiraishi Y, Matsuo H, et al. Transcriptome analysis offers a comprehensive illustration of the genetic background of pediatric acute myeloid leukemia. Blood Adv. 2019;3(20):3157–69.
Thanasopoulou A, Tzankov A, Schwaller J. Potent co-operation between the NUP98-NSD1 fusion and the FLT3-ITD mutation in acute myeloid leukemia induction. Haematologica. 2014;99(9):1465–71.
Hollink IH, van den Heuvel-Eibrink MM, Arentsen-Peters ST, Pratcorona M, Abbas S, Kuipers JE, et al. NUP98/NSD1 characterizes a novel poor prognostic group in acute myeloid leukemia with a distinct HOX gene expression pattern. Blood. 2011;118(13):3645–56.
Schmoellerl J, Barbosa IAM, Eder T, Brandstoetter T, Schmidt L, Maurer B, et al. CDK6 is an essential direct target of NUP98 fusion proteins in acute myeloid leukemia. Blood. 2020;136(4):387–400.
Jin L, Lee EM, Ramshaw HS, Busfield SJ, Peoppl AG, Wilkinson L, et al. Monoclonal antibody-mediated targeting of CD123, IL-3 receptor alpha chain, eliminates human acute myeloid leukemic stem cells. Cell Stem Cell. 2009;5(1):31–42.
El Achi H, Dupont E, Paul S, Khoury JD. CD123 as a biomarker in hematolymphoid malignancies: principles of detection and targeted therapies. Cancers (Basel). 2020;12(11):3087.
Jen EY, Gao X, Li L, Zhuang L, Simpson NE, Aryal B, et al. FDA approval summary: Tagraxofusp-erzs For treatment of blastic plasmacytoid dendritic cell neoplasm. Clin Cancer Res. 2020;26(3):532–6.
Cangini D, Silimbani P, Cafaro A, Giannini MB, Masini C, et al. Tagraxofusp and anti-CD123 in blastic plasmacytoid dendritic cell neoplasm: a new hope. Minerva Med. 2020;111(5):467–77.
Feng L, Xu X, Zhao K. NFYB potentiates STK33 activation to promote cisplatin resistance in diffuse large B-cell lymphoma. Leuk Res. 2021;111: 106708.
Babij C, Zhang Y, Kurzeja RJ, Munzli A, Shehabeldin A, Fernando M, et al. STK33 kinase activity is nonessential in KRAS-dependent cancer cells. Cancer Res. 2011;71(17):5818–26.
Acknowledgements
This study was supported by grants-in-aid for scientific research from the Japanese Ministry of Education, Culture, Sports, Science and Technology (17K10124 and 21K07759).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
12185_2023_3612_MOESM1_ESM.pdf
Supplementary file1 Fig. 1 Homology between human and murine NUP98::NSD1 amino acid sequences. Comparison between human and murine aa sequences; aa 518 of human NUP98 (Pro) is equivalent to aa 535 (Pro) in the murine Nup98, and aa 1166 of NSD1 (Ser) is equivalent to aa 1167 (Ser) in murine Nsd1. Human and murine NUP98 aa sequences fused to NSD1 exhibited approximately 98% homology, while human and murine NSD1 aa sequences fused to NUP98 had around 85% identity. Pro, Proline; Ser, Serine. Fig. 2 RT-PCR revealed expression of Nup98::Nsd1 mRNA in transduced 32D cells. Fig. 3 Western blotting did not show constitutive phosphorylation of Jak2 and Stat5 in Nup98::Nsd1+ 32D cells. Fig. 4 (a) qPCR revealed that the expression level of Meis1 was not upregulated in Nup98::Nsd1+ 32D cells. RQ, relative quantification. (b) RT-PCR did not reveal expression of Hoxa9 mRNA in either Nup98::Nsd1+ 32D or mock-transduced 32D cells. Fig. 5 Effects of Nup98::Nsd1 on colony formation. (a), (b) Neither C57BL/6 nor BALB/c Lin- bone marrow cells were immortalized by Nup98::Nsd1. KMT2A::Aff1 served as a positive control, which we previously confirmed has immortalization ability. (c), (d) RT-PCR revealed expression of Nup98::Nsd1 mRNA in murine Lin- BM cells (we used primers designed to straddle the fusion point; therefore, the size of the PCR product is 468 bp), indicating successful transduction of Nup98::Nsd1; however, Nup98::Nsd1 did not exhibit cell transformation activity. RT-PCR of Gapdh was performed using forward (5’-CATCACTGCCACCCAGAAGACTG-3’) and reverse (5’- ATGCCAGTGAGCTTCCCGTTCAG-3’) primers; therefore, the size of the PCR product is 153 bp. BM, bone marrow; PC, positive control; NTC, no template control. Fig. 6 The sensitivity of Nup98::Nsd1 to cytarabine and daunorubicin. Nup98::Nsd1+ and mock-transduced 32D cells were cultured in IL-3 containing medium at 1 × 105 cells per well with 1, 10, 100 and 1,000 nM cytarabine or daunorubicin. Viable cell numbers were measured at 24, 48, 72 and 96 hours. (a), (b) Cytarabine did not attenuate cell proliferation of Nup98::Nsd1+ 32D cells. (c), (d) Daunorubicin did not visibly attenuate cell proliferation of Nup98::Nsd1+ 32D cells. (e), (f) STK33 inhibitor had no effect to suppress the proliferation of Nup98::Nsd1+ 32D cells, because the vehicle control (indicated in green) was at the bottom. (PDF 691 KB)
12185_2023_3612_MOESM2_ESM.xlsx
Supplementary file2 Table 1 Lists of the top 10 up- and down-regulated genes in Nup98::Nsd1+ 32D cells, Table 2 Lists of the top 10 oncogenic signature gene sets (C6) enriched in Nup98::Nsd1+ 32D cells. (XLSX 15 KB)
About this article
Cite this article
Okamoto, K., Imamura, T., Tanaka, S. et al. The Nup98::Nsd1 fusion gene induces CD123 expression in 32D cells. Int J Hematol 118, 277–287 (2023). https://doi.org/10.1007/s12185-023-03612-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12185-023-03612-z