Abstract
Background
Hsa_circ_0001535 is involved in biological processes in various tumors. However, the biological effects and related mechanism of hsa_circ_0001535 in ovarian cancer (OC) is unclear. This work is aimed to probe the biological function and underlying mechanism of hsa_circ_0001535 in OC, especially sponged with mi-RNA, require further elucidation.
Methods
Hsa_circ_0001535 expression in OC tissues and cell lines were examined by qRT-PCR. Hsa_circ_0001535 overexpression model was constructed by lentivirus-mediated transfection in two OC cell lines, and the biological functions of hsa_circ_0001535 were evaluated by CCK-8, transwell assay and Western blot. Dual luciferase reporter gene assay was respectively used to explore the relationship between hsa_circ_0001535 and miR-593-3p, as well as miR-593-3p and PTEN. The expression of miR-593-3p and PTEN were detected by qRT-PCR in two OC cell lines and OC tissues.
Results
Hsa_circ_0001535 was down-regulated in OC tissues and cell lines. Hsa_circ_0001535 overexpression inhibited proliferation, migration and EMT marker expression in OC cells. Of interest, hsa_circ_0001535 targeted miR-593-3p and reduced its RNA level in OC cells. PTEN was a target gene of miR-593-3p, which was up-regulated by inhibiting miR-593-3p in OC cells. Furthermore, miR-593-3p mimic treatment reversed the up-regulation of PTEN by hsa_circ_0001535 overexpression in OC cells.
Conclusions
The above results showed that hsa_circ_0001535 acted as a molecular sponge for miR-593-3p to repress miR-593-3p expression, and promoted the expression of PTEN, thus inhibited proliferation and migration of OC cells. Our research provides a potential therapeutic target for ovarian cancer patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ovarian cancer is among the five leading causes of cancer death for female in 2021, and about 21,410 women were first diagnosed with ovarian cancer, along with around 13,770 ovarian cancer-related deaths [1]. It is reported that the development of ovarian cancer is associated with proliferation, migration and invasion of OC cells [2]. Epithelial-mesenchymal transformation (EMT) process is an early event of tumor invasion and metastasis [3], which is usually accompanied by up-regulation of mesenchymal phenotypic molecules such as N-cadherin and Slug [4]. Due to the insufficient early diagnosis, the poor prognosis, accompanied by high metastasis and recurrence, the majority of ovarian cancers are diagnosed at stages III/IV when the 5-year survival rate is 40% [5, 6]. Therefore, further elucidating the related molecular mechanisms of OC and searching for new therapeutic targets for the treatment of OC is essential.
Circular RNAs (circRNAs) are a class of highly conserved, stable and abundant noncoding RNAs (ncRNAs) [7]. Recent studies have shown that circRNAs participate in OC progression by regulating various processes, including cell proliferation, migration, invasion and apoptosis [8]. In the study of Li Ning et al. [9], they performed high-throughput sequencing of circular RNAs in pairs of ovarian cancer tumor tissues and normal ovarian tissues and found that a total of 4388 circular RNAs (2556 up-regulated and 1832 down regulated) were differentially expressed in the EOC specimens compared with the normal ovarian tissues. At the same time, circBNC2, circEXOC6B, hsa_circ_0001535, circN4BP2L2 and circRHOBTB3 were significantly decreased in EOC specimens, whereas circCELSR1 was significantly increased. However, further studies are needed to find related mechanism of hsa_circ_0001535 in OC.
Studies have found that tumor progression may be regulated by the circRNA/miRNA axis [10]. MiRNAs are short, noncoding RNAs of 19–25 nucleotides which can regulate gene expression, tumor initiation and progression [11]. MiR-593 has been observed closely involved in several kinds of human tumors, such as hepatocellular carcinoma [12], non-small cell lung cancer [13], glioblastoma multiforme [14] and breast cancer [15]. However, the exact role of miR-593 in OC development, especially the relation with hsa_circ_0001535 still is not fully understood.
Phosphatase and tensin homolog (PTEN) resides on chromosome 10q23-24. Mutations in PTEN have been detected in a wide range of cancers and are regarded as a critical step in the development of human cancers [16]. It is reported that loss of PTEN function can occur by mutations or deletions, epigenetic silencing, transcriptional repression or by micro RNA (miRNA) regulation [17]. In Ovarian Cancer cells, Wu X [18]et al. revealed that circ_0007444 expression was declined in OC and inhibited the progression of OC via mediating the miR-570-3p/PTEN Axis.
