Abstract
Hematopoietic development is governed by the coordinated expression of lineage- and differentiation stage-specific genes. Transcription factors play major roles in this process and their perturbation may underlie hematologic and immunologic disorders. Nearly 1900 transcription factors are encoded in the human genome: of these, 49 BTB (for broad-complex, tram-track and bric à brac)-zinc finger transcription factors referred to as ZBTB or POK proteins have been identified. ZBTB proteins, including BCL6, PLZF, ThPOK and LRF, exhibit a broad spectrum of functions in normal and malignant hematopoiesis. This review summarizes developmental and molecular functions of ZBTB proteins relevant to hematology.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hematological malignancies may emerge from perturbed function of transcription factors (TFs) resulting from chromosomal translocation, loss- or gain-of-function mutations, or deregulated expression. Furthermore, mutations causing aberrant DNA methylation (such as the DNMT3A mutation), which may alter the kinetics of TF binding to DNA, are critical epigenetic events that precede malignant transformation [1, 2]. Aberrant transcriptional regulation is also prevalent in non-malignant hematologic disorders. For example, germ-line GATA1 mutations are implicated in familial X-linked anemia and/or thrombocytopenia [3]. Hereditary persistence of fetal hemoglobin (HPFH), a condition characterized by persistent high levels of fetal globin in adults, is observed in individuals harboring mutations in genes encoding TFs [4]. Recent sequencing efforts have revealed mutations in epigenetic factors functioning in the pathogenesis of aplastic anemia [5].
There are 1700–1900 genes encoding TFs in the human genome [6], and at least 49 are classified as ZBTB proteins [7]. This family plays critical roles in hematopoietic differentiation, lineage fate determination and malignant transformation [8–11]. In this review, I discuss emerging themes in ZBTB protein function relevant to hematology.
ZBTB protein molecular mechanisms
ZBTB proteins are prototypical TFs consisting of an N-terminal BTB domain functioning in protein–protein interactions and C-terminal C2H2/Krüppel-type zinc fingers, which bind DNA (Fig. 1a). Between those domains lies a linker domain, whose amino acid sequence is less conserved among family proteins (Fig. 1a). The BTB domain was named after three Drosophila genes, b road-complex, t ram-track and b ric à brac, all encoding proteins essential for Drosophila development [12]. The BTB domain is also known as the POZ (for poxvirus and zinc finger) domain, as it is present in many poxvirus-related proteins [13]; thus ZBTB proteins are sometimes referred as POK (POZ and krüppel-type zinc fingers) proteins.
Structure and function of ZBTB protein domains. a The BTB domain serves as a protein/protein interaction module, while zinc fingers primarily function as a DNA binding module. The linker domain is unstructured and often targeted for posttranslational modification. The number of zinc finger motifs (ZF) varies among ZBTB proteins. Structures of the LRF-BTB domain and KAISO zinc fingers were obtained from PDB files 2NN and 4F6 N, respectively. b (i) Classical model of ZBTB/DNA interaction. Histone-binding modules of SMRT/NCOR (such as the SANT domain) or PHD domains of CHD3/4, subunits of the NuRD complex, could stabilize ZBTB/DNA binding in a chromatin context-dependent manner. (ii) A ZBTB dimer can bind DNA through one chain and interact with other proteins via the linker domain and/or zinc finger motifs. (iii) ZBTB protein may facilitate long-range interactions with regulatory regions (at promoters or enhancers)
The BTB domain
The BTB domain exerts two major functions: dimer formation and recruitment of transcriptional regulators (Fig. 1a). Biochemical and structural analyses revealed that the domain forms an obligate homodimer [7], which is essential for ZBTB protein function in some contexts. For example, wild-type LRF [for Leukemia/lymphoma-Related Factor [14] (also known as FBI-1 [15], POKEMON [16], OCZF [17]); encoded by ZBTB7A] can rescue germinal center B cell defects observed in Lrf conditional knockout (KO) mice, while a dimerization-deficient LRF mutant cannot [18]. The BTB domain can also mediate heterodimer formation. Among heterodimers identified are LRF and BCL6 (B cell Lymphoma 6; encoded by ZBTB27) [14], BCL6 and MIZ-1 (Myc-interacting zinc finger protein-1; encoded by ZBTB17) [19], BCL6 and BCL6B (a.k.a. BAZF; encoded by ZBTB28) [20] and PLZF (Promyelocytic Leukemia Zinc Finger; encoded by ZBTB16) and FAZF (also known as ROG [21], PLZP [22], or TZFP [23]; encoded by ZBTB32). It remains unclear whether only BTB domains mediate heteromeric interactions. Higher order oligomerization through the BTB domain has also been reported: PLZF-BTB domain can form oligomers through its N-terminal β-sheet [24], whereas the MIZ-1 BTB domain forms a tetramer [25].
BTB domains can also recruit transcriptional regulators (Fig. 1a). The transcriptional co-repressors NCOR (nuclear receptor co-repressor) and SMRT (silencing mediator of retinoic acid and thyroid hormone receptor) are homologous proteins originally identified as co-repressors for retinoic acid receptors (RARs) and thyroid hormone receptors [26, 27]. These co-repressors function via recruiting histone deacetylases (HDACs), namely HDAC3 [28]. The functional connection between BTB domains and co-repressors became apparent when NCOR and SMRT were identified in a complex with the leukemia-associated RAR fusion proteins, PML-RARα and PLZF-RARα [29–31]. The latter interacts with SMRT/NCOR not only through the C-terminal RAR moiety but also through the N-terminal PLZF-BTB domain [29, 32, 33]. The BCL6 BTB domain was subsequently found to associate with both SMRT and NCOR [34, 35]. Importantly, BTB homodimer formation is a prerequisite to co-repressor recruitment [36]. Structural analysis revealed that a 17 amino acid residue of SMRT, called SMRT-BBD (SMRT-BCL6 binding domain), binds to a surface created by a BTB homodimer [36]. Thus, any potential BCL6 monomer cannot recruit the co-repressor complex. A recent study suggested that BCL6 serves as a critical downstream effector of SMRT/NCOR [37]. ChIP-seq (chromatin immunoprecipitation with massively parallel DNA sequencing) analysis revealed that nearly half of SMRT/NCOR binding sites were reduced or lost in Bcl6 KO macrophages [37]. Furthermore, almost 70 % of DNA occupancy sites are shared between SMRT-BCL6 and NCOR-BCL6 complexes, indicating a high degree of overlap between both co-repressors in the context of BCL6-mediated gene silencing [37].
BTB-mediated co-repressor recruitment was initially thought to be a general mechanism shared with other ZBTB proteins; however, later biochemical and structural studies suggested it was not. BTB domains of HIC1 (hypermethylated in cancer 1; encoded by ZBTB29), MIZ1 and FAZF do not interact with SMRT/NCOR or SMRT-BBD [38, 39]. The BTB domain of KAISO (encoded by ZBTB33) associates with NCOR but not with SMRT [40]. Importantly, BCL6-BTB residues responsible for SMRT-BBD binding are not conserved among other ZBTB proteins, and SMRT-BBD does not directly interact with either LRF- or PLZF-BTB [36, 41]. Thus, reported interactions between SMRT/NCOR and LRF-BTB or PLZF-BTB [42–44] likely occur via a different binding interface (such as a charged pocket [43]), either directly or indirectly [36, 41].
The BTB domain can also associate with other transcriptional regulators, among them the co-repressors BCOR (BCL-6 interacting co-repressor) [45], SIN3A/B (SIN3 transcription regulator family member) [46] and the NuRD (nucleosome remodeling deacetylase) complex [47]. BCOR was originally identified as a BCL6-interacting protein by a yeast two-hybrid screen using BCL6-BTB as bait [45]. Remarkably, BCOR and NCOR/SMRT interact with the same surface of the BCL6-BTB homodimer [36, 45, 48]. Since one BCL6-BTB homodimer forms two co-repressor binding surfaces, two different co-repressors could associate with a dimer simultaneously [36, 48, 49].
The NuRD complex is a 1MDa multi-subunit protein complex that consists of histone deacetylases (HDAC1/2), ATP-dependent chromatin remodelers (CHD3/4, a.k.a. Mi2-α/β), methyl-CpG-binding domain protein (MBD2/3), histone chaperone protein (RBBP4/7, a.k.a. RbAp46/48), MTA1/2/3, and GATAD2A/B (a.k.a. p66α/β) [50, 51]. The complex is implicated in multiple cellular functions, including transcriptional repression or activation, DNA repair and DNA replication [50, 51]. In fact, some subunits of the complex contain motifs binding to histone tails (CHD3/4, GATAD2A/ and RBBP4/7) and/or DNA (CHD3/4 and MBD2/3) [51]. The LRF-BTB domain reportedly associates with GATAD2B and CHD3, an interaction necessary for γ-globin silencing in adult erythroid cells [47].
BTB domains of ZBTB proteins can also serve as an adaptor for protein degradation. A link between the BTB domain and the ubiquitin pathway was first reported in the BTB-BACK-Kelch protein family [52]. In this context, two protein domains, BTB and BACK, join to form a platform to recruit cullin 3 (CUL3)-based E3 ubiquitin protein ligase complexes [7, 52]. Direct interaction between CUL3 and the PLZF- or BCL6-BTB domain was subsequently reported [53]. PLZF (or BCL6) reportedly recruits the CUL3 complex via the BTB domain, and the E3-ligase complex in turn ubiquitinates PLZF-interacting proteins to promote their degradation by the proteasome. Strikingly, conditional Cul3 KO mice partially phenocopy Plzf or Bcl6 KO mice, suggesting that Cul3-mediated protein degradation is necessary for Plzf and/or Bcl6 function in vivo [53]. The structural basis for these interactions remains to be determined.
