Abstract
Microbial lipases may be produced during milk storage and processing. This can lead to organoleptic changes in the corresponding dairy products. Thus, monitoring of lipase activity in milk is desirable. Turbidity of milk prevents a direct photometric measurement of lipase activity using chromophore- or fluorophore-based assays. Laborious pretreatments or alternative analytical methods normally have to be used. With the method for the determination of lipolytic activity (MeDeLi) proposed here, it is possible to measure lipase activity directly in the natural milk utilizing tailored fluorometric substrates. Only a defatting step is carried out initially for the MeDeLi. Then, the conversion of added lipase substrate is carried out in the unmodified milk without addition of any solutions or any enzyme extraction procedure which may influence the enzyme activity. Thereafter, the milk sample is treated with two solutions to remove the turbidity of milk by dissolution. A valid and sensitive fluorometric measurement is then possible. The applicability of the MeDeLi was demonstrated in comparison with tests published previously: The limit of detection for lipolytic activity measured by MeDeLi was the lowest, with 41 pkat/mL. Raw milk, milk products, and spoiled milk samples were also investigated with the MeDeLi.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The hydrolysis of milk triacylglycerols (lipolysis) by lipases generates not only desirable flavors but also off-flavors. In general, short-chain fatty acids (SCFAs, C4:0 to C8:0) give sharp flavors, while medium-chain fatty acids (MCFAs, C10:0 and C12:0) give a soapy taste and long-chain fatty acids (LCFAs, C14:0 to C18:0) are not flavor intensive (Chen et al. 2003). Additionally, free fatty acids (FFAs) also serve as precursors for other flavor compounds, such as β-keto acids, methyl ketones, esters, and lactones (Chen et al. 2003). Off-flavors, such as rancid, bitter, soapy, and astringent, can be caused by lipolysis of the milk fat (Fox 2006). An increased concentration of FFAs produced by lipases is recognized as a rancid taste (Antonelli et al. 2002). Bovine milk contains 37 to 41 g/L lipids (Jensen et al. 1991). The threshold levels tolerated for MCFA and SCFA are about 8 mg/L (lauric acid) to 25 mg/L (butyric acid) (Chen et al. 2003). Since lipases in milk are responsible for an increased FFA concentration, the lipase activity is directly correlated to the sensory quality and the storage stability of milk (Muir 1996). Natural, indigenous milk lipase and endogenous lipases of microbial origin can cause lipolysis of milk (Antonelli et al. 2002; Blake et al. 1996). The indigenous milk lipase activity is almost inactivated by the heating process, but heat-resistant microbial lipases from psychrotrophic bacteria may resist pasteurization and ultra-high temperature (UHT) treatment (Antonelli et al. 2002). Even low remaining lipase activities can cause off-flavors in milk products (Blake et al. 1996). A study by Celestino et al. (1997), for example, showed that milk powder from stored raw milk (4 °C, 48 h) had a higher amount of FFA than that from freshly produced raw milk.
One way to determine lipolysis is the measurement of the FFAs released. The routine analysis is the determination of FFAs in milk by solvent extraction methods and subsequent titrative measurements (Verband Deutscher Landwirtschaftlicher Untersuchungs- und Forschungsanstalten (VDLUFA) 1985; Bulletin of the IDF No. 265/1991 1991). The recoveries of FFAs by these methods are not ideal, which leads to an underestimation of the actual levels of FFAs, especially for SCFAs (Bulletin of the IDF No. 265/1991 1991), and, therefore, an underestimation of lipase activity by calculations from the FFA content also occurs. The determination of FFAs by gas chromatography is also used but has the limitations that the FFAs have to be extracted and derivatized before measurement and the laboratory has to be equipped with the device to do this (Deeth et al. 1983). A further approach for the estimation of lipase activity in milk is the turnover of chromogenic (Humbert et al. 1997; Shirai et al. 1982) or fluorogenic substrates (Stead 1984). Most of these assays include a laborious sample preparation with several experimental steps for the determination of lipase activity from complex samples, such as milk. Moreover, most assays reported harbor the problem of interference with milk lipids during the assay procedure (Blake et al. 1996). In order to prevent this, a complete removal of the fat has to be carried out before determination of the lipase activity.
A reliable test system for the determination of the lipase activity is desirable to monitor and guarantee the quality of milk products. This is becoming more and more relevant, since storage periods of raw milk are increasing due to the centralization of the dairy industry and the demand for milk products with extended shelf life (Stepaniak 2002).
The aim of this study was the development of a fast, robust, and valid assay for the direct determination of lipase activity in milk and milk products. It is of utmost importance that the measurement of lipolytic activity is carried out in the original milk environment and not in modified systems which might influence the enzyme activity. Therefore, in contrast to other assays, the method for determination of lipolytic activity (MeDeLi) proposed should allow the direct determination of lipase activity in milk samples with a limited number of procedure steps by the utilization of standard lab equipment.
Materials and Methods
Chemicals, Samples, and Enzymes
All chemicals were of analytical grade or higher. Chemicals were purchased from AppliChem (Darmstadt, Germany), Carl Roth (Karlsruhe, Germany), Merck (Darmstadt, Germany), Santa Cruz Biotechnology (Dallas, TX, USA), or Sigma-Aldrich (St. Louis, MO, USA).
