Abstract
Artificial intelligence (AI) has been applied to various medical imaging tasks, such as computer-aided diagnosis. Specifically, deep learning techniques such as convolutional neural network (CNN) and generative adversarial network (GAN) have been extensively used for medical image generation. Image generation with deep learning has been investigated in studies using positron emission tomography (PET). This article reviews studies that applied deep learning techniques for image generation on PET. We categorized the studies for PET image generation with deep learning into three themes as follows: (1) recovering full PET data from noisy data by denoising with deep learning, (2) PET image reconstruction and attenuation correction with deep learning and (3) PET image translation and synthesis with deep learning. We introduce recent studies based on these three categories. Finally, we mention the limitations of applying deep learning techniques to PET image generation and future prospects for PET image generation.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Artificial intelligence (AI) is a set of powerful algorithms for realizing human-like recognition capabilities. Some early trials attempted to implement the recognition to a computer using a neural network [1,2,3], and recent developments in theories of neural networks and computer performance have made it realistic to apply the AI algorithm to practical problems [4]. It is now feasible to train a large-scale neural network. A typical application of this technology is the field of medical imaging for the diagnosis of various diseases [5, 6]. Radiologists inject their knowledge into image reading to identify hidden lesions. However, the increasing number of imaging modalities and cases to be read require increased radiologist staffing. AI is expected to help this issue by achieving some good results.
In this review, we focus on AI applications for positron emission tomography (PET). PET is a modality used in nuclear medicine and has some specific capabilities. It can visualize various functionalities in a living organ, such as glucose metabolism and a density of neuroreceptors or amyloid beta. PET realizes quantitative functional imaging, and is thus applied in various clinical applications to diagnose tumors or neurodegenerative diseases, such as Alzheimer’s disease (AD). Therefore, it is necessary to understand a wide variety of applications of AI to PET. PET also provides an absolute quantity of functionality in living organs in Bq/mL or binding potential [7].
In “Deep learning for recovering full data from noisy data”, we discuss image recovery from an acquired noisy image, which is one of the major applications of AI in PET imaging. Due to the dynamic range of the PET camera and the safety of the patient and medical staff against radiation exposure, the administered radioactive dose is limited. Therefore, a PET image is usually contaminated with noise. Some AI-based algorithms are reviewed in this section. In “Deep learning for image reconstruction and attenuation correction”, we discuss the reconstruction algorithms, including attenuation correction. Image reconstruction is a complicated mathematical process, and attenuation and scatter must be considered. AI provides some new approaches for image reconstruction. In “Deep learning for image translation and synthesis”, we present the variation of AI in the synthesis of PET images. In “Limitations and challenges”, we discuss the limitations and future challenges of AI applications in PET imaging. In “Summary”, we summarize the review. In addition, the AI algorithms are outlined in Table 1.
Deep learning for recovering full data from noisy data
One of the major applications of deep learning to PET image reconstruction and generation is the recovery of standard quality images from low-quality images. To minimize radiation exposure to the examinee and medical staff, the injection dose should be kept as low as possible. However, reducing the injection dose increases the noise on PET acquisition data and degrades the image quality. Recently, improvements in PET hardware [8, 9] and reconstruction [10, 11] have enabled us to reduce the dose while maintaining image quality. However, radiation exposure using PET scans is still significant. Techniques to suppress the noise associated with reducing the injection dose are still required to improve the comfort of the examinees of PET scans.
Deep learning techniques such as convolutional neural networks (CNNs) and generative adversarial networks (GANs) have been applied to the recovery from low-quality images to high-quality images in medical imaging areas [12] as well as natural images [13]. For example, recovery of images acquired with standard dose computed tomography (CT) scans from those acquired with low-dose scans using U-Net [14, 15] and GAN [16, 17] have been proposed. These techniques for recovering from low-dose data can be transferred to PET imaging and enable the acquisition of low-dose and short duration PET scans.
This section introduces recent studies for recovering standard dose/standard duration PET data from low-dose/short duration PET data using deep learning techniques for denoising. First, studies for recovering standard dose/duration data from low-dose/short duration data by machine learning with CNN and GAN will be reviewed. Second, we introduce a deep image prior technique and its applications in PET studies. The deep image prior technique can be performed with only single data, whereas deep learning requires a large dataset. The deep image prior technique can be important, especially in the field of PET, where collecting huge amounts of PET data is impractical because of the limited number of PET cameras compared with other modalities.
CNN
Similar to other applications of deep learning in PET studies, CNN has been the most commonly utilized for recovering standard dose/standard duration PET data. Xiang [18] applied a CNN to recover standard duration PET images from short duration PET images first. They trained a CNN that took brain fluorodeoxyglucose (FDG) PET images, acquired 3 min PET scan and MR T1 images as inputs, and the PET images for 12 min as the target with an auto-context CNN model, which consisted of three four-layered CNN blocks and skip connections of input images to each block. The images recovered by the trained auto-context CNN model corresponded to the standard duration images (peak signal-to-noise ratio [PSNR]: 24.76, normalized mean square error [NMSE]: 0.0206). These scores were comparable to those obtained using the multilevel canonical correlation analysis (PSNR: 24.67; NMSE: 0.0210), which had been the state-of-the-art algorithm at the time.
U-Net, a U-shaped CNN with skip connections [19], is useful for recovering full PET data, similar to other tasks in the medical imaging field. For example, Chen demonstrated that U-Net trained with multiple contrast MR images could recover the full-dose PET images from ultra-low-dose (1/100) PET images acquired in amyloid PET studies [20]. They demonstrated that visual evaluation for amyloid-positive/negative with the images recovered by the trained U-Net corresponded well (accuracy: 0.89) to one with the images acquired with full-dose. The U-Net was also applied to recover the full-dose images for whole-body [21] and cardiac [22] FDG PET studies.
Some groups applied neural network architecture other than the U-Net and demonstrated that these networks resolved some issues of U-Net and could recover high-quality images. Spuhler [23] proposed dilated CNN to overcome the blurring in images occurring during down-sampling in the U-Net and demonstrated that the images recovered by the dilated CNN corresponded to the full-dose images better than ones by U-Net (PSNR: 38.67 ± 0.78 [dilated CNN], 38.24 ± 0.78 [U-Net]; structural similarity index measure [SSIM]: 0.92 ± 0.01 [dilated CNN], 0.91 ± 0.01 [U-Net]). To avoid overfitting, da Costa-Luis and Reader proposed a micro-Net, a shallower network than the U-Net [24]. Wang demonstrated that a CNN trained with a loss function weighted to regions with a high prevalence of tumors could accurately recover whole-body FDG PET images [25].