In this study, we confirmed that hsa_circ_0001535 acts as a tumor suppressor to inhibit proliferation and migration in OC cells. On the basis of previous studies and online analysis of bioinformatics, hsa_circ_0001535 combined with miR-593-3p, which interacted with and inhibited PTEN expression. Thus, hsa_circ_0001535 acts as a tumor suppressor in ovarian cancer by sponging miR-593-3p, upregulating PTEN expression. Our research provides a potential therapeutic target for ovarian cancer patients.
Materials and methods
Patients and tissue samples
A total of 74 pairs of tumor tissues and adjacent normal tissues were collected from ovarian cancer patients who underwent surgical resection without previously radiotherapy or chemotherapy. All patients were identified as ovarian cancer for the first time from October 2016 to April 2021 in Second Affiliated Hospital of Nantong University, and all patients have obtained informed consent. The clinical features of patients were recorded, including age, FIGO stage, tumor grade, distant metastasis and tumor size (shown in Table 1).
Cell cultures
Normal ovarian surface epithelial cell line (IOSE80) and 6 human ovarian cancer cell lines (SKOV3, HO8910, A2780, CAOV3, OVCAR4, OVCAR3), were purchased from the institute of biovarian cancerhemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). IOSE80, A2780, CAOV3, OVCAR4 were cultured in Dulbecco's modified eagle medium (DMEM) containing 10% fetal bovine serum (FBS) and 100 mg/ml penicillin–streptomycin. SKOV3, HO8910, OVCAR3 were cultured in Roswell Park Memorial Institute-1640 (RPMI-1640) medium containing 10% FBS and 100 mg/ml penicillin–streptomycin. All cells were incubated at 37℃ in 5% CO2 in a humidified incubator.
Cell transfections
SKOV3 and HO8910 cells were transfected with the hsa_circ_0001535-overexpression or negative control lentivirus using transfectmate (Genechem) according to the manufacturer’s instructions. MiR-593-3p mimics, control mimics, miR-593-3p inhibitor and control inhibitor were synthesized by Genechem (China).
Quantitative real-time polymerase chain reaction analysis (qRT-PCR)
Total RNA was extracted from tissues and cells using TRIzol Regent (Invitrogen, Carlsbad, CA, USA). Reverse transcription into complementary DNA (cDNA) was executed using the Revertaid First Strand cDNA Synthesis Kit (Invitrogen, thermoscientific, Lithuania). QRT-PCR was performed with 2X Universal SYBR Green Fast qPCR Mix (ABclonal, Wuhan, China) according to the manufacturer’s instruction. 18S, U6 and GAPDH were used as the internal references for hsa_circ_0001535, miR-593-3p, PTEN, respectively. The relative expression was calculated using the 2 − ΔΔCt method. The specific primer sequences are displayed in Table 2.
Cell counting Kit-8(CCK-8) experiment
Cells were seeded into 96 well plates at a concentration of 1.5 × 104 cells/mL. Forty-eight hours later, cells were added with 10ul of CCK-8 solution and incubate for 2 h. The absorbance was measured at 450 nm by a microplate reader (BioTek, USA) at 0 h, 24 h, 48 h and 72 h.
Transwell experiment
The SKOV3 and HO8910 cells migration ability was examined by Transwell experiment with 8 μm pore filter membrane containing upper chamber according to the manufacturer’s instructions. The upper chamber was added with serum-free medium containing 5 − 10 × 105 cells, while the lower chamber was added with medium containing 20% fatal bovine serum (FBS). After incubating the transwell-system for 12–24 h, cells passing through the filter membrane were fixed, stained and the results were obtained by Nikon microscope (Japan).
Western blot
Samples were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene fluoride (PVDF) membrane. The membranes were blocked with 5% nonfat milk and incubated with specific primary antibodies overnight at 4℃, followed by incubation with horseradish peroxidase-conjugated secondary antibodies at room temperature (RT) for 2 h. Afterwards, protein bands were obtained with a chemiluminescence system (Bio-Rad, VT, USA).
