Abstract
Purpose
A substantial amount of evidence demonstrates suggests that long non-coding RNAs (lncRNAs) play a key role in the progression of various malignancies, cervical squamous cell carcinoma (CSCC) included. In our study, we deeply investigated the role and molecular mechanism of lncRNA NPHS2-6 in CSCC.
Methods
The expression level of gene and protein expression were measured by qRT-PCR and western blot. To test the cell proliferation and cell metastasis ability, we carried out the CCK-8 experiment, clone formation assay, transwell assay and wound healing, respectively. The interactivity among NPHS2-6, miR-1323 and SMC1B were co demonstrated using the bioinformatics tool, dual-luciferase reporter system, and RNA pulldown assay. The subcutaneous tumor model of nude mice was established to verify the results of previous studies at the in vivo. NPHS2-6 was upregulated in CSCC tissues and cells.
Results
NPHS2-6 deficiency significantly inhibited CSCC cell growth and EMT in vitro. In addition, NPHS2-6 deficiency also inhibited the growth of CSCC xenograft tumors in mice in vivo. Importantly, NPHS2-6 was a competing endogenous RNA (ceRNA) to increases SMC1B levels by binding to miR-1323, leading to activate the PI3K/Akt pathway, thereby exacerbating tumorigenesis of CSCC.
Conclusions
In conclusion, NPHS2-6/miR-1323/SMC1B/PI3K/Akt signaling accelerates the progression of CSCC, providing a new direction for the treatment strategy of CSCC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cervical cancer is a high incidence of human cancer [1]. Over the past few years, the application of the Human papillomavirus vaccine (HPV) has led to a decline in cervical cancer rates [2], however, cervical cancer remains one of the most common malignancies in women worldwide, and its incidence ranks second among female malignant tumors [3]. Cervical squamous cell carcinoma (CSCC) is the most prevalent subtype of cervical cancer, which accounts for about 90% of all cases [4]. Previous studies have revealed that most cases of CSCC are caused by HPV infection, however, HPV infection alone is not enough to cause the occurrence of tumors [5]. The occurrence and development of tumor is a multi-stage and multi-step process, and gene mutations and transcription abnormalities play an important role in this process [6].In addition, corresponding changes in the cells themselves can eventually cause the occurrence of tumors [6]. Therefore, elucidating the pathogenesis of CSCC and finding effective therapeutic strategies are the key to the treatment of CSCC.
Epithelial-mesenchymal transformation (EMT) is considered to be a key process in tumor metastasis [7]. EMTs are characterized by high levels of mesenchymal markers (vimentin and N-cadherin) and low levels of key epithelial markers (E-cadherin), which are considered to be markers of cell migration and invasion [8, 9]. Studies have shown that cancer epithelial cells are invasive and metastatic and can affect tumor progression through EMT [10].
Long non-coding RNAs (lncRNAs) are a group of RNAs with a length of more than 200 nucleotides, which have a variety of regulatory effects such as epigenetic, transcription or post-transcriptional regulation [11]. The abnormal expression of lncRNA has been confirmed to be closely related to various biological processes such as cell growth, apoptosis, metastasis and epithelial-mesenchymal transformation [12, 13]. Previous studies have revealed that lncRNAs are involved in the occurrence and progression of various tumors, including CSCC [14, 15]. For example, MAGI2-AS3 [16], ANCR [17] and PART1 [18] have been unveiled to be involved in regulating the biological behavior of CSCC cells. However, few studies have investigated the role of NPHS2-6 in the pathogenesis of CSCC. In our study, we used lncRNA microarray analysis to identify elevated lncRNA in CSCC tissue samples (Supplementary Table 1). Our clinical data suggested that level of NPHS2-6 in CSCC tissue was significantly increased compared with matched normal tissues. Therefore, we hypothesized that NPHS2-6 might be involved in regulating the progression of CSCC. However, the role and molecular mechanism of NPHS2-6 in regulating CSCC progression remains unclear.
Growing evidence suggests that lncRNAs and miRNAs act as competing endogenous RNA (ceRNAs) to inhibit each other by acting as a miRNA sponge, forming an accurate regulatory network, therefore regulating the expression of miRNA-target gene and ultimately affect the development of cancer [19]. For instance, DARS-AS1 was identified as an oncogene of cervical cancer and activates the Notch pathway through the miR-628-5p/JAG1 axis to accelerate the progression of cervical cancer [20]. MiRNAs are a class of non-coding single-stranded small molecule RNA with a length of 18–24 nucleotides [21]. MiRNAs bind to the 3'-untranslated region (3'-UTR) of mRNAs and play an important role in the post-transcriptional regulation of gene expression [22]. Furthermore, some miRNAs are believed to regulate important DNA methylator factors [23]. Among the major mechanisms involved in tumorigenesis, DNA methylation is an important epigenetic modification impacting both genomic stability and gene expression [24]. However, the role and molecular mechanisms of miR-1323 in CSCC remain uncertain.
Based on the above, our detection results revealed that miR-1323 was underexpressed in CSCC cells. Furthermore, according to the analysis of RNAhybrid, we found that there are binding relationships between NPHS2-6 and miR-1323. Therefore, we hypothesized that miR-1323 is involved in the ceRNA regulatory network as a cancer-inhibitor of CSCC. Moreover, previous studies have revealed that lncRNA can regulate the activity of PI3K/AKT pathway via regulating its receptors and ligands [25]. In this study, we found that NPHS2-6 can regulate the PI3K/AKT pathway. However, whether NPHS2-6 regulates the expression of receptors and ligands in the PI3K/AKT pathway in CSCC remains unclear.