Transfer learning, applying a pre-trained network for a targeting task, and fine-tuning with the pre-trained network can save costs for acquiring and learning huge data. Several studies have indicated that transfer learning and fine-tuning are effective for recovering PET data. Chen transferred the pre-trained U-Net to amyloid PET images acquired at another site and with a scanner other than the training data [26]. They demonstrated that the model fine-tuned with the data at the other site resulted in better performance than that without fine-tuning. They also demonstrated higher accuracy (96.3%) and sensitivity (100%) in amyloid status reading with images predicted by the fine-tuned model than those by the pre-trained model without fine-tuning (accuracy: 78.8%; sensitivity: 60.5%). Liu demonstrated no significant difference between whole-body DOTATATE PET images predicted by the model pre-trained with whole-body FDG PET data and the model fine-tuned with the DOTATATE PET data [27]. However, they also demonstrated that the model pre-trained with single-bed FDG PET resulted in a greater bias than the model fine-tuned with whole-body DOTATATE PET data. They concluded that fine-tuning of the pre-trained model is required in the case of different protocols.
Some investigators have inserted CNNs into an iterative PET reconstruction process for denoising reconstructed images [28,29,30]. Wang proposed a maximum a posteriori (MAP) reconstruction penalized with the relative difference between the ordered subset expectation maximization (OSEM) reconstruction and CNN outputs. They demonstrated that MAP with CNN can acquire images with higher contrast and less noise than MAP or CNN.
GAN
GAN frameworks have been applied to recover the standard dose/standard duration PET data. Wang first performed adversarial training between a generator to generate the brain FDG PET images acquired by a standard duration (12 min) from a short duration (3 min) and a discriminator to determine whether images were real or generated [31]. They demonstrated that the 3D conditional GAN (cGAN) resulted in higher PSNR, lower NMSE, and lower standard uptake value (SUV) bias than 3D U-Net. Lu first applied GAN to recover whole-body FDG PET images with a standard dose from images with a dose reduced by 10% [32]. They demonstrated that U-Net and GAN had comparable performances in terms of SNR and biases in the SUV.
Some techniques to improve the performance and stability of GANs have been proposed. Wang applied a locally adaptive network to generate fusion images between low-dose PET and multi-contrast MR images and regarded the fused images as inputs for the generator [33]. The GAN with images fused by the locally adaptive network resulted in a higher contrast than the GAN with multi-channel input images. Ouyang trained a classifier for amyloid-positive/negative as well as generator and discriminator and added task-specific perceptual loss, which was the loss of feature maps between real and generated images on hidden layers in the classifier, to the loss function [34]. The GAN with task-specific perceptual loss resulted in PSNR, SSIM, and root mean squared error (RMSE) comparable to the previously proposed model with PET and MR inputs, despite only PET images being regarded as an input for the GAN with perceptual loss.
CycleGAN, a cycle-consistent adversarial network for image-to-image translation [35], has been applied to recover standard dose PET images from low-dose images. In recent studies, CycleGAN has two GANs, one converts low-dose images to standard dose PET images, the other converts standard dose PET images to low-dose images. CycleGAN is trained to minimize the differences between the true images and those synthesized in a cycle fashion. Lei demonstrated that CycleGAN resulted in generated whole-body FDG PET images with higher PSNR and lower NMSE than U-Net and a single GAN [36]. Sanaat demonstrated that scores of image quality assessment and lesion detectability for whole-body FDG PET images generated by CycleGAN from 1/8 low-dose images were comparable to the standard dose images [37]. Zhao proposed CycleGAN with Wasserstein distance loss (CycleWGAN) for recovering from 10% low-dose brain [38] and whole-body PET images [39]. The CycleWGAN resulted in slightly low SSIM and PSNR, but the lowest biases in SUVmean and SUVmax relative to CNN and cGAN.
Deep image prior
The deep image prior (DIP) [40] is an important technique for denoising PET images because it can be performed only with single data, whereas deep learning requires a huge dataset, and the acquisition of a huge dataset is very difficult for PET. The DIP assumes that a CNN has the intrinsic ability to regularize ill-posed inverse problems, such as a denoising task without pre-training. In the original DIP framework, a learning process was conducted using a pair of random noise and corrupted images. The denoised image is obtained by stopping the iterations before the CNN learns the noise.
For denoising PET images, conditional DIP using prior information as input was applied instead of the original DIP with random noise as input. Cui performed DIP using CT and MR anatomical images as inputs [41]. Higher contrast, less noise, and thus better improvement of contrast-noise ratio (CNR) than conventional methods (Gaussian filter, non-local mean [NLM] filter, block-matching and 4D filtering [BM4D], and deep decoder) were demonstrated. Gong added DIP with MR images as input in the loop for penalized reconstruction (DIPRecon) [42]. The DIPRecon realized higher contrast than the DIP with MR images, CNN, kernel method with MRI, and Gaussian filter. Hashimoto performed DIP using a static image as input and dynamic frame as the target for denoising 4D dynamic PET data [43]. They demonstrated that the proposed DIP outperformed other algorithms such as the Gaussian filter, NLM filter, and image-guided filtering in terms of CNR. To accommodate the differences in tracer uptake among dynamic frames, they proposed a 4D DIP framework containing reconstruction branch networks for each frame and a feature extractor as the conventional DIP [44]. The 4D DIP resulted in lower bias and variance in radioactivity and influx rate estimated by the Patlak analysis, than the conventional DIP.
Deep learning for image reconstruction and attenuation correction
All the works introduced in the previous section are deep learning methods for image-to-image conversion, which can be viewed as post-reconstruction filtering to improve image quality. An alternative approach is direct image reconstruction, mapping from raw data (e.g., sinograms in PET imaging) to reconstruction images, a relatively new field in this category. Several proof-of-concept studies have been reported, and investigations using actual clinical PET datasets are very rare at present.
Another deep learning-based application is PET attenuation correction, which is generally required to achieve quantitative capability in PET imaging. In commercial PET/CT scanners, CT images are converted to attenuation coefficient maps and subsequently used for attenuation correction. Attenuation correction is not a trivial matter in PET-only or hybrid PET/MR scanners because of the lack of CT images, and various correction strategies, including methods using deep learning, have been developed. Deep learning-based attenuation correction is one of the applications that have been intensively researched in recent years.
This section reviews two major topics related to PET image generation: (1) deep learning-based direct image reconstruction and (2) deep learning-based attenuation correction. However, detailed reviews covering the entire aspect for attenuation correction have already been published [12, 45,46,47,48,49]. Therefore, we only provide a brief summary of the attenuation correction methods and recent updates, with the addition of related topics of scatter correction.