Dual-luciferase reporter assay
Online bioinformatics tools were used to predict the binding site fragments of miR-593-3p with hsa_circ_0001535 and PTEN, respectively. Luciferase reporter assay was then performed to research the interaction between two genes. According to the binding site, the hsa_circ_0001535-wildtype (WT) and -mutant type (Mut) fragments, as well as PTEN-WT and PTEN-Mut fragments, were designed and synthesized by Genechem (Shanghai, China). According to the instructions, these fragments were loaded onto the luciferase reporter vectors. SKOV3 cells were transfected with miR-593-3p mimics (miR-593-3p mimic group) and mimics control respectively (miR-NC group). Then these cells were cotransfected with the luciferase reporter vectors containing the four type fragments. After 48 h incubation at 37 °C, 5% CO2, the luciferase activity was investigated by the Double-Luciferase Reporter assay system (Promega, Madison, WI, USA).
Immunohistochemistry
After antigen retrieval and inhibition of endogenous peroxidase, tissue sections were incubated with anti-PTEN (Proteintech) or anti-Ki-67(EMD Millipore Corporation) at 4℃ overnight. After washed with PBS, the sections were incubated with HRP-conjugated secondary antibodies. The antigen–antibody reaction was visualized with 3, 3’-diaminobenzidine (DAB) and hematoxylin was used to stain the nucleus. The results were obtained by Nikon microscope.
Antibodies and reagents
The antibodies used in this study for immunoblot were as follows:
Rabbit polyclonal anti-PTEN (1:2000), Rabbit polyclonal anti-Slug (1:1000) and Mouse monoclonal anti-N-cadheirn (1:2000), Mouse polyclonal anti-β-Actin (1:5000) were purchased from Proteintech (USA).
Statistical analysis
All experiments were performed in triplicate. The data were analyzed using SPSS version 25.0 software and GraphPad Prism 7. The chi-square test was used to compare the distribution of categorical variables between groups. The differences between groups were analyzed by one-way analysis of variance (ANOVA) and t-test. Correlations were measured by Pearson correlation analysis. Values are presented as means ± SD, P < 0.05 was considered statistically significant.
Results
Hsa_circ_0001535 was down-regulated in ovarian cancer tissues and cells
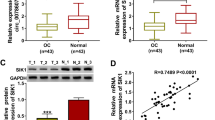
To explore the role of hsa_circ_0001535 in ovarian cancer, we collected 74 ovarian cancer patients’ tumor tissues and adjacent normal tissues, and detected the expression of hsa_circ_0001535 by qRT-PCR. The data demonstrated that hsa_circ_0001535 expression was down-regulated in ovarian cancer tissues relative to adjacent normal tissues (Fig. 1A), which was consistent with previous reports [9]. In addition, hsa_circ_0001535 expression was remarkably decreased in all six ovarian cancer cell lines relative to IOSE80 cells, especially SKOV3 and HO8910 cells (Fig. 1B). Furthermore, we analyzed the correlation between hsa_circ_0001535 expression and clinical features of OC patients. As shown in Table 1, hsa_circ_0001535 expression in cancer tissue of human ovarian cancer was negatively correlated with FIGO stage, tumor grade and tumor size. These findings implied that hsa_circ_0001535 expression was down-modulated in OC and may work as a suppressor factor.
Hsa_circ_0001535 inhibited the proliferation and migration of ovarian cancer cells. A qRT-PCR was employed to detect hsa_circ_0001535 expression in 74 OC tissues and paired adjacent tissues. B The expression of hsa_circ_0001535 in OC cells (SKOV3, HO8910, A2780, CAOV3, OVCAR4, OVCAR3) and IOSE80 cells were analyzed by qRT-PCR. C The transfection efficiency of hsa_circ_0001535 overexpression was detected by qRT-PCR in SKOV3 and HO8910 cells. D CCK-8 experiments showed that hsa_circ_0001535 overexpression inhibited cell proliferation of SKOV3 and HO8910 cells. E–F Cell migration was evaluated in OC cells overexpressed hsa_circ_0001535 via transwell assay. G–H The EMT markers (N-cad, and Slug) expression was detected by western blot in OC cells overexpressed hsa_circ_0001535. The data are means ± SD (n = 3). *P < 0.05 compared with the control group
Hsa_circ_0001535 inhibited cell proliferation and migration of ovarian cancer cells
To further determine the function of hsa_circ_0001535 in ovarian cancer, the SKOV3 and HO8910 cells were transfected with hsa_circ_0001535 overexpression lentiviral vector, and the transfection efficiency was detected by RT-qPCR (Fig. 1C). The data of CCK-8 experiments showed that hsa_circ_0001535 overexpression inhibited cell proliferation of SKOV3 and HO8910 cells (Fig. 1D). At the same time, cell migration were tested by transwell assay, and the results showed that hsa_circ_0001535 overexpression significantly inhibited cell migration of SKOV3 and HO8910 cells compared with oe-NC transfection (Fig. 1E-F). Consistently, western blot analysis discovered that oe-hsa_circ_0001535 inhibited the expression of EMT markers (N-cadherin, and Slug) compared with the oe-NC in SKOV3 and HO8910 cells (Fig. 1G and Fig. 1H). The above data indicated that the hsa_circ_0001535 overexpression repressed the proliferation and migration of ovarian cancer cells.
