Abstract
Spider dragline silk has attracted great interest due to its outstanding mechanical properties, which exceed those of man-made synthetic materials. Dragline silk, which is composed of at least major ampullate spider silk protein 1 and 2 (MaSp1 and MaSp2), contains a long repetitive domain flanked by N-terminal and C-terminal domains (NTD and CTD). Despite the small size of the CTD, this domain plays a crucial role as a molecular switch that regulates and directs spider silk self-assembly. In this study, we report the 1H, 13C, and 15N chemical shift assignments of the Latrodectus hesperus MaSp2 CTD in dimeric form at pH 7. Our solution NMR data demonstrated that this protein contains five helix regions connected by a flexible linker.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Biological context
Spider dragline silk is well recognized for its biocompatibility and unsurpassed toughness, which is a combination of superior strength and extensibility. The average toughness of dragline silk from Araneus diadematus is 180 MJ/m3, which is approximately three times tougher than that of man-made synthetic fibers, such as Kevlar 49 (Heim et al. 2009). Due to its fascinating properties, dragline silk has become a promising biomaterial for many industrial applications, such as artificial skin, drug delivery, and surgical suturing (Lammel et al. 2011; Wendt et al. 2011; Hennecke et al. 2013).
Dragline silk is a hierarchically structured fiber that is composed of several proteins, known as major ampullate spidroins (MaSps). Among spiders, these proteins are different in terms of molecular weight, amino acid composition and functional (i.e., mechanical) impact on mechanical properties. The archetypal dragline spidroins are MaSp1 and MaSp2, which mainly exhibit differences in their proline content (MaSp 1 < 0.4%, MaSp2 > 10%) (Guerette et al. 1996). Recently, several studies have demonstrated that the composition of dragline silks is more complex than previously known and includes multiple components, such as MaSp1, MaSp2, MaSp3, cysteine-rich rich protein (CRPs) and low molecular weight nonspidroin components (Pham et al. 2014; Chaw et al. 2015; Larracas et al. 2016; Kono et al. 2019, 2021). However, the transformation of soluble spidroin into insoluble fiber is still not completely understood (Malay et al. 2022).
Spidroins consist of a large core of repetitive domains flanked by small nonrepetitive N-terminal and C-terminal domains (molecular weights of NTD and CTD are 14 kDa and 12.5 kDa, respectively) (Gatesy et al. 2001; Garb et al. 2010). Each repeat contains 30–60 amino acid residues with specific amino acid motifs. The consensus motifs in the repetitive domain mainly consist of polyalanine (number of alanine residues = 4–12) and glycine-rich sequences (GGX- or GPGXX-). The β-sheet structure originating from the polyalanine region is the key motif responsible for the tensile strength of dragline silk (Keten et al. 2010). The glycine-rich region contains the GGX motif in MaSp1 and the GPGXX motif in MaSp2, which adopts a PPII helix and type II β-turn structure, respectively (Simmons et al. 1994; Kümmerlen et al. 1996; van Beek et al. 2002; Jenkins et al. 2010; Jenkins et al. 2012), and is believed to confer elasticity to silk fibers. The higher proline content in MaSp2 suggests that this protein is more elastic than MaSp1 (Rising et al. 2005). In the storage sac, spidroin is stored at high concentrations (up to 50%) (Hijirida et al. 1996) and converted into insoluble spider silks after experiencing pH and ion gradients across the major ampullate gland (Knight and Vollrath 2001; Andersson et al. 2014). Despite the predominant size of the repetitive domain, the self-assembly of spider silks is regulated by the small and highly conserved terminal domains (Askarieh et al. 2010; Hagn et al. 2010). At neutral pH and high chaotropic salt concentrations, similar to the conditions found in the storage sac, the CTD forms a folded dimer. In contrast, at acidic pH and low chaotropic salt concentration, which resembles the condition at the spinning duct, the CTD becomes unstructured and triggers spider silk self-assembly (Hagn et al. 2010; Andersson et al. 2014). Previous studies indicated that folded dimer CTD is required to maintain the solubility of spidroin, while the unfolding of CTD was believed to act as a precursor for β-sheet formation, suggesting the essential role of CTD as a molecular switch for storage and spider silk self-assembly (Hagn et al. 2010; Kronqvist et al. 2014).