Direct image reconstruction
The first achievement of deep learning-based direct reconstruction was a unified framework, AUTOMAP, proposed by Zhu [50]. This work implemented a network consisting of fully connected layers followed by a convolutional autoencoder and demonstrated that AUTOMAP is generally applicable to image reconstruction problems (i.e., domain transformation between sensor and image) for various imaging modalities, including PET. However, due to the large number of trainable parameters in the fully connected layers and hence the large memory required and overfitting problem, only a small image size (e.g., a single slice of 128 × 128) is practical, and the applicability to real PET data is very limited.
In the field of PET image reconstruction, DeepPET, developed by Haggstrom, was the first systematic work on direct image reconstruction using deep learning [51]. Instead of using fully connected layers, a deep convolutional encoder-decoder network was applied to reconstruct the PET sinogram data into images. Using simulated data pairs of true images (target) and sinograms (input) derived from a whole-body digital phantom, they demonstrated that DeepPET generates higher quality images than conventional reconstruction algorithms (OSEM/filtered back projection [FBP]) in terms of relative RMSE, SSIM, and PSNR, with a considerably shorter computation time, over 100 times faster than OSEM reconstruction, suggesting the utility of deep learning reconstruction for real-time PET imaging. They also applied DeepPET to real patient sinograms acquired on a clinical PET scanner (GE D690 PET/CT), resulting in smoother images while retaining detailed structures compared with the conventional reconstruction images (albeit no ground truth for comparison). However, the image size of outputs was still limited to a single slice of 128 × 128 in this work, and poorer performance in lower count situations has been reported [52, 53].
Instead of learning from scratch, the entire process of PET image reconstruction, data-efficient approaches that incorporate the knowledge of the sinogram-to-image relationship, an inverse Radon transform, has been developed. In the FBP-Net proposed by Wang and Liu [52], a back-projection layer that corresponds to a fully connected layer with fixed parameters (i.e., inverse Radon transform) was introduced to reduce the total number of trainable parameters and thus improve the generalization of the reconstruction network. Experiments using the simulation data set from the Zubal phantom and real in vivo data (rat) showed that FBP-Net outperforms DeepPET in terms of the overfitting problem with a small amount of training data especially for PET imaging studies. DirectPET is another example of a data-efficient network for direct reconstruction proposed by Whiteley [54], which contains a Radon inversion layer consisting of many small fully connected networks, each corresponding to the transformation between a sinusoidal mask in sinogram space and a small patch in image space. In DirectPET, the use of the Radon inversion layer enables the direct reconstruction of multi-slice image volumes (400 × 400 × 16), which is significantly larger than that in previous studies. Using 54 whole-body FDG data acquired on a clinical PET scanner (Siemens Biograph mCT), they demonstrated that the DirectPET produced images that were quantitatively and qualitatively similar to the target images reconstructed by the standard clinical protocol (OSEM with point spread function [PSF] and time-of-flight [TOF]) in a considerably shorter computation time. Interestingly, they also reported a low-dose experiment in which DirectPET was trained using low count sinograms (50% of events removed from the list-mode data) to reconstruct normal count target images, demonstrating the ability to maintain image quality under a low dose.
All the direct reconstruction networks described above are available only with non-TOF sinogram data. Sinograms acquired on a TOF-capable PET scanner are first summed over the TOF dimension and fed into the networks, sacrificing the great benefit of improved timing resolution in state-of-the-art digital PET scanners [8, 9]. FastPET [53], developed by the same group for DirectPET, is another data-efficient approach, especially for 3D TOF PET data that operates on a histo-image representation in which the TOF information of detected events is stored as position information in image space. Because of architecturally simple networks for image-to-image conversion, i.e., a U-Net style 3D CNN, the FastPET is capable of generating a near-full 3D volume reconstruction image (400 × 400 × 96, dependent on the amount of graphics processing unit memory) within a second, facilitating the use of deep learning-based direct reconstruction in clinical and research works. By analyzing the clinical whole-body FDG (standard and low-dose) and brain datasets acquired on the latest generation digital TOF PET scanner (Siemens Biograph Vision), they concluded that FastPET produces high-quality 3D volumes with lower noise and without loss of significant structural details with near real-time speed.
PET attenuation correction
Deep-learning-based attenuation correction can be classified into three categories based on the techniques used for inter/intra-modality image conversion. Many studies have reported the use of various types of networks, that is, U-Net, GAN, and CycleGAN, and most of them focused on brain PET, and so applications for whole-body PET are still scarce. Summary and discussions on deep learning-based attenuation correction have been found in several extensive reviews [12, 45, 47, 49].
-
(1)
MR to synthetic CT:
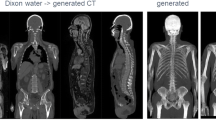
Synthetic CT images or pseudo-CT images are generated from co-registered MR images using deep learning techniques. Once the CT images are obtained, the standard reconstruction pipeline, including CT-based attenuation correction and scatter correction (usually performed with single scatter simulation), is applicable. The methods in this category have high compatibility, especially for hybrid PET/MR scanners, in which anatomical images with a wide variety of tissue contrast are simultaneously acquired using various MR sequences, ranging from conventional T1/T2-weighted acquisition to specialized sequences such as Dixon and ultrashort/zero echo time. Networks are trained using image pairs of MR and CT images, independent of PET acquisition. Collecting large training datasets is relatively easy, and thus higher accuracy is generally expected compared to other approaches.
-
(2)
Non-attenuation-corrected PET to synthetic CT:
As a substitute for anatomical MR images, non-attenuation-corrected PET images are used as input for the generation of synthetic CT images, and subsequent image reconstruction, including attenuation/scatter correction, is performed similarly to the first method. These methods are preferred when anatomical images are not provided (i.e., acquisition with a PET-only scanner). Recently, with a brain-dedicated PET scanner, it was shown that the method provides synthetic transmission CT images with high accuracy for various radiotracers that produce quite different contrasts in PET emission images, provided that the network is trained on a mixed dataset of the radiotracers [55].
-
(3)
Non-attenuation-corrected PET to attenuation-corrected PET:
An alternative to the former methods, which use synthetic CT images for attenuation correction, is a deep learning-based direct conversion from non-attenuation-corrected to attenuation-corrected PET images and is superior in terms of simplicity and computational costs because it obviates the need for processing on attenuation coefficient maps and repetitive image reconstruction. A unique and important feature of these methods is the capability of simultaneous correction for attenuation and scatter, which is very reasonable given that attenuation and scatter are physical processes that are strongly related to each other. The direct generation of attenuation/scatter-corrected PET images from uncorrected images has been actively researched for brain and whole-body PET [56,57,58,59], demonstrating that the methods are promising in general but need to be further investigated for consistency and reliability in the final PET images.