Hsa_circ_0001535 served as a sponge for miR-593-3p and had a negative correlation with miR-593-3p
To explore the mechanism of hsa_circ_0001535 regulating biological behaviors of ovarian cancer cells, the miRNA target for hsa_circ_0001535 was investigated. We performed bioinformatics analysis using Circinteractome database, and found that hsa_circ_0001535 had a putative binding site of miR-593-3p (Fig. 2A). The data of the dual-luciferase reporter gene experiment unveiled that hsa_circ_0001535 sequence contained the miR-593-3p binding sites, and miR-593-3p mimics remarkably repressed the luciferase activity of wild-type hsa_circ_0001535 reporter, while there was no remarkable effect on the luciferase activity of mutant-type hsa_circ_0001535 reporter (Fig. 2B). Markedly lower miR-593-3p expression was observed in SKOV3 and HO8910 cells of oe- hsa_circ_0001535 group when compared with oe-NC group (Fig. 2C). Then, we detected the miR-593-3p expression in ovarian cancer tissues, and the data of qRT-PCR showed that the expression of miR-593-3p was higher in ovarian cancer tissues than in adjacent normal tissues (Fig. 2D). Pearson correlation analysis indicated that the expression of hsa_circ_0001535 and miR-593-3p was negatively correlated in tumor tissues of ovarian cancer patients (Fig. 2E). These data suggested that hsa_circ_0001535 served as a molecular sponge for miR-593-3p, and hsa_circ_0001535 could repress the expression of miR-593-3p.
Hsa_circ_0001535 served as a sponge for miR-593-3p and had a negative correlation with miR-593-3p. A hsa_circ_0001535 had a putative binding site of miR-593-3p by bioinformatics analysis. B The dual-luciferase reporter gene experiment unveiled that hsa_circ_0001535 sequence contained the miR-593-3p binding sites. C MiR-593-3p expression was detected by qRT-PCR in SKOV3 and HO8910 cells which overexpressed hsa_circ_0001535. D qRT-PCR was employed to detect miR-593-3p expression in 74 OC tissues and paired adjacent tissues. E Pearson correlation analysis was carried out to assess the expression association betweenhsa_circ_0001535 and miR-593-3p in OC tissues. The data are means ± SD (n = 3). *P < 0.05 compared with the control group
PTEN was down-regulated by miR-593-3p in ovarian cancer cells
Previous study identified miRNAs as upstream mediators of PTEN can dually induce/inhibit PTEN signaling in affecting the malignant behavior of lung and breast cancer cells [19]. We performed bioinformatics analysis using targetscan database, and found that PTEN was a target of miR-593-3p, and the binding site of PTEN-WT or -Mut for miR-593-3p was exhibited in Fig. 3A. The luciferase activity of wild type PTEN reporter was decreased in miR-593-3p mimic group. However, the luciferase activity of mutant PTEN reporter remained unchanged after transfection of miR-593-3p mimic (Fig. 3B). Furthermore, we found that markedly higher PTEN expression was observed in SKOV3 and HO8910 cells of miR-593-3p inhibitor group when compared with miR-NC group (Fig. 3C). Additionally, both qRT-PCR and Immunohistochemistry signified that PTEN was remarkably down-regulated in ovarian cancer tissues relative to adjacent normal tissues (Fig. 3D and Fig. 3E). It was showed that the expression of miR-593-3p and PTEN was negatively correlated in OC tissues by Pearson correlation analysis (Fig. 3F). Altogether, we deduced that miR-593-3p directly combined with PTEN and inhibited its expression in OC cells.