Compared to the MaSp NTD from different species, which exhibits the most conserved region in silk proteins (Motriuk-Smith et al. 2005), the amino acid sequences of the CTD from different species are more varied (Fig. 1a). Nevertheless, the cysteine residue in the CTD is conserved across spider silk types and species and facilitates dimer formation (Fig. 1a). Furthermore, the CTD also contains a conserved salt bridge that is formed by the arginine side chain in helix 2 and the glutamic acid side chain in helix 4 (Fig. 1a). This salt bridge is pH sensitive and functions to stabilize the folded CTD by forming intramolecular interactions between helix 2 and helix 4 (Strickland et al. 2018). At acidic pH, the salt bridge will be disrupted, which leads to unfolding of the CTD (Hagn et al. 2010).
The dimeric form of the MaSps CTD from different species is facilitated by disulfide bonds. a Sequence alignment of the MaSps CTDs from different species. LH-M2 Latrodectus hesperus MaSp2 CTD; LH-M1 Latrodectus hesperus MaSp1 CTD; ADF-3 Araneus diadematus 3 CTD; TC-M2 Triconephila clavipes MaSp2 CTD; AV-M1 Argiope ventricosus MaSp1 CTD. The cysteine residue is conserved on those sequences (indicated by black arrow), and the arginine residue (indicated by blue arrow) and glutamic acid residue (indicated by red arrow) are also conserved and hypothesized to form salt bridges, as mentioned previously (Strickland et al. 2018). b SDS page profile of pure recombinant (13C, 15N) L. hesperus MaSp2 CTD in monomeric form (M) and dimeric form (D). The monomeric form of L. hesperus MaSp2 CTD was obtained by adding β-mercaptoethanol to the protein solution since it can reduce disulfide bonds. The molecular weights of the monomeric form and dimeric form of L. hesperus CTD are 12.2 and 24.4 kDa, respectively
In this study, we present 1H, 13C, and 15N backbone and side chain chemical shift assignments and the dynamics of soluble CTD MaSp2 from L. hesperus at pH 7 in the presence of 300 mM NaCl in dimeric form (Fig. 1b) at 25 °C. The backbone chemical shift assignment of CTD MaSp2 was also translated into a secondary structure, demonstrating that the 5 helix regions were connected by a more flexible linker.
Method and experiments
Recombinant protein expression and purification
The spidroin CTD MaSp2 used in the study was based on sequences from Latrodectus hesperus (UniProt accession code A6YIY0). Constructs encoding the WT sequences were purchased from GenScript as NdeI/XhoI inserts in the pET15b vector (Novagen). The amino acid sequence used for the L. hesperus MaSp2 CTD is shown in Fig. 2a.
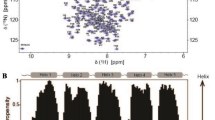
NMR assignment of L. hesperus MaSp2 CTD at pH 7 in the presence 300 mM NaCl (a). Amino acid sequences of L. hespereus CTD MaSp2 used in this construct. b 2D 1H-15N HSQC of backbone L. hesperus MaSp2 CTD signals at pH 7 in the presence 300 mM NaCl. The signals originating from unfolded populations are indicated in dashed box and glycine signal which is potentially originating from proline cis–trans isomerization is also indicated (G74*)
Doubly labeled (13C, 15N) L. hesperus CTD MaSp2 was expressed in E. coli BL21 (DE3). Cells were grown initially in 5 mL Luria Bertani (LB) medium supplemented with 100 µg/mL ampicillin sodium at 37 °C with overnight shaking at 160 rpm. The LB culture was then transferred into 100 mL of M9 minimal medium containing 2 g/L 13C-glucose and 1 g/L 15N-ammonium chloride supplemented with ampicillin. Cells were grown overnight at 37 °C with shaking at 160 rpm. This preculture was transferred into the main culture of M9 minimal medium containing 100 μg/mL ampicillin. The cells were grown until they reached OD600 ∼ 1. Then, protein expression was induced by adding 0.4 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). The cell cultures were continuously incubated overnight at 20 °C with shaking at 160 rpm.