In a nutshell, this study aimed to investigate the regulation of NPHS2-6 on proliferation, apoptosis and metastasis of CSCC cells. Data from this study confirmed that NPHS2-6 activates the PI3K-Akt pathway through miR-1323/SMC1B axis, thereby promoting the progression of CSCC, suggesting that NPHS2-6 may be a promising therapeutic target for CSCC.
Materials and methods
Clinical tissues
We collected 3 normal tissues and 13 pairs of metastatic and non-metastatic CSCC tissues from Affiliated Tumor Hospital of Xinjiang Medical University Hospital. None of the patients received preoperative chemoradiotherapy. The study protocol was approved by Affiliated Tumor Hospital of Xinjiang Medical University Hospital and received written consent from each participant.
Animal experiment
A total of ten (five each group) Balb/c nude mice (Model Animal Research Center of Nanjing University; Nanjing, China), were kept under suitable conditions of temperature and humidity, and provided with adequate food and water during this period. To conduct tumorigenicity experiments in mice, the stable C33A cells knocking down NPHS2-6 or control were injected subcutaneously into the right side of axillary of the mice [26]. Use the following formula to calculate: tumor volume (V) = (short dimension)2 × (long dimension)/2. After 28 days of experiment, the mice were sacrificed for tumor isolation, tumor size and weight were measured and photographed with digital camera. All animal experiments in this study were approved through the Ethics Committee of Affiliated Tumor Hospital of Xinjiang Medical University Hospital. All experimental procedures were in compliance with the ARRIVE (Animal Research: Reporting of In Vivo Experiments) and National Institutes of Health (NIH) guidelines for animal welfare.
Cell culture
We purchased the human CSCC cell lines (SiHa and C33A) and human normal cervical epithelial cell line (H8) from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The cells were cultured in DMEM containing with 10% fetal bovine serum (FBS; Gibco, United States) and 1% penicillin and streptomycin (Gibco, United States) at 37 °C in a humidified incubator containing 5% CO2.
Cell transfection
All the call transfections were performed by Lipofectamine 2000 reagent (Invitrogen, United States), including sh-NC, sh-NPHS2-6#1, sh-NPHS2-6#2; Vector; SMC1B OE; sh- NPHS2-6#1 + vector; sh-NPHS2-6#1 + SMC1B OE; mimic NC; miR-1323 mimic; miR-1323 mimic + vector; miR-1323 mimic + SMC1B OE; sh-NPHS2-6#1 + inhibitor NC; sh-NPHS2-6#1 + miR-1323 inhibitor; sh-NPHS2-6#1 + SMC1B OE, SMC1B OE; sh-SMC1B (GenePharma, Shanghai, China), according to the manufacturer’s protocol.
RNA extraction and qRT-PCR
We use TRIZOL reagent (Thermofisher, United States) to extract total RNA from cells and tissues. Reverse transcription of RNA into cDNA was performed using a reverse transcription kit (TaKaRa, Tokyo, Japan). Gene expression was detected using fluorescence quantitative PCR instrument (Analytick Jena A G, Germany). The reaction was conducted under the guidance of a fluorescent quantitative RT-PCR kit (SYBR Green, Bio-RAD, USA). The reaction conditions were described as: 30 s denaturation at 94 °C, followed by 35 cycles of denaturation at 95 °C for 10 s, annealing at 58 °C for 20 s and extension at 72 °C for 1 min. The primer sequences used were as follows: lncRNA NPHS2-6-F: 5′-GCGAAGACGGGAGGAAAGA-3′; lncRNA NPHS2-6-R: 5′-AGTGCAGGGTCCGAGGTATT-3′; lncRNA METTL4-2-F: 5′-TTTCTGCGTCTTCACCCT-3′; lncRNA METTL4-2-R: 5′-GTCATTTCCCATCACTTT-3′; lncRNA EEF2KMT-1-F: 5′-GCCTTCCGTTACTCCAAT-3′; lncRNA EEF2KMT-1-R: 5′-AGGGTTCTTCACCAGCAT-3′; lncRNA MIR9-3HG-F: 5′-CAGATGTTCGGTCCCCACTC-3′; lncRNA hsa_MIR9-3HG-R: 5′-TCGGCCTCCTTTGCTTAGAC-3′; SMC1B -F: 5′-AACCCACCATTACGAGTC-3′; SMC1B-R: 5′-GATTACATTTGAAACATCCC-3′; miR-1323-F: 5′-TATGCTGCGAAATGATGG-3′; miR-1323-R: 5′-TATGCTGCGAAATGATGG-3′;
U6-F: 5′-GCTTCGGCAGCACATATACTAA-3′; U6-R: 5′-AACGCTTCACGAATTTGCGT-3′; GAPDH-F: 5′-GAAGGTGAAGGTCGGAGTC-3′; GAPDH-R: 5′-GAAGATGGTGATGGGATTTC-3′; U6 was used as the internal control. All data were analyzed by adopting 2−ΔΔCT method.