PTEN was down-regulated by miR-593-3p in ovarian cancer cells. A The binding site of PTEN-WT or -Mut for miR-593-3p was exhibited. B The regulation of miR-593-3p on PTEN was detected by dual-luciferase reporter gene assay. C PTEN expression was observed in SKOV3 and HO8910 cells with or without miR-593-3p inhibitor treatment. D qRT-PCR was employed to detect PTEN expression in 74 OC tissues and paired adjacent tissues. E PTEN and Ki67 expression were detected by Immunohistochemistry in 74 OC tissues and paired adjacent tissues. F Pearson correlation analysis was carried out to assess the expression association between miR-593-3p and PTEN in OC tissues. The data are means ± SD (n = 3). *P < 0.05 compared with the control group
The regulatory effect of hsa_circ_0001535 on OC is dependent on miR-593-3p / PTEN axis
Pearson correlation analysis indicated that the expression of hsa_circ_0001535 and PTEN was positively correlated in tumor tissues of ovarian cancer patients (Fig. 4A). Furthermore, qRT-PCR and Western blot experiments were used to detect the expression of PTEN in SKOV3 and HO8910 cells. Ours results showed that the PTEN expression in oe-hsa_circ_0001535 group was higher than oe-NC group (Fig. 4B-D). To explore whether the effect of hsa_circ_0001535 on PTEN by regulating miR-593-3p in OC cells, OC cells were treated with miR-593-3p mimics after overexpressed hsa_circ_0001535, and our results discovered miR-593-3p mimic treatment reversed the up-regulation of PTEN by hsa_circ_0001535 overexpression in OC cells(Fig. 4E-F). Moreover, CCK-8 showed that hsa_circ_0001535 suppressed proliferation in OC cells, but its suppressing effect was attenuated by treatment with miR-593-3p mimic (Fig. 4G). Similarly, hsa_circ_0001535 inhibited migration by down-regulating miR-593-3p expression in OC cells (Fig. 4H-I). Thus, we concluded the regulatory effect of hsa_circ_0001535 on OC cells was dependent on miR-593-3p / PTEN axis.
The regulatory effect of hsa_circ_0001535 on OC is dependent on miR-593-3p/PTEN axis. A Pearson correlation analysis was carried out to assess the expression association between hsa_circ_0001535 and PTEN in OC tissues. B PTEN expression was detected by qRT-PCR in SKOV3 and HO8910 cells transfected with oe-NC or oe-hsa_circ_0001535. C–D Western blot was used to tested PTEN protein expression in SKOV3 and HO8910 cells transfected with oe-NC or oe-hsa_circ_0001535. SKOV3 cells were transfected with oe-hsa_circ_0001535, oe-hsa_circ_0001535 + miR-NC, oe-hsa_circ_0001535 + miR-593-3p mimic. E–F PTEN expression in OC cells was detected by Western blot. G The proliferation of OC cells was detected by CCK8. H–I The migration of OC cells was detected by transwell assay. The data are means ± SD (n = 3). *P < 0.05 compared with the control group
Discussion
Ovarian cancer is one of the major gynecologic cancers and common cause of gynecologic cancer death worldwide [20]. Research showed that hsa_circ_0001535 was significantly down-regulated in the EOC specimens and lower hsa_circ_0001535 was associated with FIGO stage [9], which is consistent with our results (Fig. 1A and Table 1). On the basis of this, we confirmed that hsa_circ_0001535 also remarkably decreased in six ovarian cancer cell lines, especially SKOV3 and HO8910 cells (Fig. 1B), and hsa_circ_0001535 overexpression repressed the proliferation and migration of ovarian cancer cells (Fig. 1C-H). Nevertheless, it is reported that hsa_circ_0001535 was the most significantly differentially expressed circRNA in HCC tissue and promoted the proliferation of HCC in vitro and in vivo [21]. Furthermore, Low expression of hsa_circ_0001535 was observed in Bladder cancer (BCa), and it was positively associated with lower tumor stage and better prognosis among patients with BCa. CCK8 assays and cloning formation assays revealed that hsa_circ_0001535 inhibited the proliferation rates of T24 and UMUC3 cells, and the function of CD8 + T cells was promoted by hsa_circ_0001535, and it could attenuate the glycolysis of BCa cells and reverse the acidic tumor microenvironment [22]. The expression and function of hsa_circ_0001535 in various tumors are different. Therefore, we will enlarge the patients’ samples to explore the closely relationship between hsa_circ_0001535 and OC.