Raw cow’s milk was obtained from the university-owned Agricultural Experiment Station. Commercial milk from cows or goats was obtained from a local supermarket: 1.5 % fat, MF (pasteurized, homogenized, microfiltered milk with 1.5 % fat; Frische Weidemilch, Schwarzwaldmilch, Freiburg, Germany); 1.5 % fat, UHT (homogenized, UHT treated milk with 1.5 % fat; Haltbare Alpenmilch, Molkerei Weihenstephan, Freising, Germany); 3.5 % fat, UHT (homogenized, UHT treated milk with 3.5 % fat; Haltbare Alpenmilch, Molkerei Weihenstephan, Freising, Germany); 3.8 % fat, MF (pasteurized, homogenized, microfiltered milk with 3.8 % fat; Frische Bio-Vollmilch, Schwarzwaldmilch, Freiburg, Germany); 0.3 % UHT (homogenized, UHT treated milk with 0.3 % fat; EDKA, Hamburg, Germany); and 1.5 % goat UHT (homogenized, UHT treated goat’s milk with 1.5 % fat (Haltbare Ziegenmilch, Andechser Molkerei Scheitz, Andechs, Germany).
Preparations of Burkholderia cepacia lipase (BcL), Pseudomonas fluorescens lipase (PfL), and Candida antarctica lipase B (CaLB) were obtained from Sigma-Aldrich (St. Louis, MO, USA). Preparations of secretory lipases from Stenotrophomonas chelatiphaga (ScL) and Pseudomonas panacis (PpL) were prepared by cultivating these microorganisms, which had been previously isolated from raw milk (von Neubeck et al. 2015). Cultivation was carried out in 100 mL of 10 % (v/v) milk in baffled shaking flasks (500 mL) at 30 °C and 120 rpm (TR-250, Infors, Bottmingen, Switzerland) for 2 days. The supernatant containing lipase was obtained by centrifugation (8000g, at 4 °C for 20 min).
MeDeLi in Original Milk Composition of Solutions
Three distinct solutions were necessary for the MeDeLi. A stop and a neutralizing solution were prepared to clarify the milk. The stop solution was composed of guanidine hydrochloride (8 M; carbamimidoylazanium chloride, GuHCl) and HCl (1 M) in water. For dissolution of GuHCl, the solution was kept at 40 °C for 30 min prior to the addition of HCl. The neutralizing solution was composed of Bis-tris (1 M; bis(2-hydroxyethyl)amino-tris(hydroxymethyl)methan), NaOH (850 mM), and EDTA disodium salt (250 mM; ethylenediaminetetraacetic acid disodium salt) in water. The synthetic substrates 4-methylumbelliferyl butyrate (4-methyl-2-oxo-2H-chromen-7-yl butyrate, 4-MUB) and 4-methylumbelliferyl laurate (4-methyl-2-oxo-2H-chromen-7-yl dodecanoate, 4-MUL) were dissolved at a concentration of 10 mM in 2-propanol for the measurement of lipase activity.
Experimental Procedure for the MeDeLi
The MeDeLi includes the following steps (Fig. 1): First, the milk sample (1.2 mL) was centrifuged at 20,000g at 4 °C for 10 min to remove the milk fat. From this sample, 250 μL defatted milk (bottom phase) was pipetted into a new reaction tube. Afterward, the original defatted milk sample was preincubated at 800 rpm at 40 °C in a ThermoMixer (Eppendorf, Hamburg, Germany) for 5 min. The measurement of lipase activity was started by the addition of 25 μL substrate solution. After a defined time period (5 min if not stated otherwise), the reaction was stopped by the addition of 150 μL stop solution. After a short mixing, the solution was neutralized and clarified by the addition of 100 μL neutralizing solution. Next, the fluorescence released was determined at the excitation at 355 nm and emission at 460 nm for the two substrates (4-MUB and 4-MUL). For this, 105 μL of the clarified test sample was transferred into a microtiter plate and measured with a fluorimeter (Fluoroskan Ascent, Thermo Fisher Scientific, Waltham, MA, USA). For milk samples containing an initial fat content higher than 1.5 %, a residual turbidity after addition of the neutralizing solution occurred. In this case, a second centrifugation step (20,000g at 4 °C for 3 min) was carried out to clarify the sample.
Workflow of the test procedure for the method for determination of lipolytic activity (MeDeLi) in milk: Milk fat is removed by centrifugation to avoid competing lipase substrates. Conversion is carried out in genuine milk using synthetic substrates. After adding stop solution and neutralizing solution, the concentration of the fluorophore released is determined spectrophotometrically
The measurement of blank values in the assay was achieved by adding the stopping solution to the milk sample before adding the substrate to the tube.
A calibration curve at concentrations between 0 and 110 μM 4-methylumbelliferone (7-hydroxy-4-methyl-2H-chromen-2-one, 4-MU) in clarified milk was prepared to calculate the enzyme activity. One katal lipase activity is defined as the release of 1 mol 4-MU within 1 s.