C2H2-type zinc fingers
The ZBTB proteins contain multiple C-terminal C2H2-type zinc fingers (Fig. 1a). The number of fingers varies [8–10]: for example, LRF harbors 4 zinc finger motifs, while BCL6 contains 6. Variations in position and number of fingers allow them to bind to DNA in a sequence-specific manner, and many of their DNA binding motifs have been identified in vitro [for example, by CAST (cyclic amplification and selection of targets) assays] [16] and/or in vivo by ChIP-seq [37, 47, 49, 54, 55] (Fig. 1a). Interestingly, DNA binding motifs may vary in a cell type-specific manner: BCL6 reportedly binds distinct sequences depending on cell type [55]. Furthermore, ZBTB proteins may compete with other TFs for DNA occupancy. For example, the consensus sequence of STAT family transcriptional activators resembles that of BCL6 [56]. BCL6 and STATs compete for the same DNA binding sites in the context of inflammatory gene regulation in macrophages [57]. Similarly, BCL6 and NFkB reportedly compete for the same DNA loci downstream of TLR (Toll-like receptor) signaling in macrophages [54].
An emerging aspect of ZBTB protein function is the capacity to bind methylcytosine (5mC) and/or oxidized methylcytosine (oxi-mCs). During DNA replication, the CpG methylation pattern is faithfully inherited by daughter cells due to activity of the DNA methyltransferase 1 (DNMT1) [58]. The de novo DNA methyltransferases, DNMT3A and DNMT3B, can establish new DNA methylation patterns catalyzing formation of 5mC at a palindromic CpG dinucleotide [58]. Members of ten–eleven-translocation (TET) family proteins are dioxygenases that catalyze active demethylation, which can occur independently of DNA replication [58]. TET proteins convert 5mC to 5-hydroxymethylcytosine (5hmC) and successively catalyze conversion of 5hmC to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC). Both 5fC and 5caC are excised by thymine DNA glycosylase (TDG) and restored to unmodified cytosines via the base excision repair pathway [58]. An outstanding question in the field is whether modified cytosine residues are recognized by specific “reader” proteins, enabling distinct functional outputs. This question is particularly relevant to hematology and oncology, as mutations in DNMT3A and TET1/2 are among the most common mutations observed in hematologic malignancies [2] and are often seen in age-related clonal hematopoiesis [59].
KAISO (encoded by ZBTB33) is one of the few factors that can preferentially bind to methylated CpG [60]. Kaiso KO mice exhibit no overt developmental phenotypes, but show resistance to tumor development when crossed with an intestinal tumor model [60]. Two other ZBTB proteins, ZBTB4 and ZBTB38, may also preferentially bind to methylated CpG [61]. Their functions in hematopoietic development remain unknown. Recently, unbiased proteomic screens focusing on reader proteins of 5hmC, 5fC and 5caC identified some ZBTB proteins as oxi-mC readers [62]. For example, ThPOK [T helper-inducing POZ/Krüppel-like factor (a.k.a. cKrox); encoded by ZBTB7B] [63, 64] was found to preferentially bind 5caC. It remains unclear whether some ZBTB proteins are indeed readers of specific DNA methylation status and execute unique functions in normal or malignant hematopoiesis.
In addition to DNA binding, zinc fingers mediate protein–protein interactions. The histone acetyltransferase EP300 reportedly interacts with the zinc fingers of PLZF and acetylates lysines in those motifs necessary for DNA binding [65]. PLZF also associates with the transcription factor GATA2 through its zinc fingers and inhibits GATA2-dependent transcriptional activation [66]. The zinc fingers of MIZ-1 interact with MYC, an activity necessary for MIZ-1-mediated gene repression [67].
The linker region
The linker (or hinge) region between the BTB domain and zinc fingers is poorly conserved among ZBTB proteins and predicted to be unstructured [7]. This region enables ZBTB proteins to flexibly bind DNA in terms of spacing and orientation (Fig. 1b) [7, 68]. In the classic model, a dimeric ZBTB protein recruits co-repressors through the BTB domain, and two zinc finger chains bind to two nearby DNA sequences in promoter or enhancer regions (Fig. 1b, i). The BCL6 linker region, called the second repression domain (RD2), is the most well characterized linker region. It contains a lysine KKYK (aa376-379) motif, in which K379 is a target for acetylation by EP300 [69]. Acetylation of this residue abrogates BCL6-mediated transcriptional repression: a mutant from mimicking a constitutively acetylated state (BCL6QQYQ) fails to recruit HDAC2 or repress target genes [69]. The KKYK motif also mediates direct interaction with metastasis-associated protein 3 (MTA3) [70], a subunit of the NuRD complex. Strikingly, mice harboring either the Bcl6 QQYQ or Bcl6 KKYQ mutation largely phenocopy Bcl6 KO mice [71, 72], suggesting that BCL6 acetylation serves as a molecular switch governing BCL6-mediated repression in vivo. The BCL6 RD2 also contains a PEST motif, which is phosphorylated by mitogen-activated protein kinase (MAPK) following B cell receptor (BCR) signaling [73]. PEST phosphorylation induces BCL6 degradation [73].
Linker regions of other ZBTB proteins are also modified post-translationally (Fig. 1a). Residues within the PLZF linker region undergo phosphorylation [74, 75] and sumoylation [76]. These modifications reportedly modulate PLZF DNA binding capacity [74, 76] or protein stability [75]. Lysine (K314) in the HIC1 linker is modified by acetylation or sumoylation, regulating HIC1 affinity to MTA1, a NuRD subunit. Similarly, ZBTB1 sumoylation in the linker alters its subcellular localization and abrogates binding to SMRT [77]. The co-repressor CtBP (C-terminal binding protein) also associates with linkers of HIC1 [78] and BCL6 [79].
In summary, the linker region could serve as a platform to recruit enzymes or complexes that induce posttranslational modification of ZBTB protein to regulate its stability or transcriptional activity. Furthermore, the linker region is predicted to be unstructured, which may allow multiple modes of DNA binding. For instance, a ZBTB dimer could bind DNA through a zinc finger chain, while another chain functions as a platform for protein/protein interactions (Fig. 1b, ii). ZBTB protein may also mediate long-range DNA interactions as reported for the Drosophila BTB domain-containing protein CP190 (Fig. 1b, iii) [80].
ZBTB function in hematopoietic development
The link between ZBTB proteins and the hematopoietic system emerged from discovery of chromosomal translocations in leukemia and lymphoma. In the early 90s, PLZF was cloned from the chromosomal translocation t(11;17) (q23; q21), which is observed in rare acute promyelocytic leukemia (APL) cases [81]. That translocation gives rise to the PLZF/RARα fusion oncoprotein [81]. Around the same time, BCL6 was cloned and characterized as a gene participating in chromosomal translocations t(3;14) (q27; q32) and t(3; 22) (q27; q11), both commonly observed in non-Hodgkin lymphoma (NHL) [82–84]. The BCL6 gene, which located on human chromosome 3, is fused downstream of the immunoglobulin heavy chain locus (IGH) or lambda locus (IGL), de-regulating BCL6 expression in mature B cells and promoting lymphoma development [85]. Since these seminal discoveries, critical roles of ZBTB proteins in hematopoietic development have been defined primarily through mouse models. At least 12 Zbtb genes are implicated in hematopoietic development in mice. Notably, each of the ZBTB proteins they encode plays a unique role in specific hematopoietic lineages or differentiation stages, as detailed below.
T and innate lymphoid development
In hematopoiesis, T cells are the most studied cell types in terms of ZBTB function [11] (Fig. 2a). ThPOK is among the first ZBTB proteins identified as a factor critical for T cell development [63, 64], and mutation of the gene that encodes it reportedly underlies phenotypes observed in a spontaneous mutant mouse strain called “helper-deficient” (HD). HD mice exhibit a selective absence of CD4+ T cells with a concomitant increase in CD8+ T cells [63]. The causal mutation was mapped to a single amino acid change in the second zinc finger of Thpok [63]. Thpok KO mice phenocopy HD mice [86–88], while forced Thpok expression in CD4/8 double-positive T cells directs MHC-class I-restricted cells to become CD4+ cells [64]. Collectively, these data indicate that Thpok is a master regulator of CD4/CD8 lineage determination in thymus (Fig. 2a). A recent study further demonstrated that Thpok and LRF cooperate to regulate T helper (Th) cell gene expression and function [89]. Thpok is also necessary for the development and function of post-thymic effector Th cells [90] and innate-like T cells such as γδT [91] and invariant natural killer T (iNKT) cells [92]. In mice, Thpok expression is positively or negatively regulated through its proximal enhancer [86], located ~3.6 kb downstream of exon 1, or through a silencer region located ~ 3 kb upstream exon 1 [93], respectively. The transcription factor Gata3 activates Thpok expression via the proximal enhancer [87], while Runx transcription factors suppress Thpok expression through the silencer [93, 94]. Mazr (also known as PATZ1; encoded by ZBTB19) reportedly represses Thpok expression [95, 96]. The T cell transcriptional network regulated by ThPOK (Fig. 2b) is reviewed in detail elsewhere [11, 97].
