Abstract
Acute promyelocytic leukemia (APL) is characterized by a series of retinoic acid receptor (RAR) fusion genes that lead to the dysregulation of RAR signaling and onset of APL. PML–RARA is the most common fusion generated from t(15;17)(q24;q21). In addition, the reciprocal fusion RARA–PML is present in over 80% of t(15;17) APL cases. The bcr3 types of RARA–PML and RARA–PLZF in particular are reciprocal fusions that contribute to leukemogenesis. Here, we report a variant APL case with t(11;17;15)(q13;q21.2;q24.1). Massive parallel sequencing of patient RNA detected the novel fusion transcripts RARA–SNX15 and SNX15–LINC02255 along with the bcr3 type of PML–RARA. Genetic analysis revealed that RARA–SNX15L is an in-frame fusion due to intron retention caused by RNA mis-splicing. RARA–SNX15L consisted mainly of SNX15 domains, including the Phox-homology domain, which has a critical role in protein–protein interactions among sorting nexins and with other partners. Co-immunoprecipitation analysis revealed that RARA–SNX15L is directly associated with SNX15 and with itself. Further studies are needed to evaluate the biological significance of RARA–SNX15L in APL. In conclusion, this is the first report of APL with a complex chromosomal rearrangement involving SNX15.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Acute promyelocytic leukemia (APL) is characterized by a series of retinoic acid receptor (RAR) fusion genes that lead to the dysregulation of RAR signaling and initiation of APL. PML–RARA is the most common fusion gene generated from the balanced chromosomal translocation t(15;17)(q24;q21) [1]. The reciprocal fusion RARA–PML is present in over 80% of t(15;17) APL cases [2]. While PML–RARA determines the disease phenotype, and is the therapeutic target of all-trans retinoic acid (ATRA) and arsenic trioxide, the bcr3 type of RARA–PML was shown to significantly increase the penetrance of APL in PML–RARA transgenic mice, and induce a less mature morphology in the APL cells [3]. In the case of PLZF–RARA, the reciprocal fusion RARA–PLZF alone induced myeloproliferative hematopoiesis in RARA–PLZF transgenic mice, and was necessary to induce the full-blown APL phenotype in PLZF–RARA and RARA–PLZF double-transgenic mice [4]. Hence, some of the reciprocal fusions in APL can affect the disease phenotype. Over the past few decades, many APL variants caused by unique chromosomal translocations have been reported, [1, 5] including NUMA1–RARA in a case of variant APL with t(11;17)(q13;q21) [6]. Here, we report novel fusions, including RARA-Sorting Nexin 15 (SNX15) and SNX15–LINC02255, in a PML–RARA-positive APL case with t(11;17;15)(q13;q21.2;q24.1).
Case presentation
A 19-year-old man was admitted due to fever and bleeding. A full blood count showed a hemoglobin level of 12.6 g/dL, a platelet count of 14 × 109/L, and a white blood cell count of 58.1 × 109/L, in which 87% of the blasts had a monocytic morphology (Fig. 1A, B) and were positive for myeloperoxidase. The morphology of the blasts was compatible with acute myeloid leukemia M3 variant in the French–American–British classification. Coagulopathy was present with an increased prothrombin time of 1.61 (international normalized ratio), an activated partial thromboplastin time of 30.2 s (normal, 26–39 s), decreased fibrinogen level of 122 mg/dL (normal, 200–400 mg/dL), and an increased level of fibrin/fibrinogen degradation products of 148.7 mg/L (normal, 0–5 mg/L). Flow cytometry analysis of the blasts revealed CD13+, CD33+, CD34+, CD117+, CD14−, and HLA-DR. Bone-marrow aspiration resulted in a dry tap, and bone-marrow biopsy showed hypercellular marrow with an excess of blasts. Computed tomography showed marked hepato-splenomegaly and tonsillar swelling. Reverse transcription quantitative polymerase chain reaction (PCR) analysis detected the chimeric PML–RARA fusion at 1.1 × 105/μg RNA. Fluorescence in situ hybridization analysis showed that the PML–RARA fusion signal was detected in 99% of the blasts (Fig. 1C). FLT3 internal tandem duplication (FLT3-ITD) was present (Fig. 1D), and the NPM1 mutation was not detected. Chromosomal analysis of the blasts revealed 46,XY,t(11;17;15)(q13;q21.2;q24.1), and spectral karyotyping analysis showed several chromosomal rearrangements among chromosomes 11, 15, and 17 (Fig. 1E). Taken together, the patient was diagnosed with the microgranular type of APL with a variant translocation, and was subsequently treated with systemic chemotherapy according to the Japan Adult Leukemia Study Group (JALSG) APL212 protocol (UMIN000008470). Administration of ATRA as induction therapy induced differentiation syndrome, but it was manageable using steroids. After a series of consolidation therapy courses according to the APL212 protocol, the patient achieved molecular complete remission.