Western blot analysis
The protein levels were detected using western blot analysis [27]. The protein samples were isolated from the cells and electrophoresed. The concentration of protein in the supernatant was examined by the BCA protein Kit (Beyotime Biotechnology, China). Approximately 50 μg of protein was firstly placed on 10% SDS-PAGE, and then shifted it to a PVDF membrane. Then the protein was incubated with the following primary antibodies at 4 °C for 16 h: Vimentin, E-Cadherin, N-Cadherin andFN1 (1:1500, SBI, USA), PI3K, P-PI3K, AKT and P-AKT (1:1500, Abcam, USA), SMC1B (1:2000, SBI, USA), GAPDH (1:1500, Santa Cruz Biotechnology, CA, USA). Washed with 1 × TBST for 3 × 5 min, then add corresponding secondary antibodies, incubate at room temperature for 1 h, and wash with 1 × TBST for 3 × 5 min. GAPDH acts as a loading control. The protein band was quantified with Image J Software (1.47 V, NIH, USA).
FISH assay
FISH assay was performed as described previously [28]. The C33A cells were placed on culture slides, subsequently fixed with 4% formaldehyde (Sigma Aldrich). The cells were then dehydrated and cultured overnight with NPHS2-6-FISH probe (Ribobio) hybridization buffer. The slides were washed with a detergent buffer containing citrate. The cell staining was performed using DAPI and then observed under a fluorescence microscope (Olympus, Tokyo, Japan).
Cell proliferation assay
The cell viability and proliferation was detected by the cell counting kit-8 (CCK-8) assay (Beyotime, Shanghai, China). In brief, C33A cells were seeded in a 96-well plate (2 × 104 cells/well), followed by adding CCK8 reagent (10 μL). After 4-h incubation, the absorbent was inspected at 450 nm wavelength with an automatic microplate reader (MolecμLar Devices, Shanghai, China).
Colony formation assay
Firstly, the C33A cells were trypsinized and centrifuged at 1500 rpm for 5 min. Then, the cells (104 per plate) were seeded on plates. After two weeks, cells were washed and fixed with methanol for 15 min, followed by staining with 0.2% crystal violet for 30 min. Colony number was manually counted and photographed.
Transwell assay
Transwell chamber (24-well chamber with an 8-µm pore) was used to evaluate the migration ability of C33A cells. The cells (1 × 104 cells/well) were seeded in the upper chamber using serum-free medium. Subsequently, add medium containing 10% FBS to the lower chamber. After being incubated overnight, we fixed the cells with methanol and stained using crystal violet. Finally, the number of migrated cells was counted under a microscope (Nikon).
Wound healing assay
CSCC cells were inoculated in 6-well plates and cultured for 24 h until the degree of fusion reached 95%. The linear wound was created by scratching cells using a micropipette tip with the same force. Then, the medium was then immediately replaced. Images were taken at 0 h and 48 h with inverted microscope (Motic Asia, Hong Kong, China).
Subcellular fractionation location
The cytoplasm and nucleus of C33A cells were separated using the PARIS Kit (Invitrogen) according to the manufacturer’s instruction. The relative expressions of GAPDH, U6 and NPHS2-6 in the nucleus and cytoplasm of SiHa cells were detected by RT-qPCR. GAPDH / U6 acted as the cytoplasmic control or the nuclear control.
HE staining
CSCC tissue specimens were collected and fixed with 4% paraformaldehyde. Following embedding by paraffin. Sections (5 μm) were stained with haematoxylin–eosin staining and dehydrated again (Olympus, Tokyo, Japan). Staining was performed according to HE staining kit, and the myocardial injury was observed under 400 times microscope (Philips, Zurich, Switzerland) for examination.
Dual-luciferase reporter assay
The pMIR-NPHS2-6-WT (or pMIR-SMC1B‑3′‑UTR‑WT) and pMIR-NPHS2-6‑MUT (or pMIR-SMC1B‑3′‑UTR‑MUT) luciferase reporter plasmids were constructed by Nanjing Synthgene Biotech. The sequences that could binding to miR-1323 were partly mutated and then inserted into the reporter plasmid to identify binding specificity. Briefly, CSCC cells were seeded in a 24-well plate until the confluence rate reached 60%. According to the instructions, using Lipofectamine 2000 (Thermofisher, USA), each well was co-transfected with a luciferase reporter plasmid (0.5 μg) and an RNA mimic (100 pmol). After 48 h transfection, the luciferase activity was examined using Dual-Luciferase Reporter Assay System (Promega).
Immunofluorescence
The C33A cells were first fixed in paraformaldehyde and then 0.1% Triton X-217100 was added to penetrate the cells. Subsequently, after 5% BSA blockade, the primary antibody against Vimentin was added at 4 °C overnight and secondary antibody was cultured in PBS for 1 h. The cells were then rinsed, followed by counterstaining with DAPI (Sigma-Aldrich). All cells were then observedby confocal microscope (Olympus, Tokyo, Japan).
Statistical analyses
All of the data were presented as mean ± SD using SPSS17.0 (SPSS, Inc.). All experiments were repeated three times. Significant differences between groups were analyzed by the student’s t test, and were assigned at *P < 0.05; **P < 0.01; ***P < 0.001, respectively.