Comparative Assays for the Measurement of Lipase Activity
Lipase Measurement by 4-Methylumbelliferyl Esters (Biochemical 4-MU Reference Assay)
Assays published previously also utilized 4-MUB and 4-MUL for the measurement of lipase activity in defined buffer systems (De Monpezat et al. 1990; Jacks and Kircher 1967; Roberts 1985). Milk samples cannot be measured directly with these assays. For this reference assay, the substrates 4-MUB and 4-MUL were prepared as 10 mM stock solutions in 2-propanol. The assay was carried out in microtiter plates. An amount of 80 μL MU buffer (250 mM MOPS (3-morpholinopropane-1-sulfonic acid), 100 mM NaCl, and 10 % (w/v) Triton X-100 (O-[4-(1,1,3,3-tetramethylbutyl)phenoxy]polyethoxyethanol); pH 6.7) were added to 100 μL lipase sample and preincubated in the temperature-controlled fluorimeter at 40 °C for 5 min. The reaction was started by the addition of 20 μL substrate solution. Fluorescence was recorded continuously at an excitation of 355 nm and emission at 460 nm for 10 min. The blank values were measured by replacing the enzyme sample with MU buffer.
A calibration curve at concentrations between 0 and 110 μM 4-MU in MU buffer was prepared for the calculation of the enzyme activity. Therefore, one katal lipase activity is defined as the release of 1 mol 4-MU within 1 s.
Lipase Measurement by Tributyrin Hydrolysis (pH-stat Procedure)
The titrative pH-stat procedure is well established for lipase measurements (Cherry and Crandall 1932; Hoppe and Theimer 1996). An emulsion of tributyrin (1,3-di(butanoyloxy)propan-2-yl butanoate) was prepared for this assay. During continuous stirring of 50 mL water, 1.0 g gum arabic and 2.5 mL tributyrin were mixed. Thereafter, emulsification was carried out by mixing at 9500 rpm for 7 min utilizing an Ultraturrax (T25, S 25 N - 18 G, IKA-Werke, Staufen, Germany). The emulsion was continuously stirred and 20 mL was used for one enzyme activity measurement. For this assay, 100 μL of enzyme solution was added to the prepared substrate emulsion, and the pH was kept constant at pH 6.7 by automatic titration of 10 mM NaOH via a pH-stat titrator (TitroLine alpha, Schott-Geräte, Hofheim am Taunus, Germany) at 22 °C over a time span of 5 min.
The lipase activity was calculated by determination of the amount of NaOH which was necessary to neutralize the butyric acid released. One katal is defined as the release of 1 mol butyric acid per 1 s.
Titrimetric Measurement of Free Fatty Acids in Milk (Method After Deeth)
A method for the determination of FFA is the discontinuous titration method after Deeth (Verband Deutscher Landwirtschaftlicher Untersuchungs- und Forschungsanstalten (VDLUFA) 1985). The method is widely used in milk analysis. In this study, the titrimetric method has been adapted to measure lipase activity. For the measurement of FFA, a milk sample (5 mL) was mixed with 10 mL of a mixture of 2-propanol/petroleum benzine/sulphuric acid (40:10:1), 6 mL petroleum benzine (30–60 °C), and 4 mL water. The mixture was shaken for 15 s and stored for 15 min to achieve a phase separation. An amount of 5 mL from the upper phase was transferred into an Erlenmeyer flask. To the latter, 1 mL 2-propanol and three drops of a methanolic phenol red (4,4-(3H-2,1-benzoxathiol-3-yliden)bisphenol-1,1-dioxid) solution were added prior to titration with 10 mM KOH in ethanol. Titration was stopped when the indicator turned a pink color.
The enzyme conversions were carried out before titrative FFA measurement for the adapted lipase activity measurement. Lipase preparation was added to a 5 mL milk sample. This milk sample was incubated at 40 °C for a reaction time of 5 min. Thereafter, the reaction was stopped by the addition of a mixture of 2-propanol/petroleum benzine/sulphuric acid, and fatty acid extraction was carried out (see above).
Similar to the activity definition for the pH-stat method, one katal is defined as the release of 1 mol of fatty acid within 1 s.
Statistics and Estimation of the LOD and LOQ
All enzymatic assays and analytic measurements were performed in triplicate, at least. The limit of detection (LOD) and the limit of quantification (LOQ) were evaluated according to DIN ISO 5725-2 and DIN 32646 (Gey 2008). Thus, the LOD of the assay is defined as six times the standard deviation of the blank values. The LOQ was calculated by the nine times the standard deviation of the blank values. Statistic analyses, calculations, and visualizations were carried out using SigmaPlot 12 (Systat Software, San Jose, CA, USA) or Microsoft Office Excel 2007 (Microsoft Corporation, Redmond, WA, USA).
Results and Discussion
Method for the Determination of Lipolytic Activity in Milk by the Conversion of Fluorogenic Substrates and Milk Clarification
A method for the determination of lipolytic activity in milk samples (MeDeLi) was developed. Therefore, the conversion of specific fluorogenic substrates (4-MUB and 4-MUL) was carried out in unmodified milk. This was followed by a clarification process, which was the essential step for spectrophotometrical determination of the lipolytic activity.
In the development of MeDeLi, it was considered that for every enzyme activity measurement it is indispensable to take account of environmental effects influencing the measured activity. The enzyme activity is dependent on the matrix which may inhibit or activate enzymes by the presence of divalent ions, other proteins, pH, or substrates or products present. Therefore, the MeDeLi was designed in such a way that the enzymatic conversion is carried out in the unmodified milk sample without any enzyme extraction procedure. This has the advantage that the lipolytic activity is measured under natural matrix conditions and not in clean biochemical buffer systems, which would create a synthetic environment which may manipulate the lipase performance. The matrix effects are enzyme-specific (Gupta et al. 2004) and unpredictable.