Cells were harvested by centrifugation, resuspended in 50 mM Tris–Cl, 10% glycerol and 0.1% Triton X-100 (pH 7.5) and stored at − 80 °C until further use. Frozen cell slurry was thawed at 37 °C and enzymatically lysed by adding hen egg lysozyme at 50 mg per 1 L culture (Wako) supplemented with 250 U of TurboNuclease (Accelagen) and complete EDTA-free protease inhibitor cocktail (Roche). After incubation at 4 °C with stirring for 4 h, the lysates were centrifuged at 8000 rpm at 4 °C for 30 min. The clear supernatant fraction was loaded onto a 5-mL HisTrap column and washed extensively with a solution containing 20 mM Tris–HCl, 20 mM imidazole, and 500 mM NaCl (pH 7.5). The His-tagged CTD MaSp2 was eluted in solution containing 20 mM Tris–HCl, 500 mM imidazole, and 500 mM NaCl (pH 7.5). The His-tag was removed by overnight digestion with thrombin at 4 °C. For NMR measurements, the buffer was exchanged into 10 mM phosphate buffer pH 7 in the presence of 300 mM NaCl. Protein concentration was determined by measuring absorbance at 280 nm using a Nanodrop instrument (Thermo Fischer Scientific). The purity of the protein was evaluated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 1b). The NMR sample contained ~ 0.8–1 mM of dimeric form of (13C, 15N) L. hesperus MaSp2 CTD in 10 mM phosphate buffer pH 7 in the presence of 300 mM NaCl supplemented with 10% D2O and 0.1 mM DSS.
NMR experiments
Sequential backbone chemical shifts of L. hesperus MaSp2 CTD at pH 7 were assigned based on several 2D and 3D NMR experiments, including 2D 1H–15N HSQC, 2D 1H–13C HSQC aliphatic, 3D HNCO/3D HN(CA)CO (Ikura et al. 1990; Yamazaki et al. 1994), 3D CBCA(CO)NH/3D HNCACB (Wittekind and Mueller 1993), 3D HNCA/3D HN(CO)CA (Bax and Ikura 1991), and 3D HBHA(CO)NH (Grzesiek and Bax 1993). Assignment of proton and carbon aliphatic and side chain chemical shifts was accomplished by assigning 3D H(CCO)NH and 3D (H)C(CO)NH TOCSY spectra (Logan et al. 1993). The side chain carboxyl group of aspartic and glutamic acids as well as the side chain carbonyl group of asparagine and glutamine chemical shift of L. hesperus MaSp2 CTD at pH 7 were assigned based on the 2D H2(C)CO spectrum (Oda et al. 1994; Oktaviani et al. 2011). The 2D correlation between the Nε and Hδ of the arginine chemical shift was assigned based on the 2D H2(C)N spectrum (André et al. 2007). All NMR data were recorded at 25 °C (298 K) using a triple resonance TCI cryogenic probe and a z-axis gradient coil with a 700 MHz Bruker spectrometer. All spectra were processed using NMRPipe (Delaglio et al. 1995) and analyzed using NMRFAM-SPARKY (Lee et al. 2015). Sodium trimethylsilylpropanesulfonate (DSS) was used as a reference standard for all NMR signals based on IUPAC recommendations (Markley et al. 1998). The structural propensities of the L. hesperus MaSp2 CTD at pH 7 and 25 °C were determined using the neighbor-corrected structural propensity calculator (ncSPC) (Tamiola and Mulder 2012).
To characterize the protein backbone dynamics on the picosecond-nanosecond time scale, {1H}-15N heteronuclear NOE experiments were measured using 700 MHz NMR machines (Bruker) by recording the following spectra: an initial spectrum recorded without the initial proton saturation and a second spectrum recorded with initial proton saturation (3 s). The steady-state NOE values were determined based on the ratios of the average intensities of the peaks with and without proton saturation. The standard deviations of the NOE values were calculated from the background noise level using the following formula:
where Isat and Iunsat are the measured intensities of the peaks in the presence and absence of proton saturation, respectively. The noise in the background regions of the spectra, which were measured with initial proton saturation and without initial proton saturation, are indicated by σIsat and σIunsat, respectively.