Results
NPHS2-6 was upregulated in CSCC tissues and cells
To explore the pathogenesis of CSCC, we selected three CSCC tissues and matched normal tissues for lncRNAs sequencing. According to the sequencing results, NPHS2-6 was upregulated the most in tumor tissues (Supplementary Table 1). Then, we detected the expression levels of NPHS2-6 in 3 normal tissues and 10 tumor tissues (5 non-metastatic and 5 metastatic) by qRT-PCR, and found that the expression level of NPHS2-6 was significantly high in CSCC tissues (Fig. 1A). In addition, qRT-PCR results revealed that the expression level of NPHS2-6 was significantly increased in CSCC metastatic tissues (Fig. 1B). Consistently, the expression of NPHS2-6 in CSCC cell lines (SiHa and C33A) was significantly higher than in normal cervical epithelial cells (H8) (Fig. 1C). In addition, subcellular fractionation and FISH assay revealed that NPHS2-6 was mainly distributed in the cytoplasm of CSCC cells (Fig. 1D). These results suggested that NPHS2-6 may be related to the occurrence and metastasis of CSCC.
NPHS2-6 was upregulated in CSCC tissues and cells. A The expression of NPHS2-6 was detected by qRT-PCR in CSCC tissues (10) and normal tissues (3). B The RT-qPCR detected NPHS2-6 expression in CSCC metastatic tissues (13) and non-metastatic tissues (13). C The expression level of NPHS2-6 was detected by qRT-PCR in CSCC cell lines (SiHa and C33A) and normal cervical epithelial cells (H8). D The subcellular localization of NPHS2-6 in CSCC cells were detected by subcellular fractionation and FISH assay. Data were presented as the mean ± SD. n = 3. *p < 0.05; **p < 0.01; ***p < 0.001
Low expression of NPHS2-6 inhibits CSCC progression
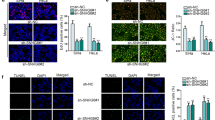
To further reveal the potential effect of NPHS2-6 on CSCC cells, we studied the biological functions of NPHS2-6 in CSCC by functional loss experiment. The knockdown efficiency of NPHS2-6 was analyzed by RT-qPCR, and the outcome revealed that the expression level of NPHS2-6 was significantly decreased in sh-LNC#1/#2 transfected cells, especially sh-LNC#1 (Fig. 2A). The proliferation curves determined by the CCK-8 assays showed that NPHS2-6 deficiency significantly attenuated cells growth compared to the sh-NC (Fig. 2B). Furthermore, colony formation assay found that NPHS2-6 deficiency impaired clonogenesis ability of cells compared to the sh-NC (Fig. 2C). Further transwell assay and wound healing assay revealed that knockdown of NPHS2-6 can attenuated the migration and invasion of cells (Fig. 2D, E). Furthermore, knockdown of NPHS2-6 induced upregulation of E-cadherin and downregulation of Vimentin, N-cadherin, FN1, suggesting that NPHS2-6 silencing can inhibit the metastasis of CSCC cells (Fig. 2F). Similarly, immunofluorescence detected decreased expression of Vimentin, which further confirmed that silenced NPHS2-6 had an inhibitory effect on EMT process of CSCC cells (Fig. 2G). All these results elucidated that downregulation of NPHS2-6 inhibits CSCC progression in vitro.
Low expression of NPHS2-6 inhibits CSCC progression. A The expression levels of NPHS2-6 were examined using qRT-PCR after transfected with sh-NC, Sh-NPHS2-6#1 and Sh-NPHS2-6#2 in C33Acells. B The cells viability was detected withCCK-8 kit and C colony formation assay when treated with the same conditions as A. D Cells migration was detected by transwell assay. Magnification, × 100, Bar scale = 20 μm. E Wound healing assay showing cells invasion. Magnification, × 10, Bar scale = 200 μm. F The expression of characteristic molecules (E-Cadherin, N-Cadherin, Vimentin and FN1) of EMT in C33A cells were detected by western blot. G The expression of Vimentin was detected by IF in C33A cells transfect with sh-NC, sh-NPHS2-6#1 and sh-NPHS2-6#2. Magnification, × 100, Bar scale = 20 μm.*p < 0.05; **p < 0.01; ***p < 0.001. Data are shown as mean ± SD (n = 3)
Low expression of NPHS2-6 was confirmed to inhibit CSCC progression in vivo
To further confirmed the effect of NPHS2-6on the CSCC progression in vivo. We firstly knockdown of NPHS2-6 in CSCC cells and analyzed its transfection efficiency using RT-qPCR (Fig. 3A). Subsequently, we subcutaneously injected CSCC cells into mice. The data showed that compared with control xenografts, the knockdown of NPHS2-6 slowed tumor growth (Fig. 3B, C). HE staining at the site of liver metastasis also suggested that the tumor mass was looser in the knockdown of NPHS2-6 group, compared to those of control xenografts (Fig. 3D). To further explore the effect of NPHS2-6 on EMT of CSCC in vivo. Western blot was used to detect the expression of EMT characteristic molecules in C33A cells, and it was found that the knockdown of NPHS2-6 reduced the protein levels of Vimentin, N-cadherin and FN1, and increased the protein levels of E-cadherin (Fig. 3E). Altogether, these data demonstrated that depletion of NPHS2-6 inhibits CSCC progression in vivo.