A centrifugal removal of fat is the first step of the MeDeLi (Fig. 1). This step is essential for obtaining valid lipolytic activities, because persisting milk fat would compete with the 4-MUX substrates added and, thus, result in apparently lower lipase activity. The utilization of 4-MUX substrates, which are commercially available with several different fatty acids of various chain lengths (De Monpezat et al. 1990; Jacks and Kircher 1967), allows the adaptation of the MeDeLi to lipases with different substrate spectra. Within this study, 4-MUB (C4) and 4-MUL (C12) were shown to be suitable.
The temperature of the lipase assay was set at 40 °C, which is a compromise between exaggerated enzyme activity and potential inactivation of the enzymes at increased temperature. Thus, the mesophilic temperature may contribute to a sensitive lipase activity measurement.
The enzymatic conversion of the substrates (4-MUB and 4-MUL) in the milk sample was terminated by the addition of the stop solution at a defined time span (Fig. 1). The stop solution shifts the pH of the milk sample to a pH < 2. The shift of the pH and the chaotropic effect of GuHCl lead to a denaturation of lipases and, thus, a stopping of the enzyme conversion at the time of the addition of the stop solution. The shift of the pH does not destroy the reaction product 4-MU, which is the fluorophore measured later. Constant fluorescence values were recorded for 50 min (fluorescence constant in the range of ±4 %) in milk with added stop solution. The substrate (4-MUL) was also stable against acidic hydrolysis. The initial background fluorescence increased by about 10 % within 10 min, at 40 °C. This allowed a series of conversions to be carried out in parallel before adding the neutralizing solution. The neutralizing solution leads to clarification and a shift to a buffered pH of 6.5 (by Bis-tris). Obtaining a constant pH is important, since the fluorescence of the 4-MU released is strongly dependent on the surrounding pH (Jacks and Kircher 1967). At pH 6.5, the product (4-MU) fluorescence is acceptable and nonenzymatic hydrolysis of the substrate is not described (Al-Kady et al. 2011; Jacks and Kircher 1967). This behavior was also observed in the MeDeLi. The substrate 4-MUL showed no increasing fluorescence at the adjusted pH of 6.5 for at least 180 min (fluorescence constant in the range of ±1 %). The hydrolysis product 4-MU formed was stable for at least 360 min (fluorescence constant in the range of ±4 %) in the neutralized, clarified milk samples. This indicates that the termination of the enzymatic conversion is sufficient under the assay conditions and that neither excess substrate nor conversion products formed are destroyed by the procedure. The stability of the clarified solution observed at pH 6.5 allows handling of a large number of samples during enzymatic conversion and gives sufficient time to read a series of samples.
In addition to the stopping of the enzymatic conversion and ensuring a stable pH value, the stop and neutralizing solutions serve synergistically as a clarifying solution. A clarification of the milk samples is necessary for a valid fluorometric measurement. If turbidity of milk is not removed, the presence of scatterers and absorbers would affect the fluorescence of the fluorophore (4-MU) released and, therefore, hinder the quantitative determination (Chen et al. 2013). The turbidity of milk is caused by emulsified micelles of phosphocaseinate and emulsified fat (Fox 2002). During the MeDeLi, the milk fat is removed by centrifugation (Fig. 1), leaving the casein micelles as turbidity-causing agent. The clarification of the milk is not achieved by separation, precipitation, extraction, or filtration, similar to some protocols for milk examination (Humbert et al. 2006). It is driven by solubilization of milk proteins by GuHCl (Bobe et al. 1998) in the stop solution and the synergistic action of EDTA in the neutralizing solution by complexing the calcium, which destabilizes the casein micelles (Humbert et al. 1995; Humbert et al. 1997). The advantage of this experimental setup is that time-consuming, preliminary steps mentioned previously are replaced by the addition of two solutions. The dilution of the initial milk sample during MeDeLi, which affects the sensitivity of the assay, is reasonably low with a factor of 2.1. Moreover, a loss of formed analyte (4-MU) before measurement, as may occur during extraction steps, is impossible.
As an alternative to the fluorometric measurement, the utilization of chromogenic substrates, such as p-nitrophenol esters which have been described for lipase measurements (Beisson et al. 2000; Hasan et al. 2009; Stoytcheva et al. 2012; Thomson et al. 1999), might be possible when adapting the protocol. Moreover, the measurement of other milk-inherent enzyme activities, such as alkaline phosphatase, N-acetyl-β-glucosaminidase, or lactoperoxidase, which are used as markers for milk quality (Blel et al. 2001; Humbert et al. 1995), with chromogenic substrates, may be possible due to the clarification during MeDeLi. Principally, MeDeLi is also applicable to measure endopeptidase activity in milk samples. However, well-established assays like the o-phthaldialdehyde (OPA) assay (Nielsen et al. 2001) may be preferable to measure an increase of free amino groups over time. In this assay, the milk protein may serve as a natural substrate. The reaction is terminated by acid precipitation of unhydrolyzed protein. Thereafter, soluble peptides are derivatized by OPA and spectrophotometrical measurement is carried out in the obtained supernatant.