Deep learning for image translation and synthesis
Intra-/inter-modality image translation and image synthesis from scratch are also important applications of deep learning in medical imaging. Deep learning techniques such as CNN and GAN have realized image translation and synthesis, which were thought to be very difficult in medical imaging [12]. For example, as mentioned in “PET attenuation correction”, CT images synthesized from MR images by deep learning have been applied for PET attenuation correction in hybrid PET/MR scanners.
Image translation and synthesis by deep learning can bring three main benefits to PET imaging: (1) supplement missing data, (2) reduction in the number of scans, and (3) data augmentation for machine learning. First, the missing data are caused by various reasons in the medical imaging area. For example, acquisitions for thin-sliced MR images are often omitted from clinical routine because of the long scan duration, even though thin-sliced MR images are mandatory for quantitative analysis of brain PET images. Therefore, the synthesis of thin-sliced MR images using deep learning can be useful for the quantitative analysis of PET images without MR acquisitions. Second, image translation and synthesis by deep learning enable us to skip acquisitions of targeting images and shorten the total acquisition time. The shortening of the total acquisition time reduces the burden on patients and the chances of radiation exposure. Finally, image translation and synthesis can perform as data augmentation to resolve the lack of training data and imbalanced data issues in classification by machine learning. Data augmentation with image translation and synthesis by deep learning can be specifically useful for computer-aided diagnosis of rare diseases, regarding which it is difficult to acquire large amounts of data.
This section introduces several applications of image translation and synthesis using deep learning in PET studies. The major application of image translation and synthesis in PET studies is supporting quantification analysis in amyloid PET studies. Thus, we first introduce the applications of amyloid PET studies. Then, we introduce several studies that applied image translation and synthesis by deep learning to targets other than amyloid PET.
Application for amyloid PET
The first application of image translation/synthesis for amyloid PET was the template-based quantification of SUVR without MR images. PET-based templates lead to bias in quantifying amyloid burden because of the different uptake patterns between healthy controls and patients with AD, and thus imprecise spatial normalization only with PET images. Choi and Lee applied GAN to generate MR T1 images, which were used for spatial normalization to the template, from 18F-florbetapir PET images [60]. They succeeded in generating MR images similar to the real ones regardless of subjects’ diagnosis (SSIM: 0.91 ± 0.04, 0.92 ± 0.04, and 0.91 ± 0.04 for AD, mild cognitive impairment, and healthy controls, respectively). The quantification with the generated MR images resulted in smaller bias in SUVR (mean absolute error [MAE]: 0.04 ± 0.03) than the quantification with PET-based methods (MAE: 0.29 ± 0.12 [normal PET template]; 0.12 ± 0.07 [multiatlas PET template]; and 0.08 ± 0.06 [PET-based segmentation]). Similarly, Kang applied a convolutional autoencoder (CAE) and GAN to generate an adaptive template from only 11C-PiB PET images [61]. SUVR quantified with the template generated by CAE and GAN corresponded to SUVR quantified with MR images (R2: 0.8901 [CAE]; 0.8919 [GAN]; and 0.8128 [template with averaged PET images]). Similar results in statistical parametric mapping (SPM) analysis were observed between the generated and MR-based templates.
Some research groups have applied CycleGAN to convert amyloid PET images to supplement missing data and/or data augmentation for ensuing machine learning. Kimura applied CycleGAN to convert images between amyloid-negative and amyloid-positive for data augmentation in machine learning for dementia other than AD [62]. They gave CycleGAN amyloid-negative images slice-by-slice to train and succeeded in generating images likely to be amyloid-positive. Their work implies the potential to apply AI-based image augmentation for diseases from which fewer patients suffer than other common diseases such as AD and Lewy body dementia. Kang applied CycleGAN to convert between 11C-PiB and 18F-florbetapir PET images to resolve inter-tracer differences in amyloid PET tracers [63]. The SUVR on generated 11C-PiB/18F-florbetapir PET images corresponded to those on real PET images (intraclass correlation coefficient [ICC] for global cerebral cortex: 0.85 [11C-PiB] and 0.87 [18F-florbetapir]).
Application for other tracers
PET with 15O-labeled water and gases (15O PET) is the second major target of image translation and synthesis. 15O PET is regarded as the gold standard method for quantifying cerebral blood flow (CBF), cerebral blood volume (CBV), oxygen extraction fraction (OEF), and cerebral metabolic rate of oxygen (CMRO2). However, 15O PET requires special instruments such as cyclotron, arterial blood sampling, and fixation of a patient for 1–2 h, including preparation and handling procedures. To outplace parts of 15O PET scans, some groups have attempted to synthesize parametric maps acquired with 15O PET. Guo synthesized CBF maps from multi-contrast MR images, including arterial spin labeling (ASL) maps, T2-weighted fluid-attenuated inversion recovery, and T1-weighted images, using a modified U-Net [64]. The synthesized CBF maps were more similar to the maps acquired from 15O-water PET than single- and multidelay ASL (SSIM: 0.854 ± 0.036 [proposed]; 0.743 ± 0.045 [single-delay ASL]; 0.732 ± 0.041 [multidelay ASL]). Chen demonstrated that the U-Net learned with multi-contrast MR images, including ASL maps, can predict maps for cerebrovascular reactivity, which are acquired from both baseline and stressed CBF maps, without vasodilator injection [65]. To skip 15O2 scans for acquiring OEF, Matsubara’s group proposed the prediction of OEF maps from CBF acquired from C15O2 scans on rest and stress, CBV acquired from C15O scans, and multi-contrast MR images (T1-, T2-, and T2*-weighted images) by U-Net [66]. The trained U-Net can predict the OEF maps similar to the real maps acquired from 15O2 scans (ICC for OEF values on cortical regions: 0.597 ± 0.082), suggesting that we can skip the 15O2 scan. However, training with a larger dataset is required to accurately predict quantitative OEF maps.
Ben-Cohen synthesized whole-body FDG PET images from CT images using a combination of a fully convolutional network (FCN) and cGAN [67]. They applied FCN to synthesize PET-like images from CT and cGAN to refine the FCN output. The combination of FCN and cGAN resulted in low MAE and high PSNR in both regions with high and low SUV, relative to single FCN and cGAN. Furthermore, they demonstrated that the PET images synthesized by the proposed method reduced the false-positive ratio on detection of lesions using detection software with CT images. Sanaat proposed a stochastic adversarial variational prediction model to predict the later frames (25–90 min) from the initial frames (0–25 min) for 18F-FDOPA PET to reduce scan duration for the acquisition of 18F-FDOPA dynamic frames [68]. They demonstrated that the time-activity curves predicted by the model on the caudate, putamen, and cerebellum corresponded to the measured curves. The influx rate constant (Ki) calculated from the predicted dynamic data also corresponded to that calculated from the measured dynamic data (bias in Ki: < 7%). Wang applied 3D U-Net to generate 11C-UCB-J, which is a tracer used to quantify synapse density, PET images from brain FDG PET images, to skip scans with tracers that are difficult to obtain [69]. They succeeded in generating 11C-UCB-J SUVR images similar to real images from FDG SUVR images (SSIM: 0.906 ± 0.032; bias in SUVR on gray matter: − 1.61% ± 4.99% [AD]; − 1.33% ± 5.61% [healthy control]).