ZBTB proteins and lymphoid development. a Schematic view of T cell and innate lymphoid cell development in mice. ZBTB proteins necessary for development and/or maintenance are indicated. HSC hematopoietic stem cell, MPP multi-potential progenitor, LMPP lymphoid-biased MPP, CLP common lymphoid progenitor, ETP early T cell precursor, DN CD4/8 double-negative T, DP CD4/8 double-positive T, iNKT invariant natural killer T, Th1 T helper 1, Th2 T helper 2, Tfh follicular helper T, Treg regulatory T, Th17 T helper 17, CILP common innate lymphoid progenitor, ILC innate lymphoid cell. b Transcriptional regulation of genes necessary for CD4/8 T (top) or Tfh cell development (bottom). ZBTB proteins are depicted in red. c B cell development in mice. MIZ1 and ZBTB1 are necessary for early B cell development in BM, while LRF, BCL6 and ZBTB20 function in mature B cell compartments. Trans. B transitional B, MZB marginal zone B, FOB follicular B, Pre-GCB pre-germinal center B, GCB germinal center B, LL Plasma long-lived plasma cell. d Transcriptional regulation of genes necessary for GCB and plasma cell development
PLZF plays a major role in development and function of innate-like T cells and innate lymphoid cells (ILCs). Unlike conventional T cells, which recognize peptide antigens bound to MHC molecules, innate-like T cells are poised to rapidly respond by producing cytokines without clonal expansion [98]. Plzf was among the first TFs identified as a factor critical for iNKT cell development [99, 100]. Plzf also regulates development or function of other innate effector cells, including γδT cells [101], mucosal-associated invariant T cells (MAIT) [100] and innate lymphoid cells (ILCs) [102, 103]. Of note, human PLZF protein is present in a greater variety of lymphocytes than is seen in mice [104]. PLZF may be a useful marker to define the subset of peripheral T cell lymphomas (PTCL), a heterogeneous disease category of mature T cell lymphoma [105]. Fazf is induced in NK cells upon viral infection and stimulates their proliferation by antagonizing Blimp1 function [106].
T follicular helper (Tfh) cells are specialized CD4+ T cells that help B cells to effectively respond to protein antigens via the germinal center (GC) reaction [107]. Bcl6 is preferentially expressed in Tfh cells among CD4+ effector T cell subsets, and its depletion in those cells leads to reduction in the number of Tfh and GCB cells [108–110]. Bcl6 maintains Tfh identity by counteracting other CD4+ effector T cell programs [111]. To do so, Bcl6 represses Blimp1 [110], Tbet/Tbx21 [108, 111] and Rorγt/Rorc [108, 111] (Fig. 2b). Interestingly, T cell-specific Bcor KO mice exhibit significantly reduced Tfh cell numbers, suggesting a functional link between Bcl6 and Bcor in Tfh development [112, 113]. The transcription factor Tcf-1, a downstream effector of Wnt signaling, reportedly induces Bcl6 expression in T cells upon viral infection [114]. Bcl6 is also implicated in maintenance of memory CD8+ T cells [115].
Three ZBTB proteins reportedly function during early T cell development. Zbtb1 mutant mice exhibit a near complete lack of T cells in thymus, suggesting a role in development of early T cell precursors [116]. Miz1 is necessary for early T cell development by functioning downstream of IL7 receptor signaling [67, 117]. Finally, Zbtb35 KO mice show severe impairment in the transition from CD4/8 double-negative to double-positive stages [118].
B lymphoid development
Following exposure to stimulation by T cell-dependent antigens, a fraction of naïve B cells is first activated within the follicle in secondary lymphoid organs and then migrates to the inter-follicular region (IFR), where they establish stable interactions with antigen-specific T cells (Tfh). This interaction further activates antigen-specific B cells, which subsequently migrate back to the follicle center and proliferate extensively [119], pushing follicular B (FOB) cells aside to form the GC, a unique histological structure observed in secondary lymphoid organs. During this process, GCB cells undergo two major modifications: somatic hypermutation (SHM) and class switch recombination (CSR) to enable efficient affinity maturation and effector function, respectively. At the final stage of the GC reaction, memory B cells and long-lived plasma cells develop. Plasma cells are effector cells producing a high level of antibodies, while memory B cells maintain a rapid immunological response upon later exposure to the same antigen. GC-independent memory B cells, which preserve germ-line antibody specificities, have also been reported [120] (Fig. 2c).
Conventional Bcl6 KO mice exhibit impaired GC responses and severe inflammatory disease [56, 121], the latter due to de-repression of genes encoding inflammatory cytokines and chemokines in macrophages [122]. B cell-specific Bcl6 KO mice (mb-1 Cre+) exhibit significantly reduced GCB cell numbers upon T cell-dependent immunization, while other B cell lineages are largely unaffected [123]. GC-specific Bcl6 deletion (through Cγ1-Cre) also promotes impaired GC formation, indicating that Bcl6 is necessary for GCB cell development in a cell intrinsic fashion. In secondary lymphoid organs, BCL6 expression increases when activated B cells associate with Tfh cells [120, 124]. Imaging studies reveal that migration of Bcl6-deficient B cells to nascent GCs is greatly impaired [120]. Since T cell-specific Bcl6 inactivation also abrogates GC formation [108–110], Bcl6 likely regulates GC formation through at least three distinct mechanisms: (1) induction of Tfh cell development; (2) initiation of pre-GCB cell migration to the follicle; and (3) support of GCB cell proliferation and survival. Notably, two groups independently showed that co-repressor recruitment via BCL6-BTB domain is necessary for GC development in vivo, although there remain discrepancies regarding the contribution of Tfhs to observed phenotypes [57, 71, 72, 113].
BCL6 regulates expression of a variety of genes during GCB cell development [49, 57, 85]. For example, at the pre-GCB cell stage, Bcl6 represses expression of the key trafficking receptors S1pr1 and Gpr183 and facilitates pre-GCB cell migration to the follicle center [71]. In GCB cells, BCL6 suppresses apoptosis and protect cells from DNA damage responses during CSR and SHM. In this context, BCL6 directly represses genes necessary for DNA damage sensing as well as checkpoint genes (such as ATR [125]) and inducers of apoptosis or cell cycle arrest (such as TP53 [126] and CDKN1A [19]). BCL6 also inhibits the plasma cell differentiation program by repressing genes necessary for development of those cells, such as BLIMP1 [70, 127, 128] (Fig. 2d).
LRF is another ZBTB protein required for GCB cell development. LRF protein is highly expressed in normal GCB cells and in a majority of diffuse large B cell lymphoma (DLBCL) and follicular lymphoma (FL) cases [16]. Of note, LRF levels are primarily regulated post-translationally in GCB cells: Lrf protein levels are greatly induced in those cells, while corresponding transcript levels decrease [18]. Both pan B cell (mb1-Cre) and GCB-specific (Cγ1-Cre) Lrf conditional KO mice exhibit a significant reduction in GCB number upon immunization [18]. Lrf is necessary for GCB cell proliferation and survival, at least in part, by directly repressing p19Arf expression [18] (Fig. 2D). As described, LRF forms an obligate homodimer essential for GCB development in vivo [18]. A yeast two-hybrid screen using the LRF-BTB domain as bait identified a series of transcriptional regulators, including NuRD subunits and chromatin remodelers, as interacting proteins [47]. Thus, NuRD recruitment via the BTB domain may modulate LRF function in GCB cells. LRF also regulates lineage fate decisions between FOB and marginal zone B (MZB) cells in secondary lymphoid organs. A slight increase in MZB cell number and a concomitant decrease in the number of FOB cells is seen in pan B cell Lrf KO mice [18].
GCB cells terminally differentiate into long-lived plasma cells that can sustain high levels of antibody production. They also produce memory B cells that respond quickly upon later exposure to the same antigen (Fig. 2c). Zbtb20 deficiency causes a defect in plasma cell development, while its exogenous expression induces plasma cell differentiation [129, 130]. Irf4, a factor necessary for GC formation and plasma cell differentiation, reportedly binds to and activates the Zbtb20 promoter [129] (Fig. 2d). Heterozygous germ-line missense mutations in human ZBTB20 were recently identified in Primrose syndrome, which is characterized by multisystem failures [131]. Interestingly, mutations observed in a ZBTB20 zinc finger have a dominant-negative effect [131]. It is unclear whether humoral immunity is altered in patients harboring these mutations.
ZBTB proteins are also implicated in early B cell development in bone marrow (BM). Miz1 reportedly plays a dual role downstream of IL7 receptor signaling in early B cells by inducing the anti-apoptotic factor Bcl2 on one hand and repressing Socs1, a negative regulator of JAK/STAT signaling, on the other [67]. ProB cell numbers are greatly reduced in Miz1 KO mice, as is also observed in Il7 or Il7r KO mice [67]. Zbtb1 KO mice exhibit a block in B cell development at around the Hardy fraction D (small PreB) [116]. Conditional depletion of Lrf in HSCs (Mx1-Cre) leads to development of CD4/8 double-positive T cells in BM at the expense of B cell development due to constitutive activation of Notch signaling [132]. This robust phenotype was initially attributed to HSC-intrinsic mechanisms; however, later it was shown that Lrf-deficient erythroblasts, rather than myeloid or lymphoid cells, aberrantly express the Notch ligand Dll4, which activates otherwise silenced Notch signaling in HSCs [133]. Lrf-mediated Dll4 silencing in erythroblasts is necessary for HSC maintenance and normal lymphoid fate determination, although its physiological significance remains unclear.
Erythroid/myeloid development
LRF is necessary for terminal erythroid differentiation. Conventional Lrf KO mice exhibit embryonic lethality at around 16.5 d.p.c. due to anemia [134]. Lrf inactivation in adult mice (through Mx1-Cre) also promotes macrocytic anemia [134]. In these cases, anemia phenotypes are attributable to increased apoptosis of mature erythroblasts (poly- and orthochromatophilic erythroblasts) and impaired terminal differentiation (Fig. 3a) [134]. LRF knockdown in human CD34+-derived hematopoietic stem progenitor cells (HPSCs) delays erythroid differentiation [47]. GATA1, a master erythroid TF, directly binds to and activates the LRF promoter [134]. EKLF (a.k.a. KLF1), a GATA1 target, also induces LRF expression [135] (Fig. 3b). LRF target genes relevant to erythroid differentiation are yet unknown.