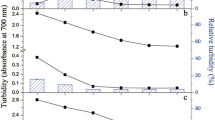
The robustness of the clarification procedure included in MeDeLi of various homogenized milk products was investigated by measuring the absorbance (A) of clarified milk samples. This was carried out at 860 nm (indicating the turbidity according to DIN/EN 27027, ISO 7027), 355 nm (the excitation wavelength for fluorometric measurement), and 460 nm (the emission wavelength measured during fluorometric measurement). The clarification procedure is visualized in Fig. 2a. It was possible to clarify cow’s and goat’s milk with the MeDeLi since the turbidity was reduced from A860 nm >3.0 (3.8 % fat UHT milk sample diluted with H2O instead of the clarification solutions) to absorbance values smaller than A860 nm 0.27 (3.5 % UHT milk sample after MeDeLi). The absorbance at the excitation and emission wavelength was consistently reduced to a level allowing fluorescence measurements in the clarified samples. In detail, milk samples with a fat content of 0.1, 0.3, and 1.5 % were clarified after the addition of the stop and neutralizing solutions, which served synergistically as clarifying reagents (A460 nm <0.80 after the clarification). Milk treatments (UHT or microfiltered (MF)) had only minor effects on the clarification process. Also, with goat’s milk (1.5 % fat, UHT) it was possible to obtain a clear solution after the clarification procedure (A460 nm 0.22 after clarification). Cow’s milk containing 3.5 and 3.8 % fat showed a residual turbidity after the addition of the stop and neutralizing solutions. This turbidity was observed in UHT and MF milk as well as in raw milk. The residual turbidity was reduced further (A460nm ≤1.20) by using a second centrifugation step (Fig. 1).
Various cows’ milk samples and one goat’s milk sample during the clarification process (a). The stop and the neutralizing solutions were replaced by water in the control sample. The samples after a first centrifugation step (top) for the removal of fat, the addition of the stop and neutralizing solutions (middle), and after the optional second centrifugation step (bottom) are illustrated. Clarification has been achieved in all samples besides the control after the clarification procedure. A linear correlation between the concentration of fluorophore (4-MU) added and the fluorescence signal measured is plotted exemplarily for an UHT milk sample of 3.5 % fat (1) and an UHT milk sample of 0.3 % fat (2) (b). Fat content is indicated as stated by the dairy company. MF microfiltered, UHT ultra high temperature
In conclusion, it was possible to render a turbid cow’s or goat’s milk sample to a clarified sample which should have an adequately low turbidity to allow spectrophotometrical measurements. In order to proof this, the fluorophore 4-MU was added in several concentrations into the clarified samples with the highest residual turbidity (3.5 % UHT milk (1) in Fig. 2) and the sample with the lowest residual turbidity (0.3 % UHT milk (2) in Fig. 2), as well as to a control sample (3.8 % MF milk, unclarified). The correlation between the 4-MU concentration added and the fluorescence measured in the clarified samples is shown in Fig. 2b. In both cases, a linear correlation, which is a precondition for valid enzyme activity measurements, was observed. No linear correlation was observed during the measurement of the unclarified control milk sample (graph not shown). As also seen here, it is commonly reported that turbidity destroys the linear correlation between concentrations of fluorophores and their fluorescence measurements (Chen et al. 2013). These findings emphasize the applicability of the MeDeLi for lipase activity measurements in milk samples.
Nevertheless, due to the high concentrations of compounds in the stop and neutralizing solutions, it is recommended to check the calibration curve for 4-MU concentrations after preparing new batches of the solutions. It is also necessary to prepare a new calibration curve for different types of milk, since the clarification is dependent on the milk sample (Fig. 2a). As a result, linear correlation between the fluorophore added and fluorescence signal measured with different slopes was observed in all clarified samples (Fig. 2b). In addition to the residual turbidity, which was reduced to an adequately low level by the clarification process, the background fluorescence in the clarified samples was also adequately low (22–31 RFU) for the preparation of calibration curves. Thus, lipase activity measurement is facilitated.
Evaluation of the MeDeLi by the 4-MU Reference Assay
The MeDeLi was evaluated by the addition of known lipase activities to MF milk, 1.5 % fat. Commercial preparations of B. cepacia lipase, P. fluorescens lipase, and C. antarctica lipase B as well as a self-made preparation of Stenotrophomonas chelatiphaga lipases were used. The added lipase activity was measured with the 4-MU reference assay in biochemical 4-MU buffer. The activity in the milk was measured by carrying out the MeDeLi procedure. The results are shown in Fig. 3. Theoretically, if the milk (in which the conversion of the lipase substrates is carried out) did not influence the lipase activity, about 100 % retrieval of the added lipase activity would be expected. This would result in a graph with the slope of one (indicated as TG in Fig. 3a).