Low expression of NPHS2-6 was confirmed to inhibit CSCC progression in vivo. A The expression levels of NPHS2-6 were examined using qRT-PCR. B Photograph of subcutaneous tumor (n = 5 each group). C The volume of CSCC subcutaneous tumors. D The H&E staining of mice liver in sh-NPHS2-6#1 group and sh-NC group. Magnification, × 100, Bar scale = 200 μm. E The expression of characteristic molecules (N-Cadherin, E-Cadherin, Vimentin and FN1) of EMT in subcutaneous tumor were detected by western blot. *p < 0.05; **p < 0.01; ***p < 0.001. The results were shown as mean ± SD; n = 3
NPHS2-6 regulates the process of tumor metastasis by regulating SMC1B
To further reveal the molecular mechanism of NPHS2-6 regulating CSCC cells, we first found that the potential target gene of NPHS2-6 was SMC1B through co-expression spectrum sequencing (Supplementary Table 2). Follow-up RT-qPCR analysis showed that SMC1B expression was significantly upregulated in CSCC tissues compared with normal tissues (Fig. 4A). Furthermore, western blot analysis showed that downregulation of NPHS2-6could significantly reduce the protein expression level ofSMC1B, while upregulation of SMC1 reversed the inhibitory effect of downregulation of NPHS2-6 on SMC1B (Fig. 4B). The analysis of CCK8 assay, Transwell, the colony formation, and wound healing revealed that NPHS2-6 silencing significantly reduced the cell growth, proliferation, migration and invasion. Further rescue experiments showed that upregulation of SMC1Bcould effectively reverse the inhibitory effect of NPHS2-6silencing on cell growth, proliferation, migration and invasion (Fig. 4C–F). Furthermore, NPHS2-6 deficiency induced an upregulation of E-cadherin and a downregulation of Vimentin, N-cadherin, FN1, while overexpression of SMC1B suppressed the effect of NPHS2-6on EMT characteristic molecules (Fig. 4G). In conclusion, these results suggest that NPHS2-6 can regulate the progression of CSCC cells via regulating the expression of SMC1B.
NPHS2-6 regulates the progression of tumor metastasis by regulating SMC1B. A The qPCR analysis of SMC1B expression in CSCC tissues (10) and normal tissues (3). B WB analysis of SMC1B expression in C33A cells transfected with Sh-NC, Sh-NPHS2-6#1, Sh-NPHS2-6#1 + vector or Sh-NPHS2-6#1 + SMC1B OE. C The proliferation of C33A cells was detected with CCK-8 kit. D Colony formation assays showing cell viability. E Transwell assay showing cells migration. Magnification, × 100, Bar scale = 20 μm. F Wound healing assay showing cells invasion. Magnification, × 10, Bar scale = 200 μm. G The expression of characteristic molecules (E-Cadherin, N-Cadherin, Vimentin and FN1) of EMT in C33A cells were detected by western blot.*p < 0.05, **p < 0.01, ***p < 0.001, n = 3 per group. The results were shown as mean ± SD
miR-1323 regulates the process of tumor metastasis by regulating SMC1B
According to the above research, found that NPHS2-6 and SMC1B were both significantly and highly expressed in CSCC tissues. Previous reports have confirmed that regulatory networks composed of lncRNAs and miRNAs play a crucial role in the occurrence and development of cancer [29, 30]. Therefore, we predicted the downstream binding miRNA of NPHS2-6 and the upstream miRNA that regulates SMC1B via starBase (http://starbase.sysu.edu.cn/), and the results revealed that miR-1323 is the target genes of NPHS2-6 and SMC1B.To further determine whether SMC1B is the target gene of miR-1323, we predict their binding sites based on TargetScan database (Fig. 5A). Then dual-luciferase reporter assay was used to verify the target binding relationship (Fig. 5B). Moreover, the protein levels of SMC1B in CSCC cell lines (SiHa and C33A) were significantly reduced compared with normal cervical epithelial cell lines (H8), especially in C33A (Fig. 5C). Subsequently, western blot analysis revealed that upregulation ofmiR-1323 significantly reduced the protein level of SMC1B, however, overexpression of SMC1B effectively reversed the inhibitory effect of overexpression of miR-1323 on SMC1B, while downregulation of miR-1323 and/or SMC1Bdid the opposite, suggesting that miR-1323could regulate the protein level of SMC1B, and was negatively correlated with the expression level of SMC1B (Fig. 5D). The analysis of CCK8 assay, transwell and wound healing showed that overexpression of miR-1323 significantly reduced the cell growth, migration and invasion, while overexpression of SMC1B effectively reversed the inhibitory effect of NPHS2-6deficiencyon cell growth, proliferation, migration and invasion, and knockdown of miR-1323 and/or SMC1B was the opposite (Fig. 5E–G). Overall, the abovementioned results demonstrated that miR-1323 can regulate the metastasis process of cancer by regulating SMC1B level.