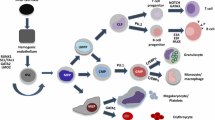
ZBTB proteins and myeloid development. a Schematic view of myeloid development in BM. ZBTB proteins necessary for the development and/or function are indicated. GMP granulocyte-macrophage progenitor, Neutro neutrophil, Baso basophil, Eosino eosinophil, MDP macrophage dendritic cell (DC) progenitor, CDPs common DC progenitors, cDC classical DC, pDC plasmacytoid DC, Mono monocyte, Mφ macrophage, OCP osteoclast progenitor, OC osteoclast, MEP megakaryocyte erythroid progenitor, BFU-E burst forming unit erythroid, CFU-E colony forming unit erythroid, ProE proerythroblast, BasoE basophilic erythroblast, PolyE polychromatophilic erythroblast, OrthoE orthochromatophilic erythroblast, RBC red blood cell, MkP megakaryocyte progenitor, Mk megakaryocyte, Plt platelet. b Transcriptional cascade regulating γ-globin expression in erythroblasts
A recent study uncovered a novel LRF function in the globin switch. Mutations in adult globin genes cause hemoglobinopathies such as sickle cell disease (SCD) and thalassemia, which are among the most common monogenic inherited human disorders [136, 137]. Induction of fetal-type globin (γ-globin) is a promising means to treat these disorders; however, precisely how γ-globin expression is silenced in adult erythroid cells is not fully understood [136]. LRF inactivation re-activates γ-globin expression in human adult erythroid cells independently of BCL11A [47], a master regulator of γ-globin expression [138]. LRF presumably represses γ-globin expression through LRF-BTB/NuRD-mediated mechanisms [47] (Fig. 3b).
ZBTB46 (a.k.a. BTBD4 or zDC) was recently identified as a TF predominantly expressed in classical dendritic cells (cDCs) and their committed progenitors, but not in plasmacytoid DCs (pDCs) [139, 140], although both of these functionally distinct DCs share a common progenitor [141] (Fig. 3a). Zbtb46-deficient CD4+ cDCs aberrantly express myeloid genes, such as G-CSF (granulocyte colony-stimulating factor) [139]. In contrast, BM myeloid progenitors preferentially differentiate into cDC cells upon Zbtb46 overexpression [139]. Although Zbtb46 depletion does not affect cDC development per se, Zbtb46 may help myeloid progenitors to acquire cDC identity [139, 142].
Other ZBTB proteins relevant to myeloid development or function include BCL6, LRF and ZBTB20. LRF protein is acutely induced upon induction of osteoclast differentiation from BM myeloid progenitors [143]. In contrast, BCL6 protein is gradually down-regulated during osteoclast differentiation [144]. LRF reportedly represses NFATc1, a positive regulator of osteoclastogenesis, and conditional inactivation of Lrf in BM progenitors (using Mx1-Cre) reduces bone mass (an osteoporotic phenotype) [143]. Bcl6 KO mice are also osteoporotic, suggesting that BCL6 negatively regulates osteoclastogenesis [144]. Remarkably, BLIMP1 represses BCL6 expression in osteoclast precursors, as it does in T and B cells [110, 144, 145].
Bcl6 conventional KO mice exhibit severe inflammation due to de-repression of cytokine and chemokine genes in macrophages [121, 122, 146–148]. Bcl6 and Nfkb reportedly antagonize downstream TLR signaling in macrophages: Bcl6-deficient macrophages are hypersensitive to TLR-mediated signals due to high Nfkb activity [37, 54]. Bcl6 knock-in mutant mice (Bcl6BTBMUT), in which the interaction between the BCL6-BTB domain and NCOR/SMRT is abrogated, do not exhibit the prominent inflammatory phenotypes seen in Bcl6 KO mice [57]. Thus, loss of Bcl6 from DNA occupancy sites might be necessary for NF-κB recruitment to overlapping DNA binding sites [57]. By contrast, Zbtb20 may function as a positive regulator of TLR signaling in macrophages [149].
PLZF is implicated in development of spermatogonial progenitor cells [150–153] and the musculoskeletal and limb system [154]. In the hematopoietic system, PLZF is expressed at high levels in HSCs/progenitors, and its expression declines as HSCs differentiate [155]. In human hematopoietic cells, PLZF prevents myeloid progenitors from differentiating by repressing myeloid-specific factors, such as GFI-1 and CEBPα, while inducing ID2, an anti-differentiation factor [156]. Plzf-deficient LT-HSCs (long-term HSCs), which are the most primitive HSCs, are prone to enter cell cycle, lose repopulating capacity, and exhibit skewed differentiation toward the myeloid over the lymphoid lineage [157]. Taken together, PLZF is necessary not only for HSC maintenance but to restrict proliferation and differentiation of myeloid progenitors [156, 157].
ZBTB proteins and hematologic malignancies
Among ZBTB proteins, BCL6 and PLZF have been studied extensively in the context of B cell malignancies and APL, respectively [85, 158–160]. During GC development, BCL6 drives GCB cell proliferation and prevents apoptosis induced by DNA damage. Although these functions are essential for GCB cells to undergo efficient SHM and CSR, BCL6 expression must be turned off when cells differentiate into long-lived memory B cells or plasma cells (Fig. 2c). Chromosomal translocations involving BCL6 are observed in 28–45 % of DLBCL and 11–14 % of FL cases [161–163]. Approximately 50 % of gene rearrangements involving BCL6 occur at immunoglobulin loci, including IgH [t(3;14)(q27;q32)], IgL [t(3;22)(q27;q11)] and IgK [t(2;3)(p12; q27)]. At least 20 genes are targeted in BCL6-involved gene rearrangements [164]. As the consequence, the BCL6 5′ regulatory region is replaced with the gene partner of the rearrangement, allowing uncontrolled BCL6 expression in mature B cells. Somatic hypermutations within the 5′ BCL6 regulatory region, through which IRF4 silences BCL6 expression, are observed in ~40 % of DLBCL cases [165]. BCL6 acetylation status may also contribute to lymphomagenesis: inactivating mutations in CREBBP or EP300, both of which encode histone acetyltransferases that also acetylate BCL6, are observed in approximately 40 % of DLBCL or FL cases [166]. These mutations lead to defective BCL6 acetylation and deregulate BCL6 transcriptional activity [166]. In Hodgkin lymphoma (HL), BCL6 protein is predominantly expressed in nodular lymphocyte-predominant HL (NLPHL), but not in classical HL, and BCL6 rearrangements are seen in ~50 % of NLPHL cases [167]. Of note, LRF protein, like BCL6, is also uniquely expressed in NLPHL cases [168].
BCL6 is an attractive target for NHL therapy, given how frequently its expression is deregulated. As described, the BCL6-BTB domain interacts with short sequence motifs in co-repressors, including SMRT, NCOR and BCOR [36, 48, 169, 170]. A cell-penetrating BCL6 peptide inhibitor containing the SMRT BCL6-BTB binding domain (BPI) inhibits BCL6 transcriptional repressor activity in vitro and in vivo [169, 170]. Furthermore, small molecules targeting the BCL6-BTB lateral groove pocket, where BPI binds, inhibit BCL6 repressor function [170]. These reports are encouraging and warrant attention as therapeutic approaches.
The translocation t(11;17) (q23; q21), which produces PLZF/RARα and RARα/PLZF fusion genes, is observed in rare APL cases [81, 158]. Resulting PLZF/RARα and RARα/ PLZF fusion proteins exert leukemogenic activity, at least in part, by disrupting normal PLZF and RARα function [158]. The PLZF/RARα fusion functions as a dominant-negative form of RARα [171, 172]. The fusion can disrupt normal RAR/RXR-mediated signaling by forming a PLZF/RARα/RXR heteromeric complex and/or PLZF/RARα homodimer through the PLZF-BTB domain [158]. The PLZF/RARα fusion could inhibit normal PLZF function by sequestering PLZF binding proteins such as co-repressors. The RARα/PLZF reciprocal fusion could also act as a dominant-negative form of PLZF by interfering with its DNA binding activity, as the fusion retains a large portion of the PLZF zinc finger motif [158].
While APLs harboring the PML/RARa fusion respond to treatment with all-trans retinoic acid (ATRA), those with the PLZF/RARα fusion exhibit poor ATRA responsiveness [173]. The latter outcome was originally attributed to the ability of the fusion’s PLZF moiety to recruit co-repressors [29, 33]; however, later studies showed that mouse APL cells harboring only PLZF/RARα could fully differentiate upon ATRA treatment [174, 175]. In agreement, transgenic mice expressing only PLZF/RARα develop a chronic myeloid leukemia (CML)-like disease, while mice expressing both PLZF/RARα and RARα/PLZF develop acute leukemia with a myeloid differentiation block, reminiscent of human APL [172]. Furthermore, a human APL case expressing only PLZF/RARα transcripts reportedly achieved complete hematologic remission upon ATRA treatment [176]. Thus, the RARα/PLZF fusion may drive ATRA resistance observed in t(11;17) (q23; q21) APL.
LRF protein is overexpressed in 60–80 % of DLBCL and FL cases, and transgenic mice, in which LRF is ectopically expressed in immature T and B cells, develop fatal acute T lymphoblastic leukemia/lymphoma [16]. Since LRF knockdown is toxic to human NHL cell lines, LRF is necessary not only for normal GCB cell development but for maintenance of B cell lymphoma cells [18]. In agreement with LRF’s proposed proto-oncogenic activity, a recent genome-wide CRISPR/Cas9-mediated screen identified LRF as an essential gene for survival in human leukemia cell lines [177].