Correlation of added lipase activities (150 ± 36, 300 ± 42, 450 ± 72, 500 ± 24 pkat/mLbuffer) measured with reference assay in biochemical 4-MU buffer (x-axis) and the lipase activity measured with the proposed method for determination of lipolytic activity (MeDeLi) in milk (y-axis). a B. cepacia lipase; b P. fluorescens lipase; c C. antarctica lipase B; d supernatant of S. chelatiphaga. Assays were carried out at 40 °C. Substrate was 4-MU laurate in (a), (b), and (d). The substrate was 4-MU butyrate in (c). TG (in (a)): theoretical graph of lipase activity without milk matrix effects
A linear correlation of added lipase activity with the retained lipase activity measured by the MeDeLi was observed in all milk samples (regression coefficient 0.973–0.997). This was demonstrated in an activity range from 150 up to 500 pkat4-MUX/mLMU buffer with all four lipase preparations (Fig. 3). Nevertheless, the total, volumetric enzyme activity observed differed between the biochemical reference assay and MeDeLi, as can be deduced from the slopes. The reason for this is most probably the influence of the milk components (e.g., ions and proteins) on the enzyme performance. About 1.5 times higher lipase activity (Fig. 3c) was measured by MeDeLi for C. antarctica lipase B. Only about 0.3 times of the enzyme activity was measured for the S. chelatiphaga lipases (Fig. 3d).
The substrates utilized, whether it was 4-MUL (Fig. 3a, b, d) or 4-MUB (Fig. 3c), had no negative contribution to the validity of the MeDeLi since a linear regression occurred in all cases.
The LOD and LOQ for the MeDeLi were determined. The LOD was determined to be 41.0 pkat4-MUL/mLmilk and the LOQ 63.0 pkat4-MUL/mLmilk in 45 independent measurements of the milk examined.
Estimation of LOD and LOQ of Alternative Methods
For comparison, two well-known tests for lipase measurement, the pH-stat method (Cherry and Crandall 1932; Hoppe and Theimer 1996) and the titrimetric method after Deeth (Verband Deutscher Landwirtschaftlicher Untersuchungs- und Forschungsanstalten (VDLUFA) 1985), were performed.
The pH-stat titration was carried out by measuring a P. fluorescens lipase solution at an added activity of 150 nkat4-MUL/mL4-MU buffer (determined by biochemical reference assay). This ~300 times higher activity than used in MeDeLi (Fig. 3b) was added to achieve a lipase activity greater than the LOQ of the pH-stat method. The measurement of the lipase sample in the tributyrin emulsion kept at pH 6.7 gave an activity of 245 ± 40 nkattributyrin/mLemulsion. The LOD and LOQ were evaluated to be 14.3 and 21.5 nkattributyrin/mLemulsion, respectively.
P. fluorescens lipase was also utilized for the titrimetric lipase measurement after Deeth. Therefore, 50 nkat4-MUL/mL4-MU buffer (determined by biochemical reference assay) was added to milk. Conversions of the original substrate, milk fat, were carried out at 40 °C for 5 min. A volumetric activity of 6.6 nkatmilk fat/mLmilk was evaluated from the amount of KOH (0.01 M) which was necessary to neutralize the liberated FFA. A corresponding LOD of 0.89 nkatmilk fat/mLmilk and a LOQ of 1.3 nkatmilk fat/mLmilk were calculated.
Comparing the limits obtained for the different methods directly (Table 1), one can see that the values for the MeDeLi with the substrate 4-MUL in milk are comparatively low. With a LOD of 41.0 pkat/mL and a LOQ of 63.0 pkat/mL, the limits of MeDeLi are ~22 times lower compared to the evaluated limits for the titration method. Compared to the pH-stat method, the LOD and LOQ are ~349 times lower. The MeDeLi only has a higher LOD and LOQ in the comparison of MeDeLi and the biochemical 4-MU reference assay (Table 1). Nevertheless, differences in the absolute values occur, since different assay types are compared with each other. In the MeDeLi and the reference standard assay, the substrate was the same, 4-MUL, but the matrix was different: milk in the MeDeLi and buffer in the reference assay. Milk was also used as the assay matrix for the titration method, but here, the substrate was different, since the milk-inherent fat was used. The pH-stat assay is different in both substrate (tributyrin) and matrix (unbuffered emulsion) from the other assays. Therefore, it is difficult to compare values directly. However, by comparing the applied lipase activities (determined by the biochemical 4-MU reference assay) in the MeDeLi evaluation with the enzyme activities applied in the other measurement allowing direct lipase activity measurement in milk (titration after Deeth), it is obvious that the sensitivity to lipase activity measurement in milk was higher for the MeDeLi. The sensitivity was higher even compared with the pH-stat titration, which does not allow the direct measurement of lipase activity in milk. A more reliable conclusion of applicability of MeDeLi can be obtained from the measurement of the genuine raw milk sample.
Measurements of Genuine Milk Samples
The lipase activity in three genuine milk samples was evaluated with the proposed MeDeLi. First, untreated raw milk was tested for lipase activity. Second, a milk sample spoiled by lipases produced by P. panacis was examined. P. panacis has been isolated from raw milk, and it has been shown that secretory enzymes can cause enzymatic deterioration of milk if this strain was present in raw milk (Baur et al. 2015). Third, a commercial UHT milk sample was measured. In all three samples, the hydrolysis of 4-MUL over time was investigated (Fig. 4). A linear behavior of the MeDeLi was demonstrated for original milk samples up to 90 min.