Morphology and cytogenetic analysis of the t(11;17;15) APL patient sample. Leukemic promyelocytes before treatment. A Monoblastic cell morphology. May-Giemsa staining. Original magnification × 100. B Microgranular pattern in the cytoplasm. May-Giemsa staining. Original magnification × 400. C Fluorescence in situ hybridization analysis. The arrow indicates a fusion signal between PML and RARA. D FLT3–ITD genotyping [15]. HEL: germline control. Pt: t(11;17;15) APL sample. The asterisk indicates hemizygous FLT3–ITD detected by PCR of genomic DNA with primers FLT3_11F (5′-GCAATTTAGGTATGAAAGCCAGC-3′) and FLT3_12R (5′-CTTTCAGCATTTTGACGGCAACC-3′). E Spectral karyotyping analysis showing t(11;17;15)
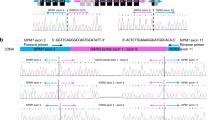
To investigate the unknown fusion transcripts, we performed massive parallel sequencing of total RNA derived from the APL cells using the Illumina platform (Illumina, San Diego, CA). The RNA sequencing protocol was approved by the Institutional Review Board, and written informed consent was obtained from the patient. There were 56.5 million total reads, and the Q20 score was 98.52%. Chimeric fusions were analyzed by the TopHat-Fusion program (Center for Computational Biology, Johns Hopkins University, Baltimore, MD). In addition to the bcr3 type of PML–RARA, we found novel fusion transcripts, including RARA–SNX15 and SNX15–LINC02255 (Fig. 2A), which were confirmed by reverse transcription-PCR (Fig. 2B). To identify junction sequences between the related genes, the PCR products were cloned into the pGEM-T Easy Vector (Promega, Madison, WI), and analyzed by Sanger sequencing (Fig. 2C, D, and E). Unexpectedly, we found three splicing variants of RARA–SNX15 (Fig. 2F). Although RARA–SNX15S and RARA–SNX15M were out-of-frame fusion transcripts, RARA–SNX15L was an in-frame fusion due to intron retention involving 68 base pairs. Colony PCR (n = 28) revealed that RARA–SNX15L, RARA–SNX15M, and RARA–SNX15S comprised 17.9%, 57.1%, and 25.0% of the transcriptional variants, respectively. A schematic diagram of the RARA–SNX15L generated by the RNA mis-splicing form of RARA–SNX15 is shown in Fig. 2G. The RARA–SNX15L consisted mainly of SNX15 domains, including the Phox-homology (PX) domain, which is a pivotal structure shared by all sorting nexins that has a critical role in protein–protein interactions among sorting nexins and with other partners [7, 8]. To investigate the protein–protein interactions of RARA–SNX15L, we performed a co-immunoprecipitation analysis in HEK293T cells. As shown in Fig. 2H, I, RARA–SNX15L associated with SNX15 and with itself.