Extent of assignment and data deposition
The 2D 1H-15N HSQC spectrum (Fig. 2b) demonstrates a wide dispersion of L. hesperus CTD MaSp2 at pH 7 on the amide proton domain, suggesting that this protein is well folded. The amide proton and nitrogen chemical shifts from the first 3 amino acids (GSH), which originated from the pET 15b vector, as well as the amide proton and nitrogen resonances of G7, S17, S18, and V19, which are part of the flexible N-terminus linker of the CTD, could not be assigned, possibly due to rapid exchange of amide protons with water. Furthermore, the amide proton and nitrogen resonances of T52, S76, S77 and G107 could not be assigned. As the structure of CTD responses to the change of pH and becomes fully unfolded at acidic pH (Hagn et al. 2010; Andersson et al. 2014), interestingly, a few visible signals originating from unfolded population of CTD (shown in dashed box) are observable, even at neutral pH (Fig. 2b). In addition, the low intensity of glycine signal which is potentially originating from proline cis–trans isomerization (G74*) is also noticeable (Fig. 2b). In total, the backbone 1HN, 15NH, 13C’, 13Cα, 13Cβ, and 1Hα chemical shift assignments were approximately 90% complete, and the completeness of the backbone chemical shift assignments of the structured region (residues 20–125) was approximately 93%. The assignment of aliphatic carbon and proton chemical shifts in the side chain based on H(CCO)NH and (H)C(CO)NH experiments was approximately 79% complete. The Cβ chemical shift of C78 was found at 44.148 ppm, suggesting that the cysteine was oxidized to form a dimer via a disulfide bond. Carboxyl and carbonyl side chain chemical shifts of Asp, Glu, Gln and Asn were achieved via the assignment of the 2D H2(C)CO spectrum, which gives the correlation between Hγ-Cδ chemical shifts for Glu and Gln and Hβ-Cγ chemical shifts for Asp and Asn. Interestingly, compared to previously reported structures on the CTD (PDB accession code 2KHM) (Hagn et al. 2010), which demonstrated two intramolecular salt bridges (R43–D93 and R52–E101), the L. hesperus MaSp2 CTD has only 1 arginine (R38), which is located in helix 2. The side chain of Nε of R38, which is located at helix 2, was assigned at 87.5 ppm, suggesting that this arginine is protonated at this pH (Platzer et al. 2014). Potentially, the R38 side chain from helix 2 forms an intramolecular salt bridge with the E87 side chain from helix 4, since these two amino acids are conserved throughout many different species and silk types and hypothesized to form a salt bridge by a previous study (Strickland et al. 2018). In agreement, the carboxyl side chain Cδ of E87 was assigned in the upfield region (177.138 ppm), suggesting that this glutamic acid forms a salt bridge. These data are similar to our recent finding on Triconephila clavipes MaSps NTD, where the carboxyl side chain chemical shift of D40 was shifted to the upfield region (approximately 175 ppm) upon salt bridge formation with its lysine counterparts (Oktaviani et al. 2023).
Translation of backbone chemical shifts (15NH, 13C’, 13Cα, 13Cβ, 1Hα) into secondary structure using neighbor-corrected structural propensity (ncSPC) (Tamiola and Mulder 2012) demonstrated that the L. hesperus MaSp2 CTD at pH 7 contains 5 helix regions (Fig. 3a). Our data also showed that helix 1 and helix 2 are kinked (Fig. 3a) due to the presence of proline, similar to the previously reported structure of the CTD of Araneus diadematus fibroin 3 (ADF-3) (PDB ID: 2KHM) (Hagn et al. 2010). Based on {1H}-15N heteronuclear data, those five helix regions were rigid, and they were connected by a more flexible linker (Fig. 3b). Here, our NMR data suggests that the CTD structures are conserved across the silk type and species, despite the amino acid sequences being relatively divergent (less than 50% sequence similarities). This study is relevant for understanding the conservation of CTD structure and mechanism that leads to the formation of stable spider dragline silks.