Time course of 4-MU liberation in UHT milk spoiled with P. panacis lipase (filled symbols) and untreated raw milk (open symbols). Values of unmodified ultra-high temperature (UHT) treated milk did not exceed the limit of quantification (LOQ) within the time investigated. The LOQ, determined in nine independent measurements, is indicated by a dashed line
A lipase activity of 19.2 pkat4-MUL/mLmilk was measured in the raw milk sample. This activity is most probably related to the lipoprotein lipase (Deeth 2006; Jensen and Pitas 1976) which is inherent in raw milk. The comparison of reported lipoprotein lipase activities is difficult due to the different utilized assays. The measured 19.2 pkat4-MUL/mLmilk (by MeDeLi) was in the same order of magnitude as reported values. However, Deeth (2006) reported about a lipoprotein lipase activity measured with a LCFA emulsion at pH 7 and 37 °C with added lipase activators of 260 pkatLCFA/mLcream. A somewhat higher activity of 6.9 nkatbutterfat/mLmilk was reported by Anderson (1982).
Measurement of the lipase activity from P. panacis in milk (spoiled milk sample) by the MeDeLi resulted in an activity of 261.2 pkat4-MUL/mLmilk. Furthermore, the repeatability was examined with this milk sample by analyzing the same sample 12 times, each in triplicate. The overall coefficient of variation, consisting of the error occurring in enzymatic conversion, clarification, and the fluorometric measurement, was satisfactory with 8.8 %.
Values measured after 30-min (raw milk) and after 5-min (spoiled milk) conversion time were above LOQ and, therefore, allowed lipase activity measurement (Fig. 4). No lipase activity was measured in a commercial UHT milk sample, which served as the control, since the 4-MU concentration did not exaggerate the LOQ after 90 min of conversion.
Conclusion
The MeDeLi proposed for the measurement of lipase activity in milk may be used to monitor the quality of milk and maintain the quality of milk products. Raw milk-inherent lipase activities as well as activities caused by bacterial contaminants can be assayed directly in the milk sample.
In comparison to other protocols published previously, the MeDeLi proposed is sensitive, reliable, and time saving. The main advantage of the test procedure protocol is the limited number of experimental steps and necessary solutions. This results in a lower LOQ and an easier procedure compared to standard protocols, such as titrative FFA measurement (Cherry and Crandall 1932; Verband Deutscher Landwirtschaftlicher Untersuchungs- und Forschungsanstalten (VDLUFA) 1985; Hoppe and Theimer 1996), or clarification protocols published (Humbert et al. 2006; Manzano et al. 2005; Nakai and Le 1970). Besides centrifugation and incubation, there are no other experimental steps, such as extraction, separation, or filtration. The demand of apparatus is also limited to a centrifuge and a fluorimeter. The MeDeLi has an adequate sensitivity to measure raw milk and spoiled samples. Moreover, it shows a good robustness. Therefore, we think that the method can be easily automated and used routinely in dairy laboratories.
References
Al-Kady AS, Ahmed EI, Gaber M, Hussein MM, Ebeid EM (2011) Kinetics of catalyzed hydrolysis of 4-methylumbelliferyl caprylate (MUCAP) salmonella reagent. Spectrochim Acta A Mol Biomol Spectrosc 79:1540–1545
Anderson M (1982) Factors affecting the distribution of lipoprotein lipase activity between serum and casein micelles in bovine milk. J Dairy Res 49:51–59
Antonelli ML, Curini R, Scricciolo D, Vinci G (2002) Determination of free fatty acids and lipase activity in milk: Quality and storage markers. Talanta 58:561–568
Baur C, Krewinkel M, Kutzli I, Kranz B, von Neubeck M, Huptas C, Wenning M, Scherer S, Stoeckel M et al (2015) Isolation and characterisation of a heat-resistant peptidase from Pseudomonas panacis withstanding general UHT processes. Int Dairy J 49:46–55
Beisson F, Tiss A, Rivière C, Verger R (2000) Methods for lipase detection and assay: A critical review. Eur J Lipid Sci Technol 102:133–153
Blake MR, Koka R, Weimer BC (1996) A semiautomated reflectance colorimetric method for the determination of lipase activity in milk. J Dairy Sci 79:1164–1171
Blel M, Guingamp M, Gaillard J, Humbert G (2001) Improvement of a method for the measurement of lactoperoxidase activity in milk. Int Dairy J 11:795–799
Bobe G, Beitz DC, Freeman AE, Lindberg GL (1998) Separation and quantification of bovine milk proteins by reversed-phase high-performance liquid chromatography. J Agric Food Chem 46:458–463
Bulletin of the IDF No. 265/1991 (1991) Determination of free fatty acids in milk and milk products. 265:1-52
Celestino EL, Iyer M, Roginski H (1997) Reconstituted UHT-treated milk: Effects of raw milk, powder quality and storage conditions of UHT milk on its physicochemical attributes and flavour. Int Dairy J 7:129–140
Chen L, Daniel RM, Coolbear T (2003) Detection and impact of protease and lipase activities in milk and milk powders. Int Dairy J 13:255–275
Chen Y, Chen Z, Yang J, Jin J, Zhang J, Yu R (2013) Quantitative fluorescence spectroscopy in turbid media: A practical solution to the problem of scattering and absorption. Anal Chem 85:2015–2020