LRF function in myeloid development is not well understood. Lrf conditional knockout mice do not exhibit gross defects in myeloid development (except for erythroid cells and osteoclasts) [132–134]. Of note, recent sequencing studies reveal LRF mutations in human AML cases. Ivey et al. reported 3 cases with LRF mutations out of 223 NPM1-mutated AML cases (1.3 %) [178]. Furthermore, LRF frameshift mutations, which generate a truncated LRF protein, were recently reported in 3 of 20 cases of AML exhibiting t(8;21) (q22;q22); RUNX1-RUNX1T1 [179]. It remains unclear whether and how these mutations contribute to leukemogenesis.
Future perspective
Recent progress in sequencing technology has greatly advanced our understanding of basic biology and human genetics. It is now possible to systemically analyze genome-wide profiles of DNA, RNA or the epigenome; protein-bound DNA (or RNA); long-range genome interactions; chromatin structure. These new technologies will greatly further our understanding of molecular function of ZBTB proteins in normal and malignant hematopoiesis. Furthermore, current efforts in human genome sequencing will certainly reveal novel mutations and/or variants relevant to hematology or oncology within ZBTB genes. From a clinical standpoint, the biggest challenge lies in pharmacological targeting of ZBTB protein. As described, BCL6 inhibition is a promising strategy for NHL therapy, while LRF or the LRF–NuRD complex may warrant attention as a drug target for NHL or hemoglobinopathies. Team efforts by scientists in the fields of hematology and oncology and medicinal chemistry should pave the way for future drug development.
References
Oshima M, Iwama A. Epigenetics of hematopoietic stem cell aging and disease. Int J Hematol. 2014;100:326–34.
Guillamot M, Cimmino L, Aifantis I. The impact of DNA methylation in hematopoietic malignancies trends in cancer. 2016;2:70–83.
Nichols KE, Crispino JD, Poncz M, White JG, Orkin SH, Maris JM, et al. Familial dyserythropoietic anaemia and thrombocytopenia due to an inherited mutation in GATA1. Nat Genet. 2000;24:266–70.
Bauer DE, Kamran SC, Orkin SH. Reawakening fetal hemoglobin: prospects for new therapies for the β-globin disorders. Blood. 2012;120:2945–53.
Yoshizato T, Dumitriu B, Hosokawa K, Makishima H, Yoshida K, Townsley D, et al. Somatic mutations and clonal hematopoiesis in aplastic anemia. N Engl J Med. 2015;373:35–47.
Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat Rev Genet. 2009;10:252–63.
Stogios PJ, Downs GS, Jauhal JJ, Nandra SK, Privé GG. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005;6:R82.
Lee SU, Maeda T. POK/ZBTB proteins: an emerging family of proteins that regulate lymphoid development and function. Immunol Rev. 2012;247:107–19.
Siggs OM, Beutler B. The BTB-ZF transcription factors. Cell Cycle. 2012;11:3358–69.
Chevrier S, Corcoran LM. BTB-ZF transcription factors, a growing family of regulators of early and late B-cell development. Immunol Cell Biol. 2014;92:481–8.
Ellmeier W, Taniuchi I. The role of BTB-zinc finger transcription factors during T cell development and in the regulation of T cell-mediated immunity. Curr Top Microbiol Immunol. 2014;381:21–49.
Zollman S, Godt D, Privé GG, Couderc JL, Laski FA. The BTB domain, found primarily in zinc finger proteins, defines an evolutionarily conserved family that includes several developmentally regulated genes in Drosophila. Proc Natl Acad Sci USA. 1994;91:10717–21.
Bardwell VJ, Treisman R. The POZ domain: a conserved protein-protein interaction motif. Genes Dev. 1994;8:1664–77.
Davies JM, Hawe N, Kabarowski J, Huang QH, Zhu J, Brand NJ, et al. Novel BTB/POZ domain zinc-finger protein, LRF, is a potential target of the LAZ-3/BCL-6 oncogene. Oncogene. 1999;18:365–75.
Pessler F, Pendergrast PS, Hernandez N. Purification and characterization of FBI-1, a cellular factor that binds to the human immunodeficiency virus type 1 inducer of short transcripts. Mol Cell Biol. 1997;17:3786–98.
Maeda T, Hobbs RM, Merghoub T, Guernah I, Zelent A, Cordon-Cardo C, et al. Role of the proto-oncogene Pokemon in cellular transformation and ARF repression. Nature. 2005;433:278–85.
Kukita A, Kukita T, Ouchida M, Maeda H, Yatsuki H, Kohashi O. Osteoclast-derived zinc finger (OCZF) protein with POZ domain, a possible transcriptional repressor, is involved in osteoclastogenesis. Blood. 1999;94:1987–97.
Sakurai N, Maeda M, Lee SU, Ishikawa Y, Li M, Williams JC, et al. The LRF transcription factor regulates mature B cell development and the germinal center response in mice. J Clin Invest. 2011;121:2583–98.
Phan RT, Saito M, Basso K, Niu H, Dalla-Favera R. BCL6 interacts with the transcription factor Miz-1 to suppress the cyclin-dependent kinase inhibitor p21 and cell cycle arrest in germinal center B cells. Nat Immunol. 2005;6:1054–60.
Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. ‘Green mice’ as a source of ubiquitous green cells. FEBS Lett. 1997;407:313–9.
Miaw SC, Choi A, Yu E, Kishikawa H, Ho IC. ROG, repressor of GATA, regulates the expression of cytokine genes. Immunity. 2000;12:323–33.
Piazza F, Costoya JA, Merghoub T, Hobbs RM, Pandolfi PP. Disruption of PLZP in mice leads to increased T-lymphocyte proliferation, cytokine production, and altered hematopoietic stem cell homeostasis. Mol Cell Biol. 2004;24:10456–69.
Lin W, Lai CH, Tang CJ, Huang CJ, Tang TK. Identification and gene structure of a novel human PLZF-related transcription factor gene, TZFP. Biochem Biophys Res Commun. 1999;264:789–95.
Li X, Peng H, Schultz DC, Lopez-Guisa JM, Rauscher FJ, Marmorstein R. Structure-function studies of the BTB/POZ transcriptional repression domain from the promyelocytic leukemia zinc finger oncoprotein. Cancer Res. 1999;59:5275–82.
Stead MA, Trinh CH, Garnett JA, Carr SB, Baron AJ, Edwards TA, Wright SC. A beta-sheet interaction interface directs the tetramerisation of the Miz-1 POZ domain. J Mol Biol. 2007;373:820–6.
Hörlein AJ, Näär AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404.
Chen JD, Evans RM. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–7.
Perissi V, Jepsen K, Glass CK, Rosenfeld MG. Deconstructing repression: evolving models of co-repressor action. Nat Rev Genet. 2010;11:109–23.
Hong SH, David G, Wong CW, Dejean A, Privalsky ML. SMRT corepressor interacts with PLZF and with the PML-retinoic acid receptor alpha (RARalpha) and PLZF-RARalpha oncoproteins associated with acute promyelocytic leukemia. Proc Natl Acad Sci USA. 1997;94:9028–33.
Grignani F, De Matteis S, Nervi C, Tomassoni L, Gelmetti V, Cioce M, et al. Fusion proteins of the retinoic acid receptor-alpha recruit histone deacetylase in promyelocytic leukaemia. Nature. 1998;391:815–8.
Lin RJ, Nagy L, Inoue S, Shao W, Miller WH, Evans RM. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature. 1998;391:811–4.
He LZ, Guidez F, Tribioli C, Peruzzi D, Ruthardt M, Zelent A, Pandolfi PP. Distinct interactions of PML-RARalpha and PLZF-RARalpha with co-repressors determine differential responses to RA in APL. Nat Genet. 1998;18:126–35.
Guidez F, Ivins S, Zhu J, Söderström M, Waxman S, Zelent A. Reduced retinoic acid-sensitivities of nuclear receptor corepressor binding to PML- and PLZF-RARalpha underlie molecular pathogenesis and treatment of acute promyelocytic leukemia. Blood. 1998;91:2634–42.
Dhordain P, Albagli O, Lin RJ, Ansieau S, Quief S, Leutz A, et al. Corepressor SMRT binds the BTB/POZ repressing domain of the LAZ3/BCL6 oncoprotein. Proc Natl Acad Sci USA. 1997;94:10762–7.
Huynh KD, Bardwell VJ. The BCL-6 POZ domain and other POZ domains interact with the co-repressors N-CoR and SMRT. Oncogene. 1998;17:2473–84.
Ahmad KF, Melnick A, Lax S, Bouchard D, Liu J, Kiang CL, et al. Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain. Mol Cell. 2003;12:1551–64.
Barish GD, Yu RT, Karunasiri MS, Becerra D, Kim J, Tseng TW, et al. The Bcl6-SMRT/NCoR cistrome represses inflammation to attenuate atherosclerosis. Cell Metab. 2012;15:554–62.
Deltour S, Guerardel C, Leprince D. Recruitment of SMRT/N-CoR-mSin3A-HDAC-repressing complexes is not a general mechanism for BTB/POZ transcriptional repressors: the case of HIC-1 and gammaFBP-B. Proc Natl Acad Sci USA. 1999;96:14831–6.
Stogios PJ, Cuesta-Seijo JA, Chen L, Pomroy NC, Privé GG. Insights into strand exchange in BTB domain dimers from the crystal structures of FAZF and Miz1. J Mol Biol. 2010;400:983–97.
Yoon HG, Chan DW, Reynolds AB, Qin J, Wong J. N-CoR mediates DNA methylation-dependent repression through a methyl CpG binding protein Kaiso. Mol Cell. 2003;12:723–34.