miR-1323 regulates the progression of tumor metastasis via regulating SMC1B. A 3′-UTR base pairing diagram of miR-1323 and SMC1B. B The target relationship between miR-1323 and SMC1B was verified by dual-luciferase reporter assay. C RT-qPCR detected miR-1323 expression in H8, SiHa and C33A. D The protein expression of SMC1B in C33A cells by Western blot when transfected with mimic NC/inhibitor NC, miR-1323 mimic/miR-1323 inhibitor, miR-1323 mimic + vector/miR-1323 inhibitor + vector and miR-1323 mimic + SMC1B OE/ miR-1323 inhibitor + sh-SMC1B. E CCK-8 assay showing cells proliferation. F Cells migration was detected by transwell assay. Magnification, × 100, Bar scale = 20 μm. G Wound healing assay showing cells invasion. Magnification, × 10, Bar scale = 200 μm.*p < 0.05, **p < 0.01, ***p < 0.001, n = 3 per group. Data are mean ± SD
NPHS2-6can regulate the process of tumor metastasis by miR-1323/ SMC1B pathway
To determine whether miR-1323 can target NPHS2-6, we predict their binding sites based on RNAhybrid (Fig. 6A). Then dual-luciferase reporter assay was used to verify the target binding relationship (Fig. 6B). Subsequently, Western blot analysis showed that knockdown ofNPHS2-6 significantly reduced the protein level of SMC1B, however, downregulation ofmiR-1323 effectively reversed the inhibitory effect of NPHS2-6deficiencyon SMC1B, suggesting that NPHS2-6 regulates SMC1B through miR-1323 (Fig. 6C). The analysis of colony formation, transwell and wound healing showed that NPHS2-6deficiency significantly reduced the cell proliferation, migration and invasion, while knockdown ofmiR-1323 effectively reversed this effect (Fig. 6D–F). Furthermore, NPHS2-6 deficiency induced a downregulation of Vimentin, N-cadherin and FN1 and an upregulation of E-cadherin, while miR-1323deficiency suppressed the effect of NPHS2-6 on EMT characteristic molecules (Fig. 6G). Overall, these results indicated that NPHS2-6 regulates the growth and EMT of CSCC cells via miR-1323/SMC1B pathway.
NPHS2-6 regulates the progression of tumor metastasis by miR-1323/ SMC1B pathway. A The binding sites between NPHS2-6 and miR-1323 were predicted by RNAhybrid. B The target relationship between NPHS2-6 and miR-1323 was verified by dual-luciferase reporter assay. C Western blot was used to analyze the levels of SMC1B in C33A cells transfected with NC, Sh-NPHS2-6#1and Sh-NPHS2-6#1 + miR-1323 inhibitor. D Colony formation assay showing cells proliferation. E Transwell assay showing cells migration. Magnification, × 100, Bar scale = 20 μm. F Wound healing assay showing cells invasion. Magnification, × 10, Bar scale = 200 μm. G The expression of characteristic molecules (E-Cadherin, N-Cadherin, Vimentin and FN1) of EMT in C33A cells were detected by western blot.*p < 0.05; **p < 0.01; ***p < 0.001 comparing to NC. Data were shown as mean ± SD (n = 3)
SMC1B regulates EMT of CSCC cells via PI3K-AKT pathway
To further determine the molecular mechanism by which SMC1B regulates EMT in CSCC cells, we firstly constructed cell transfected with NC, SMC1B OE and sh-SMC1Btreatment. The results revealed that upregulation of SMC1Bsignificantly increased the protein levels of P-PI3K, P-Akt, Vimentin, N-cadherin and FN1, and reduced the level of E-cadherin, while knockdown of SMC1B was the opposite (Fig. 7A). Further transwell and wound healing assay test revealed that upregulation of SMC1B significantly increased the ability of cell migration and invasion, while downregulation of SMC1B was the opposite (Fig. 7B, C). Then we constructed cell transfected with sh-NC, sh-NPHS2-6 and sh-NPHS2-6 + SMC1B OE treatment to further investigate the effect of SMC1B on PI3K-Akt pathway. Western blot analysis revealed that NPHS2-6 silencing could significantly reduce the protein levels of P-PI3K and P-Akt, while overexpression of SMC1B could rescue the inhibitory effect of downregulation of NPHS2-6 (Fig. 7D). In summary, these data strong evidence that the SMC1B regulates EMT of CSCC cells via PI3K-AKT pathway.
SMC1B regulates EMT of CSCC cells via PI3K-AKT pathway. A Western blot analysis of P-PI3K, P-AKT, N-Cadherin, E-Cadherin, Vimentin and FN1 in C33A cells with different level of SMC1B. B The cells migration was detected by transwell assay in C33A cells with different level of SMC1B. Magnification, × 100, Bar scale = 20 μm. C The cells invasion was detected using wound healing assay in C33A cells with different level of SMC1B. Magnification, × 10, Bar scale = 200 μm. D Western blot detected P-PI3K or P-AKT expression in C33A cells transfected with sh-NC, sh-NPHS2-6#1 and sh-NPHS2-6#1 + SMC1B-OE.*p < 0.05, **p < 0.01, ***p < 0.001, n = 3 per group. Data are mean ± SD
Discussion
CSCC is a common tumor of female reproductive system, with high recurrence rate and metastasis rate,and its prognosis is very dismal [4]. Therefore, new biomarkers and therapeutic targets are urgently needed for CSCC. This study aims to elucidate the biological functions and possible molecular mechanisms of NPHS2-6 in the progression of CSCC.