Cherry IS, Crandall LA (1932) The specifity of pancreatic lipase: Its appearance in the blood after pancreatic injury. Am J Physiol 100:266–273
De Monpezat TDL, De Jeso B, Butour J, Chavant L, Sancholle M (1990) A fluorimetric method for measuring lipase activity based on umbelliferyl esters. Lipids 25:661–664
Deeth HC (2006) Lipoprotein lipase and lipolysis in milk. Int Dairy J 16:555–562
Deeth HC, Fitz-Gerald CH, Snow AJ (1983) A gas chromatographic method for the quantitative determination of free fatty acids in milk and milk products. NZ J Dairy Sci Technol 18:13–20
Fox PF (2002) Milk introduction. In: Roginski H (ed) Encyclopedia of dairy sciences. Elsevier, Oxford, pp 1805–1812
Fox P (ed) (2006) Advanced dairy chemistry, 3rd edn. Springer, New York
Gey MH (2008) Instrumentelle analytik und bioanalytik. Springer, Berlin, pp 166–172
Gupta R, Gupta N, Rathi P (2004) Bacterial lipases: An overview of production, purification and biochemical properties. Appl Microbiol Biotechnol 64:763–781
Hasan F, Shah AA, Hameed A (2009) Methods for detection and characterization of lipases: A comprehensive review. Biotechnol Adv 27:782–798
Hoppe A, Theimer RR (1996) Titrimetric test for lipase activity using stabilized triolein emulsions. Phytochemistry 42:973–978
Humbert G, Guingamp MF, Choukri A, Linden G (1995) Method for the measurement of N-acetyl-beta-glucosaminidase activity in bovine milk. J Dairy Res 62:369–372
Humbert G, Guingamp M-F, Linden G (1997) Method for the measurement of lipase activity in milk. J Dairy Res 64:465
Humbert G, Guingamp M, Linden G, Gaillard J (2006) The clarifying reagent, or how to make the analysis of milk and dairy products easier. J Dairy Res 73:464
Jacks TJ, Kircher HW (1967) Fluorometric assay for the hydrolytic activity of lipase using fatty acyl esters of 4-methylumbelliferone. Anal Biochem 21:279–285
Jensen PG, Pitas RE (1976) Milk lipoprotein lipases: A review. J Dairy Sci 59:1203–1214
Jensen RG, Ferris AM, Lammi-Keefe CJ (1991) The composition of milk fat. J Dairy Sci 74:3228–3243
Manzano S, Antonio Ordóñez J, de la Hoz L, Fernández M (2005) A rapid method for the estimation of the microbiological quality of refrigerated raw milk based on the aminopeptidase activity of gram-negative bacteria. Int Dairy J 15:79–84
Muir DD (1996) The shelf-life of dairy products: 3. factors influencing intermediate and long life dairy products. Int J Dairy Technol 49:67–72
Nakai S, Le AC (1970) Spectrophotometric determination of protein and fat in milk simultaneously. J Dairy Sci 53(3):276–278
Nielsen PM, Petersen D, Dambmann C (2001) Improved method for determining food protein degree of hydrolysis. J Food Sci 66:642–646
Roberts IM (1985) Hydrolysis of 4-methylumbelliferyl butyrate: A convenient and sensitive fluorescent assay for lipase activity. Lipids 20:243–247
Shirai K, Jackson RL, Quinn DM (1982) Reciprocal effect of apolipoprotein C-II on the lipoprotein lipase-catalyzed hydrolysis of p-nitrophenyl butyrate and trioleoylglycerol. J Biol Chem 257:10200–10203
Stead D (1984) A fluorimetric method for determination of Pseudomonas fluorescens AR11 lipase in skim milk powder, whey powder and whey protein concentrate. J Dairy Res 51:623–628
Stepaniak L (2002) Psychortophic bacteria: Bacteria other than Pseudomonas spp. In: Roginski H (ed) Encyclopedia of dairy dciences. Elsevier, Oxford, pp 2345–2351
Stoytcheva M, Montero G, Zlatev R, LeónJÁ GV (2012) Analytical methods for lipases activity determination: A review. Curr Anal Chem 8:400–407
Thomson CA, Delaquis PJ, Mazza G (1999) Detection and measurement of microbial lipase activity: A review. Crit Rev Food Sci Nutr 39:165–187
Verband Deutscher Landwirtschaftlicher Untersuchungs- und Forschungsanstalten (VDLUFA) (1985) Methodenbuch Band VI. C 35.3 ed
von Neubeck M, Baur C, Krewinkel M, Stoeckel M, Kranz B, Stressler T, Fischer L, Hinrichs J, Scherer S, Wenning M (2015) Biodiversity of refrigerated raw milk microbiota and their enzymatic spoilage potential. Int J Food Microbiol submitted manuscript
Compliance with Ethical Standards
Funding
This research project was supported by the Federal Ministry for Economic Affairs and Energy (via AiF) and the FEI (Forschungskreis für Ernährungsindustrie e.V., Bonn): Project AiF 16588 N.
Conflict of Interest
Manuel Krewinkel declares that he has no conflict of interest. Claudia Baur declares that she has no conflict of interest. Bertolt Kranz declares that he has no conflict of interest. Mario von Neubeck declares that he has no conflict of interest. Mareike Wenning declares that she has no conflict of interest. Siegfried Scherer declares that he has no conflict of interest . Marina Stoeckel declares that she has no conflict of interest. Jörg Hinrichs declares that he has no conflict of interest. Lutz Fischer declares that he has no conflict of interest. This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
Not applicable.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Krewinkel, M., Baur, C., Kranz, B. et al. A Sensitive and Robust Method for Direct Determination of Lipolytic Activity in Natural Milk Environment. Food Anal. Methods 9, 646–655 (2016). https://doi.org/10.1007/s12161-015-0233-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12161-015-0233-4