Stogios PJ, Chen L, Prive GG. Crystal structure of the BTB domain from the LRF/ZBTB7 transcriptional regulator. Protein Sci. 2007;16:336–42.
Melnick A, Ahmad KF, Arai S, Polinger A, Ball H, Borden KL, et al. In-depth mutational analysis of the promyelocytic leukemia zinc finger BTB/POZ domain reveals motifs and residues required for biological and transcriptional functions. Mol Cell Biol. 2000;20:6550–67.
Melnick A, Carlile G, Ahmad KF, Kiang CL, Corcoran C, Bardwell V, et al. Critical residues within the BTB domain of PLZF and Bcl-6 modulate interaction with corepressors. Mol Cell Biol. 2002;22:1804–18.
Jeon BN, Yoo JY, Choi WI, Lee CE, Yoon HG, Hur MW. Proto-oncogene FBI-1 (Pokemon/ZBTB7A) represses transcription of the tumor suppressor Rb gene via binding competition with Sp1 and recruitment of co-repressors. J Biol Chem. 2008;283:33199–210.
Huynh KD, Fischle W, Verdin E, Bardwell VJ. BCoR, a novel corepressor involved in BCL-6 repression. Genes Dev. 2000;14:1810–23.
David G, Alland L, Hong SH, Wong CW, DePinho RA, Dejean A. Histone deacetylase associated with mSin3A mediates repression by the acute promyelocytic leukemia-associated PLZF protein. Oncogene. 1998;16:2549–56.
Masuda T, Wang X, Maeda M, Canver MC, Sher F, Funnell AP, et al. Transcription factors LRF and BCL11A independently repress expression of fetal hemoglobin. Science. 2016;351:285–9.
Ghetu AF, Corcoran CM, Cerchietti L, Bardwell VJ, Melnick A, Privé GG. Structure of a BCOR corepressor peptide in complex with the BCL6 BTB domain dimer. Mol Cell. 2008;29:384–91.
Hatzi K, Jiang Y, Huang C, Garrett-Bakelman F, Gearhart MD, Giannopoulou EG, et al. A Hybrid Mechanism of Action for BCL6 in B Cells Defined by Formation of Functionally Distinct Complexes at Enhancers and Promoters. Cell Rep. 2013;4:578–88.
Lai AY, Wade PA. Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat Rev Cancer. 2011;11:588–96.
Torchy MP, Hamiche A, Klaholz BP. Structure and function insights into the NuRD chromatin remodeling complex. Cell Mol Life Sci. 2015;72:2491–507.
Pintard L, Willems A, Peter M. Cullin-based ubiquitin ligases: Cul3-BTB complexes join the family. EMBO J. 2004;23:1681–7.
Mathew R, Seiler MP, Scanlon ST, Mao AP, Constantinides MG, Bertozzi-Villa C, et al. BTB-ZF factors recruit the E3 ligase cullin 3 to regulate lymphoid effector programs. Nature. 2012;491:618–21.
Barish GD, Yu RT, Karunasiri M, Ocampo CB, Dixon J, Benner C, et al. Bcl-6 and NF-kappaB cistromes mediate opposing regulation of the innate immune response. Genes Dev. 2010;24:2760–5.
Liu X, Lu H, Chen T, Nallaparaju KC, Yan X, Tanaka S, et al. Genome-wide analysis identifies Bcl6-controlled regulatory networks during T follicular helper cell differentiation. Cell Rep. 2016;14:1735–47.
Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276:589–92.
Huang C, Hatzi K, Melnick A. Lineage-specific functions of Bcl-6 in immunity and inflammation are mediated by distinct biochemical mechanisms. Nat Immunol. 2013;14:380–8.
Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45–68.
Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–98.
Prokhortchouk A, Sansom O, Selfridge J, Caballero IM, Salozhin S, Aithozhina D, et al. Kaiso-deficient mice show resistance to intestinal cancer. Mol Cell Biol. 2006;26:199–208.
Filion GJ, Zhenilo S, Salozhin S, Yamada D, Prokhortchouk E, Defossez PA. A family of human zinc finger proteins that bind methylated DNA and repress transcription. Mol Cell Biol. 2006;26:169–81.
Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, Bauer C, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–59.
He X, He X, Dave VP, Zhang Y, Hua X, Nicolas E, et al. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–33.
Sun G, Liu X, Mercado P, Jenkinson SR, Kypriotou M, Feigenbaum L, et al. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat Immunol. 2005;6:373–81.
Guidez F, Howell L, Isalan M, Cebrat M, Alani RM, Ivins S, et al. Histone acetyltransferase activity of p300 is required for transcriptional repression by the promyelocytic leukemia zinc finger protein. Mol Cell Biol. 2005;25:5552–66.
Tsuzuki S, Enver T. Interactions of GATA-2 with the promyelocytic leukemia zinc finger (PLZF) protein, its homologue FAZF, and the t(11;17)-generated PLZF-retinoic acid receptor alpha oncoprotein. Blood. 2002;99:3404–10.
Möröy T, Saba I, Kosan C. The role of the transcription factor Miz-1 in lymphocyte development and lymphomagenesis-Binding Myc makes the difference. Semin Immunol. 2011;23:379–87.
Pessler F, Hernandez N. Flexible DNA binding of the BTB/POZ-domain protein FBI-1. J Biol Chem. 2003;278:29327–35.
Bereshchenko OR, Gu W, Dalla-Favera R. Acetylation inactivates the transcriptional repressor BCL6. Nat Genet. 2002;32:606–13.
Fujita N, Jaye DL, Geigerman C, Akyildiz A, Mooney MR, Boss JM, Wade PA. MTA3 and the Mi-2/NuRD complex regulate cell fate during B lymphocyte differentiation. Cell. 2004;119:75–86.
Huang C, Gonzalez DG, Cote CM, Jiang Y, Hatzi K, Teater M, et al. The BCL6 RD2 Domain Governs Commitment of Activated B Cells to Form Germinal Centers. Cell Rep. 2014;8:1497–508.
Nance JP, Bélanger S, Johnston RJ, Hu JK, Takemori T, Crotty S. Bcl6 middle domain repressor function is required for T follicular helper cell differentiation and utilizes the corepressor MTA3. Proc Natl Acad Sci USA. 2015;112:13324–9.
Niu H, Ye BH, Dalla-Favera R. Antigen receptor signaling induces MAP kinase-mediated phosphorylation and degradation of the BCL-6 transcription factor. Genes Dev. 1998;12:1953–61.
Ball HJ, Melnick A, Shaknovich R, Kohanski RA, Licht JD. The promyelocytic leukemia zinc finger (PLZF) protein binds DNA in a high molecular weight complex associated with cdc2 kinase. Nucleic Acids Res. 1999;27:4106–13.
Costoya JA, Hobbs RM, Pandolfi PP. Cyclin-dependent kinase antagonizes promyelocytic leukemia zinc-finger through phosphorylation. Oncogene. 2008;27:3789–96.
Kang SI, Chang WJ, Cho SG, Kim IY. Modification of promyelocytic leukemia zinc finger protein (PLZF) by SUMO-1 conjugation regulates its transcriptional repressor activity. J Biol Chem. 2003;278:51479–83.
Matic I, Schimmel J, Hendriks IA, van Santen MA, van de Rijke F, van Dam H, et al. Site-specific identification of SUMO-2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Mol Cell. 2010;39:641–52.
Deltour S, Pinte S, Guerardel C, Wasylyk B, Leprince D. The human candidate tumor suppressor gene HIC1 recruits CtBP through a degenerate GLDLSKK Motif. Mol Cell Biol. 2002;22:4890–901.
Mendez LM, Polo JM, Yu JJ, Krupski M, Ding BB, Melnick A, Ye BH. CtBP is an essential corepressor for BCL6 autoregulation. Mol Cell Biol. 2008;28:2175–86.
Vogelmann J, Le Gall A, Dejardin S, Allemand F, Gamot A, Labesse G, et al. Chromatin insulator factors involved in long-range DNA interactions and their role in the folding of the Drosophila genome. PLoS Genet. 2014;10:e1004544.
Chen Z, Brand NJ, Chen A, Chen SJ, Tong JH, Wang ZY, et al. Fusion between a novel Krüppel-like zinc finger gene and the retinoic acid receptor-alpha locus due to a variant t(11;17) translocation associated with acute promyelocytic leukaemia. EMBO J. 1993;12:1161–7.
Baron BW, Nucifora G, McCabe N, Espinosa R, Le Beau MM, McKeithan TW. Identification of the gene associated with the recurring chromosomal translocations t(3;14)(q27;q32) and t(3;22)(q27;q11) in B-cell lymphomas. Proc Natl Acad Sci USA. 1993;90:5262–6.
Ye BH, Rao PH, Chaganti RS, Dalla-Favera R. Cloning of bcl-6, the locus involved in chromosome translocations affecting band 3q27 in B-cell lymphoma. Cancer Res. 1993;53:2732–5.
Kerckaert JP, Deweindt C, Tilly H, Quief S, Lecocq G, Bastard C. LAZ3, a novel zinc-finger encoding gene, is disrupted by recurring chromosome 3q27 translocations in human lymphomas. Nat Genet. 1993;5:66–70.
Basso K, Dalla-Favera R. Roles of BCL6 in normal and transformed germinal center B cells. Immunol Rev. 2012;247:172–83.
Muroi S, Naoe Y, Miyamoto C, Akiyama K, Ikawa T, Masuda K, et al. Cascading suppression of transcriptional silencers by ThPOK seals helper T cell fate. Nat Immunol. 2008;9:1113–21.
Wang L, Wildt KF, Zhu J, Zhang X, Feigenbaum L, Tessarollo L, et al. Distinct functions for the transcription factors GATA-3 and ThPOK during intrathymic differentiation of CD4(+) T cells. Nat Immunol. 2008;9:1122–30.