Novel chimeric fusions RARA–SNX15 and SNX15–LINC02255 in t(11;17;15) APL. A Schematic diagram of the three-way translocation in t(11;17;15) APL. Cen centromere. Tel telomere. B Chimeric fusions detected by RT-PCR. The following primer pairs were used: RARA–SNX15, R18F (5′-TGGACAGCAGCTCCAGGACA-3′) and SNX15e5R (5′-ATCAGAGGGGGTGGCAGGATGTGTA-3′); SNX15–LINC02255, SNX15e1F (5′-CCCCAAGGGCTACACCGAGTACAAA-3′) and LINC2255R (5′-ACCATCGTGGGCTTCCTCATTCTTG-3′). RT reverse transcription. C Sequence analysis of the RARA–SNX15 transcript at the junction site. The junction of the RARA and SNX15 transcripts is indicated by a bold arrowhead. The DNA and amino acid sequences spanning the junction are shown below. D Sequence analysis of the mis-spliced RARA–SNX15 transcript between a part of intron 2 and exon 3 of SNX15 that resulted in an in-frame RARA–SNX15L fusion protein. E Sequence analysis of the SNX15–LINC02255 transcript at the junction site. F Schematic diagram of SNX15 and the three variants of the RARA–SNX15 transcript. The blue box indicates the SNX15 exon. The violet box indicates the RARA exon. The orange box indicates the novel domain created by the mis-spliced sequence of SNX15 intron 2. Single asterisks indicate the location of the sequence in (C). Double asterisks indicate the location of the sequence in (D). G Schematic diagram of SNX15, RARA, and the RARA–SNX15L fusion protein. The breakpoint is indicated by the black line. The domains of RARA–SNX15L and RARA are indicated as follows: Phox homology (PX) domain, microtubule interacting and trafficking (MIT) domain, transactivation domain (TAD), DNA-binding domain (DBD), and ligand-binding domain (LBD). The numbers indicate the position of the amino acid sequence. H RARA–SNX15L associated with SNX15. Co-immunoprecipitation in HEK293T cells between MYC-tagged and FLAG-tagged proteins as previously described [5]. IP immunoprecipitation. IB immunoblotting. I Identification of RARA–SNX15L self-association. Co-immunoprecipitation in HEK293T cells
Discussion
In this report, we described the discovery of the novel fusions RARA–SNX15 and SNX15–LINC02255 in a PML–RARA-positive t(11;17;15)(q13;q21.2;q24.1) APL case. The current dogma is that PML–RARA primarily interferes with retinoic acid signaling, leading to a differentiation block. In addition, some gene mutations, such as FLT3–ITD [9, 10] or reciprocal fusions, influence the APL phenotype [3, 4]. In the present t(11;17;15) APL case, we found an in-frame fusion, RARA–SNX15L, generated by intron retention. Intron retention is primarily caused by RNA mis-splicing, which is a major mechanism of the pathogenesis of many inheritable diseases and cancers [11]. Of note, we did not detect any recurrent somatic mutations in the RNA-splicing machinery in this case.
The SNX15 located on 11q13.1 is involved in intracellular trafficking, including the trafficking of platelet-derived growth factor receptor, insulin receptor (IR), and hepatocyte growth factor receptor [7]. SNX15 associates with SNX1, SNX2, SNX4, and SNX15 through the PX domain. SNX15 not only directly regulates the surface localization of platelet-derived growth factor receptor, but it also impairs the post-translational processing of IR, resulting in negative regulation of IR signaling [7]. Although insulin is an important element for culturing cell lines in vitro [12], it also constitutively activates phosphatidylinositol-3 kinase–AKT–mammalian target of rapamycin signaling in acute myeloid leukemia cells [13, 14]. In Fig. 2H, I, we showed that RARA–SNX15L was directly associated with SNX15 and RARA–SNX15L in HEK293T cells. This indicates that the novel fusion RARA–SNX15L might have some kind of biological interaction with wild-type SNX15. Further studies are needed to evaluate the biological significance of RARA–SNX15L in APL. The third fusion partner, LINC02255 located on 15q24.1, is a long non-coding RNA, and its biological role has not yet been reported. However, SNX15–LINC02255 was over-expressed when compared to LINC02255 in normal bone marrow (n = 9) and acute myeloid leukemia (n = 30) samples.
In conclusion, this is the first report of APL with a complex chromosomal rearrangement involving SNX15.
References
Geoffroy MC, de The H. Classic and variants APLs, as viewed from a therapy response. Cancers (Basel). 2020;12(4):967.
Walz C, Grimwade D, Saussele S, Lengfelder E, Haferlach C, Schnittger S, et al. Atypical mRNA fusions in PML-RARA positive, RARA-PML negative acute promyelocytic leukemia. Genes Chromosomes Cancer. 2010;49(5):471–9.