Secondary structure and dynamics of L. hesperus MaSp2 CTD in 10 mM phosphate buffer pH 7.0 and 300 mM NaCl. a The secondary structure of the L. hesperus MaSp2 CTD at pH 7 demonstrated that it has five helical regions connected by a linker. b {1 H}-15N heteronuclear NOE measurements for CTD MaSp2. High NOE values (> 0.5) indicate a relatively rigid structure, while low NOE values indicate a more flexible conformation
Data availability
The 1H, 13C, and 15N chemical shift assignments of the dimeric form of the L. hesperus MaSp2 CTD at pH 7 in the presence of 300 mM NaCl and 25 °C have been deposited in the Biological Magnetic Resonance Data Bank (BMRB) with accession code 51991.
References
Andersson M, Chen G, Otikovs M et al (2014) Carbonic anhydrase generates CO2 and H+ that drive spider silk formation via opposite effects on the terminal domains. PLOS Biol 12:e1001921. https://doi.org/10.1371/journal.pbio.1001921
André I, Linse S, Mulder FAA (2007) Residue-specific pKa determination of lysine and arginine side chains by indirect 15N and 13C NMR spectroscopy: application to apo calmodulin. J Am Chem Soc 129:15805–15813. https://doi.org/10.1021/ja0721824
Askarieh G, Hedhammar M, Nordling K et al (2010) Self-assembly of spider silk proteins is controlled by a pH-sensitive relay. Nature 465:236–238. https://doi.org/10.1038/nature08962
Bax A, Ikura M (1991) An efficient 3D NMR technique for correlating the proton and 15N backbone amide resonances with the alpha-carbon of the preceding residue in uniformly 15N/13C enriched proteins. J Biomol NMR 1:99–104. https://doi.org/10.1007/BF01874573
Chaw RC, Correa-Garhwal SM, Clarke TH et al (2015) Proteomic evidence for components of spider silk synthesis from black widow silk glands and fibers. J Proteome Res 14:4223–4231. https://doi.org/10.1021/acs.jproteome.5b00353
Delaglio F, Grzesiek S, Vuister GW et al (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293. https://doi.org/10.1007/BF00197809
Garb JE, Ayoub NA, Hayashi CY (2010) Untangling spider silk evolution with spidroin terminal domains. BMC Evol Biol 10:243. https://doi.org/10.1186/1471-2148-10-243
Gatesy J, Hayashi C, Motriuk D et al (2001) Extreme diversity, conservation, and convergence of spider silk fibroin sequences. Science 291:2603–2605. https://doi.org/10.1126/science.1057561
Grzesiek S, Bax A (1993) Amino acid type determination in the sequential assignment procedure of uniformly 13C/15N-enriched proteins. J Biomol NMR 3:185–204. https://doi.org/10.1007/BF00178261
Guerette PA, Ginzinger DG, Weber BH, Gosline JM (1996) Silk properties determined by gland-specific expression of a spider fibroin gene family. Science 272:112–115. https://doi.org/10.1126/science.272.5258.112
Hagn F, Eisoldt L, Hardy JG et al (2010) A conserved spider silk domain acts as a molecular switch that controls fibre assembly. Nature 465:239–242. https://doi.org/10.1038/nature08936
Heim M, Keerl D, Scheibel T (2009) Spider silk: from soluble protein to extraordinary fiber. Angew Chem Int Ed 48:3584–3596. https://doi.org/10.1002/anie.200803341
Hennecke K, Redeker J, Kuhbier JW et al (2013) Bundles of spider silk, braided into sutures, resist basic cyclic tests: potential use for flexor tendon repair. PLoS ONE 8:e61100. https://doi.org/10.1371/journal.pone.0061100
Hijirida DH, Do KG, Michal C et al (1996) 13C NMR of Nephila clavipes major ampullate silk gland. Biophys J 71:3442–3447. https://doi.org/10.1016/S0006-3495(96)79539-5