Accumulating evidence indicates that LncRNAs are involved in the occurrence and progression of different cancers, including CSCC [31, 32]. Some lncRNAs have been reported to act as oncogenes or cancer suppressors in CSCC in earlier studies [33, 34]. For example, Cheng et al. found that lncRNA-BLACAT1 was abnormally overexpressed in CSCC cells, indicating poor prognosis of CSCC patients [32]. In our study, we found that NPHS2-6 was significantly upregulated in CSCC tissues and cells.
Moreover, we found that the depletion of NPHS2-6 significantly inhibited the migration, proliferation, invasion and EMT of CSCC cells. Similarly, in vivo experiments uncovered that knockdown of NPHS2-6 repressed the growth and metastasis of xenografts. In conclusion, NPHS2-6 played an oncogenic role in the progression of CSCC and could be developed as a new therapeutic target for CSCC.
Growing evidence has indicated that lncRNAs are mostly act as miRNAs sponges and form a lncRNA-miRNA-mRNA axis, thereby playing their biological role in gene regulation [35, 36]. For instance, lncRNA MAGI2-AS3 can affect the development and progression of cervical squamous cell carcinoma (CSCC) by the sponge miRNA-233/EPB41L3 axis [16]. Based on this, in this study, we explore the underlying ceRNA network of NPHS2-6. Initially, our results revealed that miR-1323 was significantly downregulated in CSCC cells. Subsequently, the dual-luciferase assay confirmed that the NPHS2-6 and SMC1B were verified to directly target miR-1323. Moreover, we observed NPHS2-6 negatively regulate the expression of miR-1323, and miR-1323 negatively regulates the expression of SMC1B. Importantly, functional assays confirmed that NPHS2-6 regulates the expression of SMC1B via sponging miR-1323, thereby regulating proliferation, metastasis, and EMT of CSCC cells. In current study, SMC1B was confirmed to be a target of miR-1323. In addition, the data from this study revealed that NPHS2-6 activated the PI3K/AKT signaling pathway of CSCC cells through positive regulation of SMC1B. Moreover, the results of the rescue experiment verified that the inhibitory effect of NPHS2-6 silencing on CSCC cell growth could be counteract by SMC1B overexpression. Further studies showed that SMC1B could regulate EMT of CSCC cells by activating the PI3K/Akt pathway. Taken together, NPHS2-6 accelerated the CSCC tumorigenesis by targeting the miR-1323/SMC1B/PI3K/AKT pathway.
In conclusion, our study explored the regulatory role of NPHS2-6 in the progression of CSCC. All data from this study illuminate that NPHS2-6 activates the PI3K/AKT signaling pathway through miR-1323/SMC1B pathway, thereby driving CSCC progression. Based on this, we believe that NPHS2-6 is expected to become a novel target for clinical treatment of CSCC.
Data availability
Not applicable.
References
Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–86.
Hildesheim A, Gonzalez P, Kreimer AR, Wacholder S, Schussler J, Rodriguez AC, et al. Impact of human papillomavirus (HPV) 16 and 18 vaccination on prevalent infections and rates of cervical lesions after excisional treatment. Am J Obstet Gynecol. 2016;215(2):212.e1-212.e15.
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34.
Groves IJ, Coleman N. Pathogenesis of human papillomavirus-associated mucosal disease. J Pathol. 2015;235(4):527–38.
Crosbie EJ, Einstein MH, Franceschi S, Kitchener HC. Human papillomavirus and cervical cancer. Lancet. 2013;382(9895):889–99.
Qin W, Dong P, Ma C, Mitchelson K, Deng T, Zhang L, et al. MicroRNA-133b is a key promoter of cervical carcinoma development through the activation of the ERK and AKT1 pathways. Oncogene. 2012;31(36):4067–75.
Brabletz T, Kalluri R, Nieto MA, Weinberg RA. EMT in cancer. Nat Rev Cancer. 2018;18(2):128–34.
Wrighton KH. Cell migration: EMT promotes contact inhibition of locomotion. Nat Rev Mol Cell Biol. 2015;16(9):518.
Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. J Clin Invest. 2009;119(6):1429–37.
Márton É, Lukács J, Penyige A, Janka E, Hegedüs L, Soltész B, et al. Circulating epithelial-mesenchymal transition-associated miRNAs are promising biomarkers in ovarian cancer. J Biotechnol. 2019;297:58–65.
Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteom Bioinform. 2016;14(1):42–54.
Wu Q, Xiang S, Ma J, Hui P, Wang T, Meng W, et al. Long non-coding RNA CASC15 regulates gastric cancer cell proliferation, migration and epithelial mesenchymal transition by targeting CDKN1A and ZEB1. Mol Oncol. 2018;12(6):799–813.
Huang Y, Xiang B, Liu Y, Wang Y, Kan H. LncRNA CDKN2B-AS1 promotes tumor growth and metastasis of human hepatocellular carcinoma by targeting let-7c-5p/NAP1L1 axis. Cancer Lett. 2018;437:56–66.
Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. 2014;1839(11):1097–109.
Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71(1):3–7.
Hou A, Zhang Y, Fan Y, Zheng Y, Zhou X, Liu H. LncRNA MAGI2-AS3 affects cell invasion and migration of cervical squamous cell carcinoma (CSCC) via sponging miRNA-233/EPB41L3 axis. Cancer Manag Res. 2020;12:4209–16.