Pollock JL, Westervelt P, Kurichety AK, Pelicci PG, Grisolano JL, Ley TJ. A bcr-3 isoform of RARalpha-PML potentiates the development of PML-RARalpha-driven acute promyelocytic leukemia. Proc Natl Acad Sci USA. 1999;96(26):15103–8.
He LZ, Bhaumik M, Tribioli C, Rego EM, Ivins S, Zelent A, et al. Two critical hits for promyelocytic leukemia. Mol Cell. 2000;6(5):1131–41.
Yamamoto Y, Tsuzuki S, Tsuzuki M, Handa K, Inaguma Y, Emi N. BCOR as a novel fusion partner of retinoic acid receptor alpha in a t(X;17)(p11;q12) variant of acute promyelocytic leukemia. Blood. 2010;116(20):4274–83.
Wells RA, Catzavelos C, Kamel-Reid S. Fusion of retinoic acid receptor alpha to NuMA, the nuclear mitotic apparatus protein, by a variant translocation in acute promyelocytic leukaemia. Nat Genet. 1997;17(1):109–13.
Phillips SA, Barr VA, Haft DH, Taylor SI, Haft CR. Identification and characterization of SNX15, a novel sorting nexin involved in protein trafficking. J Biol Chem. 2001;276(7):5074–84.
Hanley SE, Cooper KF. Sorting nexins in protein homeostasis. Cells. 2020;10(1):17.
Madan V, Shyamsunder P, Han L, Mayakonda A, Nagata Y, Sundaresan J, et al. Comprehensive mutational analysis of primary and relapse acute promyelocytic leukemia. Leukemia. 2016;30(8):1672–81.
Callens C, Chevret S, Cayuela JM, Cassinat B, Raffoux E, de Botton S, et al. Prognostic implication of FLT3 and Ras gene mutations in patients with acute promyelocytic leukemia (APL): a retrospective study from the European APL Group. Leukemia. 2005;19(7):1153–60.
Scotti MM, Swanson MS. RNA mis-splicing in disease. Nat Rev Genet. 2016;17(1):19–32.
Sinclair J, McClain D, Taetle R. Effects of insulin and insulin-like growth factor I on growth of human leukemia cells in serum-free and protein-free medium. Blood. 1988;72(1):66–72.
Wahner Hendrickson AE, Haluska P, Schneider PA, Loegering DA, Peterson KL, Attar R, et al. Expression of insulin receptor isoform A and insulin-like growth factor-1 receptor in human acute myelogenous leukemia: effect of the dual-receptor inhibitor BMS-536924 in vitro. Cancer Res. 2009;69(19):7635–43.
Nepstad I, Hatfield KJ, Gronningsaeter IS, Aasebo E, Hernandez-Valladares M, Hagen KM, et al. Effects of insulin and pathway inhibitors on the PI3K-Akt-mTOR phosphorylation profile in acute myeloid leukemia cells. Signal Transduct Target Ther. 2019;4:20.
Yamamoto Y, Kiyoi H, Nakano Y, Suzuki R, Kodera Y, Miyawaki S, et al. Activating mutation of D835 within the activation loop of FLT3 in human hematologic malignancies. Blood. 2001;97(8):2434–9.
Acknowledgements
We are grateful to Dr. Nobuhiko Emi (Toyohashi Medical Center) for helpful discussions. This work was supported by grants-in-aid from Chubu University, Kyowa Kirin, and Daiichi-Sankyo.
Author information
Authors and Affiliations
Contributions
YY designed the research; YY and HN performed the research; YY, HN, and AA analyzed the data; KH, SK, AI, MR, HK, and SI collected the clinical data; YY wrote the manuscript.
Corresponding author
Ethics declarations
Conflicts of interest
None of the authors have any relevant conflicts of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
About this article
Cite this article
Hirade, K., Kusumoto, S., Abe, A. et al. A novel RARA–SNX15 fusion in PML–RARA-positive acute promyelocytic leukemia with t(11;17;15)(q13;q21.2;q24.1). Int J Hematol 116, 956–960 (2022). https://doi.org/10.1007/s12185-022-03421-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12185-022-03421-w