Limitations and challenges
One of the concerns in PET image generation using deep learning is the evaluation of the quantitative accuracy of PET images generated by deep learning. In most PET image generation studies, including the study presented in this review, the PSNR and SSIM, commonly used in deep learning of general images, are used to evaluate the generated images. However, these measures reflect human perceptual similarity and do not reflect the quantitative accuracy of PET images. In addition, determining some settings for these indices is difficult for PET images. For example, the dynamic range of images given in the calculation of SSIM cannot be strictly determined for PET images. Default parameters defined for natural images may be applied in many PET studies. Validation of these parameters for PET images is required. The studies presented in this review compared radioactivity concentration, and SUV by scatter plot, Bland–Altman plot, joint histogram, and calculated ICC of quantitative values, as well as SSIM and PSNR. To evaluate conventional PET images, some studies have calculated the contrast recovery, contrast-noise property, and bias-variance property. Although there is still a lack of consensus on assessing the quantitative accuracy of PET images generated by deep learning, it is essential to assess whether quantitative accuracy can be ensured by using conventional methods as well as evaluating the similarity to the correct image using PSNR and SSIM.
In most of the papers introduced in this review, insufficient training data are one of the limitations. In general, deep learning requires thousands to hundreds of thousands of data, and it is almost impossible to collect such a large amount of data in a single PET facility. The amount of data in the studies presented in this review ranged from tens to hundreds. As described in “Deep learning for recovering full data from noisy data”, it is possible to train a PET task on a small amount of data by fine-tuning a network that has been trained on a large amount of data with PET data. However, fine-tuning may not be successful in some cases, such as when there is a difference in the data distribution between the data used for training the original network and the data used for fine-tuning, when there is a difference between the task used for training the original network and the task to be fine-tuned, or in both cases. In such cases, it is necessary to train on large datasets. Therefore, if large-scale PET data or learned networks trained on such data were made available to everyone, problem-solving by transfer learning would become more active. However, in the field of PET, there is no database except ADNI. Therefore, such a database should be developed. Techniques such as unsupervised learning, self-supervised learning, and weakly supervised learning, which do not require large amounts of data, can be alternative solutions for the lack of training data. These techniques have been applied for image registration [70, 71] and segmentation [72,73,74] in medical imaging, as well as clustering [75] and feature extraction [76] in computer vision. In PET image denoising and reconstruction, DIP, which is one of the techniques for unsupervised learning, has been utilized in image denoising.
Transformer [77], a breakthrough in natural language processing, is also very likely to have a significant impact on nuclear medicine image generation. The transformer is an attention-based encoder-decoder model that can avoid gradient vanishing and accelerate massive parallelization. Some accurate language translation models using transformers have been proposed, such as BERT [78] and XLNet [79]. The transformer is also highly versatile for other tasks. For example, several applications of the transformer in image processing have been reported [80, 81] and have been applied to SPECT images [82]. The application of transformer to image transformation and image generation in PET imaging will be an exciting topic.
Another limitation is multimodal data handling. Unless an integrated PET/MR system is used, the alignment between PET and MR is assumed, and the influence of alignment error is a concern. However, few studies have been conducted to evaluate the effect of alignment errors in various tasks of PET image generation by deep learning. Therefore, a systematic evaluation of the effect of the PET-MR alignment error is desirable to evaluate the accuracy of deep learning in various tasks.
Summary
This paper describes deep learning techniques that have brought significant advances in full data recovery, image reconstruction, and image translation focused on PET imaging. In addition, deep learning techniques are rapidly advancing. Advanced unsupervised learning techniques and the transformer are typical examples of such techniques. Although most of these techniques are still in the research stage, they will be commonly used in clinical practice soon. We can easily imagine a future in which deep learning-based algorithms replace existing image generation and quality improvement algorithms. It is also possible that deep learning techniques will bring new images, which have never existed before, to clinical sites. Deep learning also has the potential to help reduce the burden on patients and clinical staff. For deep learning-based algorithms to be widely used in clinical practice, it is essential to increase the understanding of deep learning among clinical staff. Against this background, there have been many symposiums and seminars on deep learning at various conferences and research groups. The quantitative nature of the generated images is required for clinical use of the technology. Discussions and technologies are expected to progress in the future.
References
Fukushima K, Miyake S. Neocognitron: a self-organizing neural network model for a mechanism of visual pattern recognition. Competition and cooperation in neural nets. Lecture notes in biomathematics. Springer; 1982. p. 267–85.
Rumelhart DE, Hinton GE, Williams RJ. Learning representations by back-propagating errors. Nature. 1986;323(6088):533–6. https://doi.org/10.1038/323533a0.
Hinton GE, Sejnowski TJ. Learning and relearning in Boltzmann machines Parallel distributed processing: explorations in the microstructure of cognition. MIT Press; 1986.
Goodfellow I, Bengio Y, Courville A. Deep learning. MIT Press; 2016.
Doi K. Computer-aided diagnosis in medical imaging historical review, current status and future potential. Comput Med Imaging Gr. 2007;31(4–5):198–211. https://doi.org/10.1016/j.compmedimag.2007.02.002.
Fujita H. AI-based computer-aided diagnosis (AI-CAD): the latest review to read first. Radiol Phys Technol. 2020;13(1):6–19. https://doi.org/10.1007/s12194-019-00552-4.
Innis RB, Cunningham VJ, Delforge J, Fujita M, Gjedde A, Gunn RN, et al. Consensus nomenclature for in vivo imaging of reversibly binding radioligands. J Cereb Blood Flow Metab. 2007;27(9):1533–9. https://doi.org/10.1038/sj.jcbfm.9600493.
Hsu DFC, Ilan E, Peterson WT, Uribe J, Lubberink M, Levin CS. Studies of a next-generation silicon-photomultiplier-based time-of-flight PET/CT system. J Nucl Med. 2017;58(9):1511–8. https://doi.org/10.2967/jnumed.117.189514.
van Sluis J, de Jong J, Schaar J, Noordzij W, van Snick P, Dierckx R, et al. Performance characteristics of the digital biograph Vision PET/CT system. J Nucl Med. 2019;60(7):1031–6. https://doi.org/10.2967/jnumed.118.215418.