Ikura M, Kay LE, Bax A (1990) A novel approach for sequential assignment of proton, carbon-13, and nitrogen-15 spectra of larger proteins: heteronuclear triple-resonance three-dimensional NMR spectroscopy. Application to Calmodulin. Biochemistry 29:4659–4667. https://doi.org/10.1021/bi00471a022
Jenkins JE, Creager MS, Butler EB et al (2010) Solid-state NMR evidence for elastin-like β-turn structure in spider dragline silk. Chem Commun 46:6714–6716. https://doi.org/10.1039/C0CC00829J
Jenkins EJ, Holland PG, Yarger LJ (2012) High resolution magic angle spinning NMR investigation of silk protein structure within major ampullate glands of orb weaving spiders. Soft Matter 8:1947–1954. https://doi.org/10.1039/C2SM06462F
Keten S, Xu Z, Ihle B, Buehler MJ (2010) Nanoconfinement controls stiffness, strength and mechanical toughness of β-sheet crystals in silk. Nat Mater 9:359–367. https://doi.org/10.1038/nmat2704
Knight DP, Vollrath F (2001) Changes in element composition along the spinning duct in a Nephila spider. Naturwissenschaften 88:179–182. https://doi.org/10.1007/s001140100220
Kono N, Nakamura H, Ohtoshi R et al (2019) Orb-weaving spider Araneus ventricosus genome elucidates the spidroin gene catalogue. Sci Rep 9:8380. https://doi.org/10.1038/s41598-019-44775-2
Kono N, Nakamura H, Mori M et al (2021) Multicomponent nature underlies the extraordinary mechanical properties of spider dragline silk. PNAS. https://doi.org/10.1073/pnas.2107065118
Kronqvist N, Otikovs M, Chmyrov V et al (2014) Sequential pH-driven dimerization and stabilization of the N-terminal domain enables rapid spider silk formation. Nat Commun 5:3254. https://doi.org/10.1038/ncomms4254
Kümmerlen J, van Beek JD, Vollrath F, Meier BH (1996) Local structure in spider dragline silk investigated by two-dimensional spin-diffusion nuclear magnetic resonance. Macromolecules 29:2920–2928. https://doi.org/10.1021/ma951098i
Lammel A, Schwab M, Hofer M et al (2011) Recombinant spider silk particles as drug delivery vehicles. Biomaterials 32:2233–2240. https://doi.org/10.1016/j.biomaterials.2010.11.060
Larracas C, Hekman R, Dyrness S et al (2016) Comprehensive proteomic analysis of spider dragline silk from black widows: a recipe to build synthetic silk fibers. Int J Mol Sci 17:1537. https://doi.org/10.3390/ijms17091537
Lee W, Tonelli M, Markley JL (2015) NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31:1325–1327. https://doi.org/10.1093/bioinformatics/btu830
Logan TM, Olejniczak ET, Xu RX, Fesik SW (1993) A general method for assigning NMR spectra of denatured proteins using 3D HC(CO)NH-TOCSY triple resonance experiments. J Biomol NMR 3:225–231. https://doi.org/10.1007/BF00178264
Malay AD, Craig HC, Chen J et al (2022) Complexity of spider dragline silk. Biomacromol 23:1827–1840. https://doi.org/10.1021/acs.biomac.1c01682
Markley JL, Bax A, Arata Y et al (1998) Recommendations for the presentation of NMR structures of proteins and nucleic acids. Eur J Biochem 256:1–15. https://doi.org/10.1046/j.1432-1327.1998.2560001.x
Motriuk-Smith D, Smith A, Hayashi CY, Lewis RV (2005) Analysis of the conserved N-terminal domains in major ampullate spider silk proteins. Biomacromol 6:3152–3159. https://doi.org/10.1021/bm050472b
Oda Y, Yamazaki T, Nagayama K et al (1994) Individual ionization constants of all the carboxyl groups in ribonuclease HI from Escherichia coli determined by NMR. Biochemistry 33:5275–5284. https://doi.org/10.1021/bi00183a034
Oktaviani NA, Otten R, Dijkstra K et al (2011) 100% complete assignment of non-labile (1)H, (13)C, and (15)N signals for calcium-loaded Calbindin D(9k) P43G. Biomol NMR Assign 5:79–84. https://doi.org/10.1007/s12104-010-9272-3