Egawa T, Littman DR. ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nat Immunol. 2008;9:1131–9.
Carpenter AC, Grainger JR, Xiong Y, Kanno Y, Chu HH, Wang L, et al. The Transcription Factors Thpok and LRF Are Necessary and Partly Redundant for T Helper Cell Differentiation. Immunity. 2012;37:622–33.
Vacchio MS, Wang L, Bouladoux N, Carpenter AC, Xiong Y, Williams LC, et al. A ThPOK-LRF transcriptional node maintains the integrity and effector potential of post-thymic CD4(+) T cells. Nat Immunol. 2014;15:947–56.
Park K, He X, Lee HO, Hua X, Li Y, Wiest D, Kappes DJ. TCR-mediated ThPOK induction promotes development of mature (CD24−) gammadelta thymocytes. EMBO J. 2010;29:2329–41.
Engel I, Hammond K, Sullivan BA, He X, Taniuchi I, Kappes D, Kronenberg M. Co-receptor choice by V alpha14i NKT cells is driven by Th-POK expression rather than avoidance of CD8-mediated negative selection. J Exp Med. 2010;207:1015–29.
Setoguchi R, Tachibana M, Naoe Y, Muroi S, Akiyama K, Tezuka C, et al. Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science. 2008;319:822–5.
He X, Park K, Wang H, He X, Zhang Y, Hua X, et al. CD4-CD8 lineage commitment is regulated by a silencer element at the ThPOK transcription-factor locus. Immunity. 2008;28:346–58.
Kobayashi A, Yamagiwa H, Hoshino H, Muto A, Sato K, Morita M, et al. A combinatorial code for gene expression generated by transcription factor Bach2 and MAZR (MAZ-related factor) through the BTB/POZ domain. Mol Cell Biol. 2000;20:1733–46.
Sakaguchi S, Hombauer M, Bilic I, Naoe Y, Schebesta A, Taniuchi I, Ellmeier W. The zinc-finger protein MAZR is part of the transcription factor network that controls the CD4 versus CD8 lineage fate of double-positive thymocytes. Nat Immunol. 2010;11:442–8.
Egawa T. Continued mission of ThPOK. Nat Immunol. 2014;15:900–2.
De Obaldia ME, Bhandoola A. Transcriptional regulation of innate and adaptive lymphocyte lineages. Annu Rev Immunol. 2015;33:607–42.
Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008;9:1055–64.
Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403.
Kreslavsky T, Savage AK, Hobbs R, Gounari F, Bronson R, Pereira P, et al. TCR-inducible PLZF transcription factor required for innate phenotype of a subset of gammadelta T cells with restricted TCR diversity. Proc Natl Acad Sci USA. 2009;106:12453–8.
Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nature. 2014;508:397–401.
Ishizuka IE, Chea S, Gudjonson H, Constantinides MG, Dinner AR, Bendelac A, Golub R. Single-cell analysis defines the divergence between the innate lymphoid cell lineage and lymphoid tissue-inducer cell lineage. Nat Immunol. 2016;17:269–76.
Eidson M, Wahlstrom J, Beaulieu AM, Zaidi B, Carsons SE, Crow PK, et al. Altered development of NKT cells, γδ T cells, CD8 T cells and NK cells in a PLZF deficient patient. PLoS ONE. 2011;6:e24441.
McGregor S, Shah A, Raca G, Mirza MK, Smith SM, Anastasi J, et al. PLZF staining identifies peripheral T-cell lymphomas derived from innate-like T-cells with TRAV1-2-TRAJ33 TCR-α rearrangement. Blood. 2014;123:2472–3.
Beaulieu AM, Zawislak CL, Nakayama T, Sun JC. The transcription factor Zbtb32 controls the proliferative burst of virus-specific natural killer cells responding to infection. Nat Immunol. 2014;15:546–53.
Vinuesa CG, Linterman MA, Yu D, MacLennan IC. Follicular helper T cells. Annu Rev Immunol. 2016;34:335–68.
Yu D, Rao S, Tsai LM, Lee SK, He Y, Sutcliffe EL, et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–68.
Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, et al. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–5.
Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–10.
Hatzi K, Nance JP, Kroenke MA, Bothwell M, Haddad EK, Melnick A, Crotty S. BCL6 orchestrates Tfh cell differentiation via multiple distinct mechanisms. J Exp Med. 2015;212:539–53.
Yang JA, Tubo NJ, Gearhart MD, Bardwell VJ, Jenkins MK. Cutting edge: Bcl6-interacting corepressor contributes to germinal center T follicular helper cell formation and B cell helper function. J Immunol. 2015;194:5604–8.
Nance JP, Bélanger S, Johnston RJ, Takemori T, Crotty S. Cutting edge: T follicular helper cell differentiation is defective in the absence of Bcl6 BTB repressor domain function. J Immunol. 2015;194:5599–603.
Choi YS, Gullicksrud JA, Xing S, Zeng Z, Shan Q, Li F, et al. LEF-1 and TCF-1 orchestrate T(FH) differentiation by regulating differentiation circuits upstream of the transcriptional repressor Bcl6. Nat Immunol. 2015;16:980–90.
Ichii H, Sakamoto A, Hatano M, Okada S, Toyama H, Taki S, et al. Role for Bcl-6 in the generation and maintenance of memory CD8+ T cells. Nat Immunol. 2002;3:558–63.
Siggs OM, Li X, Xia Y, Beutler B. ZBTB1 is a determinant of lymphoid development. J Exp Med. 2012;209:19–27.
Saba I, Kosan C, Vassen L, Möröy T. IL-7R-dependent survival and differentiation of early T-lineage progenitors is regulated by the BTB/POZ domain transcription factor Miz-1. Blood. 2011;117:3370–81.
Iguchi T, Aoki K, Ikawa T, Taoka M, Taya C, Yoshitani H, et al. BTB-ZF protein Znf131 regulates cell growth of developing and mature T cells. J Immunol. 2015;195:982–93.
Okada T, Moriyama S, Kitano M. Differentiation of germinal center B cells and follicular helper T cells as viewed by tracking Bcl6 expression dynamics. Immunol Rev. 2012;247:120–32.
Kitano M, Moriyama S, Ando Y, Hikida M, Mori Y, Kurosaki T, Okada T. Bcl6 protein expression shapes pre-germinal center B cell dynamics and follicular helper T cell heterogeneity. Immunity. 2011;34:961–72.
Ye BH, Cattoretti G, Shen Q, Zhang J, Hawe N, de Waard R, et al. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat Genet. 1997;16:161–70.
Toney LM, Cattoretti G, Graf JA, Merghoub T, Pandolfi PP, Dalla-Favera R, et al. BCL-6 regulates chemokine gene transcription in macrophages. Nat Immunol. 2000;1:214–20.
Kaji T, Ishige A, Hikida M, Taka J, Hijikata A, Kubo M, et al. Distinct cellular pathways select germline-encoded and somatically mutated antibodies into immunological memory. J Exp Med. 2012;209:2079–97.
Kerfoot SM, Yaari G, Patel JR, Johnson KL, Gonzalez DG, Kleinstein SH, Haberman AM. Germinal center B cell and T follicular helper cell development initiates in the interfollicular zone. Immunity. 2011;34:947–60.
Ranuncolo SM, Polo JM, Dierov J, Singer M, Kuo T, Greally J, et al. Bcl-6 mediates the germinal center B cell phenotype and lymphomagenesis through transcriptional repression of the DNA-damage sensor ATR. Nat Immunol. 2007;8:705–14.
Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432:635–9.
Shaffer AL, Yu X, He Y, Boldrick J, Chan EP, Staudt LM. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 2000;13:199–212.
Tunyaplin C, Shaffer AL, Angelin-Duclos CD, Yu X, Staudt LM, Calame KL. Direct repression of prdm1 by Bcl-6 inhibits plasmacytic differentiation. J Immunol. 2004;173:1158–65.
Chevrier S, Emslie D, Shi W, Kratina T, Wellard C, Karnowski A, et al. The BTB-ZF transcription factor Zbtb20 is driven by Irf4 to promote plasma cell differentiation and longevity. J Exp Med. 2014;211:827–40.
Wang Y, Bhattacharya D. Adjuvant-specific regulation of long-term antibody responses by ZBTB20. J Exp Med. 2014;211:841–56.
Cordeddu V, Redeker B, Stellacci E, Jongejan A, Fragale A, Bradley TE, et al. Mutations in ZBTB20 cause primrose syndrome. Nat Genet. 2014;46:815–7.
Maeda T, Merghoub T, Hobbs RM, Dong L, Maeda M, Zakrzewski J, et al. Regulation of B versus T lymphoid lineage fate decision by the proto-oncogene LRF. Science. 2007;316:860–6.
Lee SU, Maeda M, Ishikawa Y, Li SM, Wilson A, Jubb AM, et al. LRF-mediated Dll4 repression in erythroblasts is necessary for hematopoietic stem cell maintenance. Blood. 2013;121:918–29.
Maeda T, Ito K, Merghoub T, Poliseno L, Hobbs RM, Wang G, et al. LRF is an essential downstream target of GATA1 in erythroid development and regulates BIM-dependent apoptosis. Dev Cell. 2009;17:527–40.
Hodge D, Coghill E, Keys J, Maguire T, Hartmann B, McDowall A, et al. A global role for EKLF in definitive and primitive erythropoiesis. Blood. 2006;107:3359–70.
Stamatoyannopoulos G. Control of globin gene expression during development and erythroid differentiation. Exp Hematol. 2005;33:259–71.
Pleasants S. Epidemiology: a moving target. Nature. 2014;515:S2.
Orkin SH. Recent advances in globin research using genome-wide association studies and gene editing. Ann N Y Acad Sci. 2016;1368:5–10.