Teoh EJ, McGowan DR, Macpherson RE, Bradley KM, Gleeson FV. Phantom and clinical evaluation of the Bayesian Penalized likelihood reconstruction algorithm Q.Clear on an LYSO PET/CT system. J Nucl Med. 2015;56(9):1447–52. https://doi.org/10.2967/jnumed.115.159301.
Lee YS, Kim JS, Kim KM, Kang JH, Lim SM, Kim HJ. Performance measurement of PSF modeling reconstruction (True X) on Siemens Biograph TruePoint TrueV PET/CT. Ann Nucl Med. 2014;28(4):340–8. https://doi.org/10.1007/s12149-014-0815-z.
Wang T, Lei Y, Fu Y, Wynne JF, Curran WJ, Liu T, et al. A review on medical imaging synthesis using deep learning and its clinical applications. J Appl Clin Med Phys. 2021;22(1):11–36. https://doi.org/10.1002/acm2.13121.
Tian C, Fei L, Zheng W, Xu Y, Zuo W, Lin CW. Deep learning on image denoising: an overview. Neural Netw. 2020;131:251–75. https://doi.org/10.1016/j.neunet.2020.07.025.
Kang E, Min J, Ye JC. A deep convolutional neural network using directional wavelets for low-dose X-ray CT reconstruction. Med Phys. 2017;44(10):e360–75. https://doi.org/10.1002/mp.12344.
Shan H, Padole A, Homayounieh F, Kruger U, Khera RD, Nitiwarangkul C, et al. Competitive performance of a modularized deep neural network compared to commercial algorithms for low-dose CT image reconstruction. Nat Mach Intell. 2019;1(6):269–76. https://doi.org/10.1038/s42256-019-0057-9.
Wolterink JM, Leiner T, Viergever MA, Isgum I. Generative adversarial networks for noise reduction in low-dose CT. IEEE Trans Med Imaging. 2017;36(12):2536–45. https://doi.org/10.1109/TMI.2017.2708987.
Yi X, Babyn P. Sharpness-aware low-dose CT denoising using conditional generative adversarial network. J Digit Imaging. 2018;31(5):655–69. https://doi.org/10.1007/s10278-018-0056-0.
Xiang L, Qiao Y, Nie D, An L, Wang Q, Shen D. Deep auto-context convolutional neural networks for standard-dose PET image estimation from low-dose PET/MRI. Neurocomputing. 2017;267:406–16. https://doi.org/10.1016/j.neucom.2017.06.048.
Ronneberger O, Fischer P, Brox T. U-Net convolutional networks for biomedical image segmentation. Medical Image Computing and Computer-Assisted Intervention-MICCAI 2015. Springer International Publishing; 2015. p. 234–41.
Chen KT, Gong E, de Carvalho Macruz FB, Xu J, Boumis A, Khalighi M, et al. Ultra-low-dose (18)F-Florbetaben amyloid PET imaging using deep learning with multi-contrast MRI inputs. Radiology. 2019;290(3):649–56. https://doi.org/10.1148/radiol.2018180940.
Schaefferkoetter J, Yan J, Ortega C, Sertic A, Lechtman E, Eshet Y, et al. Convolutional neural networks for improving image quality with noisy PET data. EJNMMI Res. 2020;10(1):105. https://doi.org/10.1186/s13550-020-00695-1.
Ladefoged CN, Hasbak P, Hornnes C, Hojgaard L, Andersen FL. Low-dose PET image noise reduction using deep learning: application to cardiac viability FDG imaging in patients with ischemic heart disease. Phys Med Biol. 2021;66(5): 054003. https://doi.org/10.1088/1361-6560/abe225.
Spuhler K, Serrano-Sosa M, Cattell R, DeLorenzo C, Huang C. Full-count PET recovery from low-count image using a dilated convolutional neural network. Med Phys. 2020;47(10):4928–38. https://doi.org/10.1002/mp.14402.
da Costa-Luis CO, Reader AJ. Micro-networks for robust MR-guided low count PET imaging. IEEE Trans Radiat Plasma Med Sci. 2021;5(2):202–12. https://doi.org/10.1109/TRPMS.2020.2986414.
Wang YJ, Baratto L, Hawk KE, Theruvath AJ, Pribnow A, Thakor AS, et al. Artificial intelligence enables whole-body positron emission tomography scans with minimal radiation exposure. Eur J Nucl Med Mol Imaging. 2021;48(9):2771–81. https://doi.org/10.1007/s00259-021-05197-3.
Chen KT, Schurer M, Ouyang J, Koran MEI, Davidzon G, Mormino E, et al. Generalization of deep learning models for ultra-low-count amyloid PET/MRI using transfer learning. Eur J Nucl Med Mol Imaging. 2020;47(13):2998–3007. https://doi.org/10.1007/s00259-020-04897-6.
Liu H, Wu J, Lu W, Onofrey JA, Liu YH, Liu C. Noise reduction with cross-tracer and cross-protocol deep transfer learning for low-dose PET. Phys Med Biol. 2020;65(18): 185006. https://doi.org/10.1088/1361-6560/abae08.
Kim K, Wu D, Gong K, Dutta J, Kim JH, Son YD, et al. Penalized PET reconstruction using deep learning prior and local linear fitting. IEEE Trans Med Imaging. 2018;37(6):1478–87. https://doi.org/10.1109/TMI.2018.2832613.
Lv Y, Xi C. PET image reconstruction with deep progressive learning. Phys Med Biol. 2021;66(10): 105016. https://doi.org/10.1088/1361-6560/abfb17.
Wang X, Zhou L, Wang Y, Jiang H, Ye H. Improved low-dose positron emission tomography image reconstruction using deep learned prior. Phys Med Biol. 2021;66(11): 115001. https://doi.org/10.1088/1361-6560/abfa36.
Wang Y, Yu B, Wang L, Zu C, Lalush DS, Lin W, et al. 3D conditional generative adversarial networks for high-quality PET image estimation at low dose. Neuroimage. 2018;174:550–62. https://doi.org/10.1016/j.neuroimage.2018.03.045.
Lu W, Onofrey JA, Lu Y, Shi L, Ma T, Liu Y, et al. An investigation of quantitative accuracy for deep learning based denoising in oncological PET. Phys Med Biol. 2019;64(16): 165019. https://doi.org/10.1088/1361-6560/ab3242.
Wang Y, Zhou L, Wang L, Yu B, Zu C, Lalush DS, et al. Locality adaptive multi-modality GANs for high-quality PET image synthesis. Medical image computing and computer assisted intervention–MICCAI 201. Springer International Publishing; 2018. p. 329–37.