Recent studies have demonstrated circRNAs exert biological functions by acting as microRNA sponges [23]. For instance, hsa_circ_0001535, a sponge of miR-212, is involved in the regulation of E2F5 gene expression by competitively binding to miR-212, inhibits the activation of the P53 signaling pathway, and promotes the proliferation of HCC cells [21]. The miR-593-3p showed different and multiple functions via target different substrate protein in various tumors cells. It can target the FGFR3 to hinder the breast cancer cell malignant progression [24], target CCND2 to inhibit non-small cell lung cancer cell growth [25], target FOXK1 to suppress gastric cancer cells invasion [26], and also target P38/JNK to suppress oral squamous cell carcinoma cells proliferation migration and invasion [27]. But, in our study, we found miR-593-3p could promote OC progression which showed opposite results with above mentioned. We think maybe there exist two possibilities to explain this phenomenon. First, as we all known, about 20,000 genes and about 1000 miRNAs are in the human body. Also, one miRNA can target several substrate to affect cell behaviour which means that the function of substrate proteins decide the miRNA function. Second, it is very complex in the tumor tissue and different tumor have different tumor environment which showed completely function even if same miRNA、protein and signaling pathway. Combined with our experimental data, it is possible miR-593-3P promote the OC progression. In the meantime, research showed that miR-593-3p promoted Prostate cancer (PCa) cell proliferation, colony formation, spheroid formation, migration and invasion [28].
MiRNAs/PTEN axis has also been proved to regulate the cell progress in multiple tumors [29]. Loss or mutation of PTEN has been found in ovarian cancer and is associated with the worse prognosis of cancer patients [30]. In this study, we found PTEN was a target of miR-593-3p and revealed that the luciferase activity of wild type PTEN reporter was decreased in miR-593-3p mimic group (Fig. 3A-B). Additionally, both qRT-PCR and Immunohistochemistry signified that PTEN was remarkably down-regulated in ovarian cancer tissues relative to adjacent normal tissues, which is consistent with a previous research [31]. Meanwhile, the expression of miR-593-3p and PTEN was negatively correlated in OC tissues (Fig. 3F). The above evidence indicated that miR-593-3p directly combined with PTEN and inhibited its expression in OC cells.
Actually, the expression of hsa_circ_0001535 and PTEN was positively correlated in OC tissues. To confirm whether the effect of hsa_circ_0001535 on PTEN by regulating miR-593-3p in OC cells, OC cells were treated with miR-593-3p mimics after overexpressed hsa_circ_0001535, and our results discovered miR-593-3p mimic treatment reversed the up-regulation of PTEN by hsa_circ_0001535 overexpression in OC cells (Fig. 4E-F). Furthermore, hsa_circ_0001535 inhibited proliferation and migration, but its suppressing effect was attenuated by treatment with miR-593-3p mimic (Fig. 4G-I). Based on this, extensive studies are needed to further illustrate the involvement of hsa_circ_0001535 on ovarian cancer by down-regulating miR-593-3p/PTEN in vivo.
Conclusions
In summary, our present data indicated the expression level of hsa_circ_0001535 down-regulated in ovarian cancer, and overexpression of hsa_circ_0001535 could suppress proliferation and migration of OC cells by inhibiting miR-593-3p/PTEN axis. Further studies are required to confirm hsa_circ_0001535 overexpression weakened OC cells growth in vivo by tumor implantations experiment. Therefore, hsa_circ_0001535 may be considered as a cancer-suppressor gene and might serve as a promising biomarker for the treatment of ovarian cancer.
Data availability
All data generated or analyzed during this study are included in this published article.
References
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33.
Qiao ZW, Jiang Y, Wang L, Wang L, Jiang J, Zhang JR, et al. LINC00852 promotes the proliferation and invasion of ovarian cancer cells by competitively binding with miR-140-3p to regulate AGTR1 expression. BMC Cancer. 2021;21(1):1004.
Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7(2):131–42.
Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. J Clin Invest. 2009;119(6):1429–37.
Sheng R, Li X, Wang Z, Wang X. Circular RNAs and their emerging roles as diagnostic and prognostic biomarkers in ovarian cancer. Cancer Lett. 2020;473:139–47.
Torre LA, Trabert B, DeSantis CE, Miller KD, Samimi G, Runowicz CD, et al. Ovarian cancer statistics, 2018. CA Cancer J Clin. 2018;68(4):284–96.