Oktaviani NA, Malay AD, Matsugami A et al (2023) Unusual pKa values mediate the self-assembly of spider dragline silk proteins. Biomacromol 24:1604–1616. https://doi.org/10.1021/acs.biomac.2c01344
Pham T, Chuang T, Lin A et al (2014) Dragline silk: a fiber assembled with low-molecular-weight cysteine-rich proteins. Biomacromol 15:4073–4081. https://doi.org/10.1021/bm5011239
Platzer G, Okon M, McIntosh LP (2014) pH-dependent random coil 1H, 13C, and 15N chemical shifts of the ionizable amino acids: a guide for protein pKa measurements. J Biomol NMR 60:109–129. https://doi.org/10.1007/s10858-014-9862-y
Rising A, Nimmervoll H, Grip S et al (2005) Spider silk proteins—mechanical property and gene sequence. Zool Sci 22:273–281. https://doi.org/10.2108/zsj.22.273
Simmons A, Ray E, Jelinski LW (1994) Solid-State 13C NMR of Nephila clavipes dragline silk establishes structure and identity of crystalline regions. Macromolecules 27:5235–5237. https://doi.org/10.1021/ma00096a060
Strickland M, Tudorica V, Řezáč M et al (2018) Conservation of a pH-sensitive structure in the C-terminal region of spider silk extends across the entire silk gene family. Heredity 120:574–580. https://doi.org/10.1038/s41437-018-0050-9
Tamiola K, Mulder FAA (2012) Using NMR chemical shifts to calculate the propensity for structural order and disorder in proteins. Biochem Soc Trans 40:1014–1020. https://doi.org/10.1042/BST20120171
van Beek JD, Hess S, Vollrath F, Meier BH (2002) The molecular structure of spider dragline silk: folding and orientation of the protein backbone. Proc Natl Acad Sci 99:10266–10271. https://doi.org/10.1073/pnas.152162299
Wendt H, Hillmer A, Reimers K et al (2011) Artificial skin—culturing of different skin cell lines for generating an artificial skin substitute on cross-weaved spider silk fibres. PLoS ONE 6:e21833. https://doi.org/10.1371/journal.pone.0021833
Wittekind M, Mueller L (1993) HNCACB, a high-sensitivity 3D NMR experiment to correlate amide-proton and nitrogen resonances with the alpha- and beta-carbon resonances in proteins. J Magn Reson, Ser B 101:201–205. https://doi.org/10.1006/jmrb.1993.1033
Yamazaki T, Lee W, Arrowsmith CH et al (1994) A suite of triple resonance NMR experiments for the backbone assignment of 15N, 13C, 2H labeled proteins with high sensitivity. J Am Chem Soc 116:11655–11666. https://doi.org/10.1021/ja00105a005
Acknowledgements
Not applicable
Funding
This work was supported by Naito foundation subsidy for female researchers (to N.A.O), Japan Society for Promotion of Science (JSPS) Kakenhi for early career scientists (to N.A.O) and SPDR fellowship (to N.A.O) and Grants-in-Aid from the Japan Science and Technology Agency Exploratory Research for Advanced Technology (JST-ERATO; Grant No. JPMJER1602 to K.N.), COI-NEXT (to K.N.), and MEXT Data Creation and Utilization-type MaTerial R&D project (to K.N.).
Author information
Authors and Affiliations
Contributions
NAO performed the NMR experiments, analyzed the data and wrote the main manuscript; MG prepared the NMR samples; TN and FH performed the NMR experiments; All authors wrote and reviewed the manuscript; Resources: NAO and KN.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethical approval
Not applicable.
Consent for publication
All authors have agreed to the publication of the manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Oktaviani, N.A., Malay, A.D., Goto, M. et al. NMR assignment and dynamics of the dimeric form of soluble C-terminal domain major ampullate spidroin 2 from Latrodectus hesperus. Biomol NMR Assign 17, 249–255 (2023). https://doi.org/10.1007/s12104-023-10150-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12104-023-10150-6