Ouyang J, Chen KT, Gong E, Pauly J, Zaharchuk G. Ultra-low-dose PET reconstruction using generative adversarial network with feature matching and task-specific perceptual loss. Med Phys. 2019;46(8):3555–64. https://doi.org/10.1002/mp.13626.
Zhu J-Y, Park T, Isola P, Efros AA. Unpaired image-to-image translation using cycle-consistent adversarial networks. 2017 IEEE International Conference on Computer Vision (ICCV). Venice, Italy: IEEE; 2017. p. 2242–51.
Lei Y, Dong X, Wang T, Higgins K, Liu T, Curran WJ, et al. Whole-body PET estimation from low count statistics using cycle-consistent generative adversarial networks. Phys Med Biol. 2019;64(21): 215017. https://doi.org/10.1088/1361-6560/ab4891.
Sanaat A, Shiri I, Arabi H, Mainta I, Nkoulou R, Zaidi H. Deep learning-assisted ultra-fast/low-dose whole-body PET/CT imaging. Eur J Nucl Med Mol Imaging. 2021;48(8):2405–15. https://doi.org/10.1007/s00259-020-05167-1.
Zhao K, Zhou L, Gao S, Wang X, Wang Y, Zhao X, et al. Study of low-dose PET image recovery using supervised learning with CycleGAN. PLoS ONE. 2020;15(9): e0238455. https://doi.org/10.1371/journal.pone.0238455.
Zhou L, Schaefferkoetter JD, Tham IWK, Huang G, Yan J. Supervised learning with cyclegan for low-dose FDG PET image denoising. Med Image Anal. 2020;65: 101770. https://doi.org/10.1016/j.media.2020.101770.
Ulyanov D, Vedaldi A, Lempitsky V. Deep Image Prior [https://arxiv.org/abs/quant-ph/1711.10925]. 2017.
Cui J, Gong K, Guo N, Wu C, Meng X, Kim K, et al. PET image denoising using unsupervised deep learning. Eur J Nucl Med Mol Imaging. 2019;46(13):2780–9. https://doi.org/10.1007/s00259-019-04468-4.
Gong K, Catana C, Qi J, Li Q. PET image reconstruction using deep image prior. IEEE Trans Med Imaging. 2019;38(7):1655–65. https://doi.org/10.1109/TMI.2018.2888491.
Hashimoto F, Ohba H, Ote K, Teramoto A, Tsukada H. Dynamic PET image denoising using deep convolutional neural networks without prior training datasets. IEEE Access. 2019;7:96594–603. https://doi.org/10.1109/access.2019.2929230.
Hashimoto F, Ohba H, Ote K, Kakimoto A, Tsukada H, Ouchi Y. 4D deep image prior: dynamic PET image denoising using an unsupervised four-dimensional branch convolutional neural network. Phys Med Biol. 2021;66(1): 015006. https://doi.org/10.1088/1361-6560/abcd1a.
Wang T, Lei Y, Fu Y, Curran WJ, Liu T, Nye JA, et al. Machine learning in quantitative PET: a review of attenuation correction and low-count image reconstruction methods. Phys Med. 2020;76:294–306. https://doi.org/10.1016/j.ejmp.2020.07.028.
Shiyam Sundar LK, Muzik O, Buvat I, Bidaut L, Beyer T. Potentials and caveats of AI in hybrid imaging. Methods. 2021;188:4–19. https://doi.org/10.1016/j.ymeth.2020.10.004.
Arabi H, AkhavanAllaf A, Sanaat A, Shiri I, Zaidi H. The promise of artificial intelligence and deep learning in PET and SPECT imaging. Phys Med. 2021;83:122–37. https://doi.org/10.1016/j.ejmp.2021.03.008.
Zaharchuk G, Davidzon G. Artificial Intelligence for Optimization and Interpretation of PET/CT and PET/MR Images. Semin Nucl Med. 2021;51(2):134–42. https://doi.org/10.1053/j.semnuclmed.2020.10.001.
Lee JS. A review of deep-learning-based approaches for attenuation correction in positron emission tomography. IEEE Trans Rad Plasma Med Sci. 2021;5(2):160–84. https://doi.org/10.1109/trpms.2020.3009269.
Zhu B, Liu JZ, Cauley SF, Rosen BR, Rosen MS. Image reconstruction by domain-transform manifold learning. Nature. 2018;555(7697):487–92. https://doi.org/10.1038/nature25988.
Haggstrom I, Schmidtlein CR, Campanella G, Fuchs TJ. DeepPET: a deep encoder-decoder network for directly solving the PET image reconstruction inverse problem. Med Image Anal. 2019;54:253–62. https://doi.org/10.1016/j.media.2019.03.013.
Wang B, Liu H. FBP-Net for direct reconstruction of dynamic PET images. Phys Med Biol. 2020. https://doi.org/10.1088/1361-6560/abc09d.
Whiteley W, Panin V, Zhou C, Cabello J, Bharkhada D, Gregor J. FastPET: near real-time reconstruction of PET histo-image data using a neural network. IEEE Trans Rad Plasma Med Sci. 2021;5(1):65–77. https://doi.org/10.1109/trpms.2020.3028364.
Whiteley W, Luk WK, Gregor J. DirectPET: full-size neural network PET reconstruction from sinogram data. J Med Imaging (Bellingham). 2020;7(3): 032503. https://doi.org/10.1117/1.JMI.7.3.032503.
Hashimoto F, Ito M, Ote K, Isobe T, Okada H, Ouchi Y. Deep learning-based attenuation correction for brain PET with various radiotracers. Ann Nucl Med. 2021;35(6):691–701. https://doi.org/10.1007/s12149-021-01611-w.
Shiri I, Arabi H, Geramifar P, Hajianfar G, Ghafarian P, Rahmim A, et al. Deep-JASC: joint attenuation and scatter correction in whole-body (18)F-FDG PET using a deep residual network. Eur J Nucl Med Mol Imaging. 2020;47(11):2533–48. https://doi.org/10.1007/s00259-020-04852-5.
Arabi H, Bortolin K, Ginovart N, Garibotto V, Zaidi H. Deep learning-guided joint attenuation and scatter correction in multitracer neuroimaging studies. Hum Brain Mapp. 2020;41(13):3667–79. https://doi.org/10.1002/hbm.25039.
Yang J, Park D, Gullberg GT, Seo Y. Joint correction of attenuation and scatter in image space using deep convolutional neural networks for dedicated brain (18)F-FDG PET. Phys Med Biol. 2019;64(7): 075019. https://doi.org/10.1088/1361-6560/ab0606.