Satpathy AT, Kc W, Albring JC, Edelson BT, Kretzer NM, Bhattacharya D, et al. Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. J Exp Med. 2012;209:1135–52.
Meredith MM, Liu K, Darrasse-Jeze G, Kamphorst AO, Schreiber HA, Guermonprez P, et al. Expression of the zinc finger transcription factor zDC (Zbtb46, Btbd4) defines the classical dendritic cell lineage. J Exp Med. 2012;209:1153–65.
Satpathy AT, Wu X, Albring JC, Murphy KM. Re(de)fining the dendritic cell lineage. Nat Immunol. 2012;13:1145–54.
Meredith MM, Liu K, Kamphorst AO, Idoyaga J, Yamane A, Guermonprez P, et al. Zinc finger transcription factor zDC is a negative regulator required to prevent activation of classical dendritic cells in the steady state. J Exp Med. 2012;209:1583–93.
Tsuji-Takechi K, Negishi-Koga T, Sumiya E, Kukita A, Kato S, Maeda T, et al. Stage-specific functions of leukemia/lymphoma-related factor (LRF) in the transcriptional control of osteoclast development. Proc Natl Acad Sci USA. 2012;109:2561–6.
Miyauchi Y, Ninomiya K, Miyamoto H, Sakamoto A, Iwasaki R, Hoshi H, et al. The Blimp1-Bcl6 axis is critical to regulate osteoclast differentiation and bone homeostasis. J Exp Med. 2010;207:751–62.
Shaffer AL, Lin KI, Kuo TC, Yu X, Hurt EM, Rosenwald A, et al. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17:51–62.
Takeda N, Arima M, Tsuruoka N, Okada S, Hatano M, Sakamoto A, et al. Bcl6 is a transcriptional repressor for the IL-18 gene. J Immunol. 2003;171:426–31.
Yu RY, Wang X, Pixley FJ, Yu JJ, Dent AL, Broxmeyer HE, et al. BCL-6 negatively regulates macrophage proliferation by suppressing autocrine IL-6 production. Blood. 2005;105:1777–84.
Mondal A, Sawant D, Dent AL. Transcriptional repressor BCL6 controls Th17 responses by controlling gene expression in both T cells and macrophages. J Immunol. 2010;184:4123–32.
Liu X, Zhang P, Bao Y, Han Y, Wang Y, Zhang Q, et al. Zinc finger protein ZBTB20 promotes Toll-like receptor-triggered innate immune responses by repressing IκBα gene transcription. Proc Natl Acad Sci USA. 2013;110:11097–102.
Buaas FW, Kirsh AL, Sharma M, McLean DJ, Morris JL, Griswold MD, et al. Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet. 2004;36:647–52.
Costoya JA, Hobbs RM, Barna M, Cattoretti G, Manova K, Sukhwani M, et al. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet. 2004;36:653–9.
Hobbs RM, Seandel M, Falciatori I, Rafii S, Pandolfi PP. Plzf regulates germline progenitor self-renewal by opposing mTORC1. Cell. 2010;142:468–79.
Hobbs RM, Fagoonee S, Papa A, Webster K, Altruda F, Nishinakamura R, et al. Functional antagonism between Sall4 and Plzf defines germline progenitors. Cell Stem Cell. 2012;10:284–98.
Barna M, Hawe N, Niswander L, Pandolfi PP. Plzf regulates limb and axial skeletal patterning. Nat Genet. 2000;25:166–72.
Reid A, Gould A, Brand N, Cook M, Strutt P, Li J, et al. Leukemia translocation gene, PLZF, is expressed with a speckled nuclear pattern in early hematopoietic progenitors. Blood. 1995;86:4544–52.
Doulatov S, Notta F, Rice KL, Howell L, Zelent A, Licht JD, Dick JE. PLZF is a regulator of homeostatic and cytokine-induced myeloid development. Genes Dev. 2009;23:2076–87.
Vincent-Fabert C, Platet N, Vandevelde A, Poplineau M, Koubi M, Finetti P, et al. PLZF mutation alters mouse hematopoietic stem cell function and cell cycle progression. Blood. 2016;127:1881–5.
Melnick A, Licht JD. Deconstructing a disease: RARalpha, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood. 1999;93:3167–215.
Ablain J, de The H. Revisiting the differentiation paradigm in acute promyelocytic leukemia. Blood. 2011;117:5795–802.
Bunting KL, Melnick AM. New effector functions and regulatory mechanisms of BCL6 in normal and malignant lymphocytes. Curr Opin Immunol. 2013;25:339–46.
Lo Coco F, Ye BH, Lista F, Corradini P, Offit K, Knowles DM, et al. Rearrangements of the BCL6 gene in diffuse large cell non-Hodgkin’s lymphoma. Blood. 1994;83:1757–9.
Bastard C, Deweindt C, Kerckaert JP, Lenormand B, Rossi A, Pezzella F, et al. LAZ3 rearrangements in non-Hodgkin’s lymphoma: correlation with histology, immunophenotype, karyotype, and clinical outcome in 217 patients. Blood. 1994;83:2423–7.
Otsuki T, Yano T, Clark HM, Bastard C, Kerckaert JP, Jaffe ES, Raffeld M. Analysis of LAZ3 (BCL-6) status in B-cell non-Hodgkin’s lymphomas: results of rearrangement and gene expression studies and a mutational analysis of coding region sequences. Blood. 1995;85:2877–84.
Huret JL, Ahmad M, Arsaban M, Bernheim A, Cigna J, Desangles F et al. Atlas of genetics and cytogenetics in oncology and haematology in 2013. Nucleic Acids Res. 2013; 41(D1):D920–4.
Pasqualucci L, Migliazza A, Basso K, Houldsworth J, Chaganti RS, Dalla-Favera R. Mutations of the BCL6 proto-oncogene disrupt its negative autoregulation in diffuse large B-cell lymphoma. Blood. 2003;101:2914–23.
Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, Trifonov V, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471:189–95.
Schmitz R, Stanelle J, Hansmann ML, Küppers R. Pathogenesis of classical and lymphocyte-predominant Hodgkin lymphoma. Annu Rev Pathol. 2009;4:151–74.
Bohn O, Maeda T, Filatov A, Lunardi A, Pandolfi PP, Teruya-Feldstein J. Utility of LRF/Pokemon and NOTCH1 protein expression in the distinction between nodular lymphocyte-predominant Hodgkin lymphoma and classical Hodgkin lymphoma. Int J Surg Pathol. 2014;22:6–11.
Polo JM, Dell’Oso T, Ranuncolo SM, Cerchietti L, Beck D, Da Silva GF, et al. Specific peptide interference reveals BCL6 transcriptional and oncogenic mechanisms in B-cell lymphoma cells. Nat Med. 2004;10:1329–35.
Cerchietti LC, Ghetu AF, Zhu X, Da Silva GF, Zhong S, Matthews M, et al. A small-molecule inhibitor of BCL6 kills DLBCL cells in vitro and in vivo. Cancer Cell. 2010;17:400–11.
Ruthardt M, Testa U, Nervi C, Ferrucci PF, Grignani F, Puccetti E, et al. Opposite effects of the acute promyelocytic leukemia PML-retinoic acid receptor alpha (RAR alpha) and PLZF-RAR alpha fusion proteins on retinoic acid signalling. Mol Cell Biol. 1997;17:4859–69.
He LZ, Bhaumik M, Tribioli C, Rego EM, Ivins S, Zelent A, Pandolfi PP. Two critical hits for promyelocytic leukemia. Mol Cell. 2000;6:1131–41.
Licht JD, Chomienne C, Goy A, Chen A, Scott AA, Head DR, et al. Clinical and molecular characterization of a rare syndrome of acute promyelocytic leukemia associated with translocation (11;17). Blood. 1995;85:1083–94.
Nasr R, Guillemin MC, Ferhi O, Soilihi H, Peres L, Berthier C, et al. Eradication of acute promyelocytic leukemia-initiating cells through PML-RARA degradation. Nat Med. 2008;14:1333–42.
Rice KL, Hormaeche I, Doulatov S, Flatow JM, Grimwade D, Mills KI, et al. Comprehensive genomic screens identify a role for PLZF-RARalpha as a positive regulator of cell proliferation via direct regulation of c-MYC. Blood. 2009;114:5499–511.
Petti MC, Fazi F, Gentile M, Diverio D, De Fabritiis P, De Propris MS, et al. Complete remission through blast cell differentiation in PLZF/RARalpha-positive acute promyelocytic leukemia: in vitro and in vivo studies. Blood. 2002;100:1065–7.
Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, et al. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–101.
Ivey A, Hills RK, Simpson MA, Jovanovic JV, Gilkes A, Grech A, et al. Assessment of minimal residual disease in standard-risk AML. N Engl J Med. 2016;374:422–33.
Lavallée VP, Lemieux S, Boucher G, Gendron P, Boivin I, Armstrong RN, et al. RNA-sequencing analysis of core binding factor AML identifies recurrent ZBTB7A mutations and defines RUNX1-CBFA2T3 fusion signature. Blood. 2016;127:2498–501.
Acknowledgments
I am grateful to current and former members of the Maeda lab for their contributions to the work reviewed here. I also thank Elise Lamar for critical reading of the manuscript. This work was supported by American Cancer Society (RSG-13-379-01-LIB), NIH (R56 DK105001) and an American Society of Hematology Bridge Grant Program to T.M.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author has no conflict of interest to declare.
About this article
Cite this article
Maeda, T. Regulation of hematopoietic development by ZBTB transcription factors. Int J Hematol 104, 310–323 (2016). https://doi.org/10.1007/s12185-016-2035-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12185-016-2035-x