Ta W, Zhang Y, Zhang S, Sun P. LncRNA ANCR downregulates hypoxia-inducible factor 1α and inhibits the growth of HPV-negative cervical squamous cell carcinoma under hypoxic conditions. Mol Med Rep. 2020;21(1):413–9.
Liu H, Zhu C, Xu Z, Wang J, Qian L, Zhou Q, et al. lncRNA PART1 and MIR17HG as ΔNp63α direct targets regulate tumor progression of cervical squamous cell carcinoma. Cancer Sci. 2020;111(11):4129–41.
Kartha RV, Subramanian S. Competing endogenous RNAs (ceRNAs): new entrants to the intricacies of gene regulation. Front Genet. 2014;5:8.
Chen Y, Wu Q, Lin J, Wei J. DARS-AS1 accelerates the proliferation of cervical cancer cells via miR-628-5p/JAG1 axis to activate Notch pathway. Cancer Cell Int. 2020;20(1):535.
Xu P, Li X, Liang Y, Bao Z, Zhang F, Gu L, et al. PmiRtarbase: a positive miRNA-target regulations database. Comput Biol Chem. 2022;98: 107690.
Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–79.
Zare M, Bastami M, Solali S, Alivand MR. Aberrant miRNA promoter methylation and EMT-involving miRNAs in breast cancer metastasis: diagnosis and therapeutic implications. J Cell Physiol. 2018;233(5):3729–44.
Jurisic V, Obradovic J, Nikolic N, Javorac J, Perin B, Milasin J. Analyses of P16(INK4a) gene promoter methylation relative to molecular, demographic and clinical parameters characteristics in non-small cell lung cancer patients: a pilot study. Mol Biol Rep. 2023;50(2):971–9.
Shi WJ, Liu H, Ge YF, Wu D, Tan YJ, Shen YC, et al. LINC00673 exerts oncogenic function in cervical cancer by negatively regulating miR-126-5p expression and activates PTEN/PI3K/AKT signaling pathway. Cytokine. 2020;136: 155286.
Li R, Huang D, Ju M, Chen HY, Luan C, Zhang JA, et al. The long non-coding RNA PVT1 promotes tumorigenesis of cutaneous squamous cell carcinoma via interaction with 4EBP1. Cell Death Discov. 2023;9(1):101.
Jurisic V, Srdic-Rajic T, Konjevic G, Bogdanovic G, Colic M. TNF-α induced apoptosis is accompanied with rapid CD30 and slower CD45 shedding from K-562 cells. J Membr Biol. 2011;239(3):115–22.
Liu Y, Shi M, He X, Cao Y, Liu P, Li F, et al. LncRNA-PACERR induces pro-tumour macrophages via interacting with miR-671-3p and m6A-reader IGF2BP2 in pancreatic ductal adenocarcinoma. J Hematol Oncol. 2022;15(1):52.
Liang WC, Fu WM, Wong CW, Wang Y, Wang WM, Hu GX, et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6(26):22513–25.
Wang L, Cho KB, Li Y, Tao G, Xie Z, Guo B. Long noncoding RNA (lncRNA)-mediated competing endogenous RNA networks provide novel potential biomarkers and therapeutic targets for colorectal cancer. Int J Mol Sci. 2019;20(22):5758.
Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–81.
Cheng H, Tian J, Wang C, Ren L, Wang N. LncRNA BLACAT1 is upregulated in cervical squamous cell carcinoma (CSCC) and predicts poor survival. Reprod Sci. 2020;27(2):585–91.
Dong J, Su M, Chang W, Zhang K, Wu S, Xu T. Long non-coding RNAs on the stage of cervical cancer. Oncol Rep. 2017;38(4):1923–31.
No authors listed. Expression of concern: LncRNA SRA1 is downregulated in HPV-negative cervical squamous cell carcinoma (CSCC) and regulates cancer cell behaviors. Biosci Rep. 2020;40(7):BSR-20191226_EOC.
Zhu J, Zhang X, Gao W, Hu H, Wang X, Hao D. lncRNA/circRNA-miRNA-mRNA ceRNA network in lumbar intervertebral disc degeneration. Mol Med Rep. 2019;20(4):3160–74.
Zhou RS, Zhang EX, Sun QF, Ye ZJ, Liu JW, Zhou DH, et al. Integrated analysis of lncRNA-miRNA-mRNA ceRNA network in squamous cell carcinoma of tongue. BMC Cancer. 2019;19(1):779.
Funding
This work is supported by the project of Science and Technology Department of Xinjiang Uygur Autonomous Region (No. 2020E02125 and No. 2020D01C204).
Author information
Authors and Affiliations
Contributions
FL and YW conceived and designed the study, and drafted the manuscript. MW, GA and JY collected, analyzed and interpreted the experimental data. TZ and GS revised the manuscript for important intellectual content. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The study was approved by Ethical Committee of Affiliated Tumor Hospital of Xinjiang Medical University and conducted in accordance with the ethical standards.
Informed consent
Subjects signed the informed consent.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, F., Wang, Y., Wen, M. et al. NPHS2-6 drives cervical squamous cell carcinoma (CSCC) progression via hsa-miR-1323/SMC1B axis to activate PI3K-Akt pathway. Clin Transl Oncol 26, 245–259 (2024). https://doi.org/10.1007/s12094-023-03248-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-023-03248-9