Yang J, Sohn JH, Behr SC, Gullberg GT, Seo Y. CT-less direct correction of attenuation and scatter in the image space using deep learning for whole-body FDG PET: potential benefits and pitfalls. Radiol Artif Intell. 2021;3(2): e200137. https://doi.org/10.1148/ryai.2020200137.
Choi H, Lee DS. Alzheimer’s disease neuroimaging I. Generation of structural MR images from Amyloid PET: application to MR-less quantification. J Nucl Med. 2018;59(7):1111–7. https://doi.org/10.2967/jnumed.117.199414.
Kang SK, Seo S, Shin SA, Byun MS, Lee DY, Kim YK, et al. Adaptive template generation for amyloid PET using a deep learning approach. Hum Brain Mapp. 2018;39(9):3769–78. https://doi.org/10.1002/hbm.24210.
Kimura Y, Watanabe A, Yamada T, Watanabe S, Nagaoka T, Nemoto M, et al. AI approach of cycle-consistent generative adversarial networks to synthesize PET images to train computer-aided diagnosis algorithm for dementia. Ann Nucl Med. 2020;34(7):512–5. https://doi.org/10.1007/s12149-020-01468-5.
Kang SK, Choi H, Lee JS. Alzheimer’s disease neuroimaging initiative translating amyloid PET of different radiotracers by a deep generative model for interchangeability. Neuroimage. 2021;232:117890. https://doi.org/10.1016/j.neuroimage.2021.117890.
Guo J, Gong E, Fan AP, Goubran M, Khalighi MM, Zaharchuk G. Predicting (15)O-Water PET cerebral blood flow maps from multi-contrast MRI using a deep convolutional neural network with evaluation of training cohort bias. J Cereb Blood Flow Metab. 2020;40(11):2240–53. https://doi.org/10.1177/0271678X19888123.
Chen DYT, Ishii Y, Fan AP, Guo J, Zhao MY, Steinberg GK, et al. Predicting PET Cerebrovascular Reserve with Deep Learning by Using Baseline MRI: a pilot investigation of a drug-free brain stress test. Radiology. 2020;296(3):627–37. https://doi.org/10.1148/radiol.2020192793.
Matsubara K, Ibaraki M, Shinohara Y, Takahashi N, Toyoshima H, Kinoshita T. Prediction of an oxygen extraction fraction map by convolutional neural network: validation of input data among MR and PET images. Int J Comput Assist Radiol Surg. 2021;16(11):1865–74. https://doi.org/10.1007/s11548-021-02356-7.
Ben-Cohen A, Klang E, Raskin SP, Soffer S, Ben-Haim S, Konen E, et al. Cross-modality synthesis from CT to PET using FCN and GAN networks for improved automated lesion detection. Eng Appl Artif Intell. 2019;78:186–94. https://doi.org/10.1016/j.engappai.2018.11.013.
Sanaat A, Mirsadeghi E, Razeghi B, Ginovart N, Zaidi H. Fast dynamic brain PET imaging using stochastic variational prediction for recurrent frame generation. Med Phys. 2021;48(9):5059–71. https://doi.org/10.1002/mp.15063.
Wang R, Liu H, Toyonaga T, Shi L, Wu J, Onofrey JA, et al. Generation of synthetic PET images of synaptic density and amyloid from (18) F-FDG images using deep learning. Med Phys. 2021;48(9):5115–29. https://doi.org/10.1002/mp.15073.
Balakrishnan G, Zhao A, Sabuncu MR, Guttag J, Dalca AV. An unsupervised learning model for deformable medical image registration. In: 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition. Salt Lake City, UT, USA: IEEE; 2018. p. 9252–60.
Hering A, Kuckertz S, Heldmann S, Heinrich MP. Memory-efficient 2.5D convolutional transformer networks for multi-modal deformable registration with weak label supervision applied to whole-heart CT and MRI scans. Int J Comput Assist Radiol Surg. 2019;14(11):1901–12. https://doi.org/10.1007/s11548-019-02068-z.
Yang G, Wang C, Yang J, Chen Y, Tang L, Shao P, et al. Weakly-supervised convolutional neural networks of renal tumor segmentation in abdominal CTA images. BMC Med Imaging. 2020;20(1):37. https://doi.org/10.1186/s12880-020-00435-w.
Kallenberg M, Petersen K, Nielsen M, Ng AY, Pengfei D, Igel C, et al. Unsupervised deep learning applied to breast density segmentation and mammographic risk scoring. IEEE Trans Med Imaging. 2016;35(5):1322–31. https://doi.org/10.1109/TMI.2016.2532122.
Spitzer H, Kiwitz K, Amunts K, Harmeling S, Dickscheid T. Improving Cytoarchitectonic Segmentation of Human Brain Areas with Self-supervised Siamese Networks [https://arxiv.org/abs/quant-ph/1806.05104]. 2018.
Caron M, Bojanowski P, Joulin A, Douze M. Deep Clustering for Unsupervised Learning of Visual Features [https://arxiv.org/abs/quant-ph/1807.05520]. 2018.
Pathak D, Krahenbuhl P, Donahue J, Darrell T, Efros AA. Context Encoders: Feature Learning by Inpainting [https://arxiv.org/abs/quant-ph/1604.07379]. 2016.
Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, et al. Attention Is All You Need [https://arxiv.org/abs/quant-ph/1706.03762]. 2017.
Devlin J, Chang M-W, Lee K, Toutanova K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding [https://arxiv.org/abs/quant-ph/1810.04805]. 2018.
Yang Z, Dai Z, Yang Y, Carbonell J, Salakhutdinov R, Le QV. XLNet: Generalized Autoregressive Pretraining for Language Understanding [https://arxiv.org/abs/quant-ph/1906.08237]. 2019.
Dosovitskiy A, Beyer L, Kolesnikov A, Weissenborn D, Zhai X, Unterthiner T, et al. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale [https://arxiv.org/abs/quant-ph/2010.11929]. 2020.
Jiang Y, Chang S, Wang Z. TransGAN: Two Pure Transformers Can Make One Strong GAN, and That Can Scale Up [https://arxiv.org/abs/quant-ph/2102.07074]. 2021.
Watanabe S, Ueno T, Kimura Y, Mishina M, Sugimoto N. Generative image transformer (GIT): unsupervised continuous image generative and transformable model for [(123)I]FP-CIT SPECT images. Ann Nucl Med. 2021;35(11):1203–13. https://doi.org/10.1007/s12149-021-01661-0.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Matsubara, K., Ibaraki, M., Nemoto, M. et al. A review on AI in PET imaging. Ann Nucl Med 36, 133–143 (2022). https://doi.org/10.1007/s12149-021-01710-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12149-021-01710-8