Sheng JQ, Liu L, Wang MR, Li PY. Circular RNAs in digestive system cancer: potential biomarkers and therapeutic targets. Am J Cancer Res. 2018;8(7):1142–56.
Chen Q, Zhang J, He Y, Wang Y. hsa_circ_0061140 knockdown reverses FOXM1-mediated cell growth and metastasis in ovarian cancer through miR-370 sponge activity. Mol Ther Nucleic Acids. 2018;13:55–63.
Ning L, Long B, Zhang W, Yu M, Wang S, Cao D, et al. Circular RNA profiling reveals circEXOC6B and circN4BP2L2 as novel prognostic biomarkers in epithelial ovarian cancer. Int J Oncol. 2018;53(6):2637–46.
Chan JJ, Tay Y. Noncoding RNA:RNA regulatory networks in cancer. Int J Mol Sci. 2018;19(5):1310.
Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011;223(2):102–15.
Gong XY, Huang AL. Regulatory roles of miR-593 in the proliferation and invasion of human hepatic carcinoma. Eur Rev Med Pharmacol Sci. 2020;24(12):6657–64.
Yan L, Zhang Y, Li K, Wang M, Li J, Qi Z, et al. miR-593-5p inhibit cell proliferation by targeting PLK1 in non small cell lung cancer cells. Pathol Res Pract. 2020;216(2):152786.
Zhou F, Wang B, Wang H, Hu L, Zhang J, Yu T, et al. circMELK promotes glioblastoma multiforme cell tumorigenesis through the miR-593/EphB2 axis. Mol Ther Nucleic Acids. 2021;25:25–36.
Song L, Xiao Y. Downregulation of hsa_circ_0007534 suppresses breast cancer cell proliferation and invasion by targeting miR-593/MUC19 signal pathway. Biochem Biophys Res Commun. 2018;503(4):2603–10.
Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15(4):356–62.
Gong ZH, Zhou F, Shi C, Xiang T, Zhou CK, Wang QQ, et al. miRNA-221 promotes cutaneous squamous cell carcinoma progression by targeting PTEN. Cell Mol Biol Lett. 2019;24:9.
Wu X, Liu D, Wang S, Liu J. Circ_0007444 inhibits the progression of ovarian cancer via mediating the miR-570-3p/PTEN axis. Onco Targets Ther. 2021;14:97–110.
Abadi AJ, Zarrabi A, Gholami MH, Mirzaei S, Hashemi F, Zabolian A, et al. Small in size, but large in action: microRNAs as potential modulators of PTEN in breast and lung cancers. Biomolecules. 2021;11(2):304.
Lheureux S, Braunstein M, Oza AM. Epithelial ovarian cancer: evolution of management in the era of precision medicine. CA Cancer J Clin. 2019;69(4):280–304.
Xie Y, Hang X, Xu W, Gu J, Zhang Y, Wang J, et al. CircFAM13B promotes the proliferation of hepatocellular carcinoma by sponging miR-212, upregulating E2F5 expression and activating the P53 pathway. Cancer Cell Int. 2021;21(1):410.
Lv J, Li K, Yu H, Han J, Zhuang J, Yu R, et al. HNRNPL induced circFAM13B increased bladder cancer immunotherapy sensitivity via inhibiting glycolysis through IGF2BP1/PKM2 pathway. J Exp Clin Cancer Res. 2023;42(1):41.
Huang A, Zheng H, Wu Z, Chen M, Huang Y. Circular RNA-protein interactions: functions, mechanisms, and identification. Theranostics. 2020;10(8):3503–17.
Xie J, Wan Y, Zhang M, Jin Z, Yao Y. Circ_0061825 acts as a miR-593-3p sponge to promote breast cancer progression by regulating FGFR3 expression. Cancer Manag Res. 2020;12:11243–55.
Han W, Wang L, Zhang L, Wang Y, Li Y. Circular RNA circ-RAD23B promotes cell growth and invasion by miR-593-3p/CCND2 and miR-653-5p/TIAM1 pathways in non-small cell lung cancer. Biochem Biophys Res Commun. 2019;510(3):462–6.
Dong L, Hong H, Chen X, Huang Z, Wu W, Wu F. LINC02163 regulates growth and epithelial-to-mesenchymal transition phenotype via miR-593-3p/FOXK1 axis in gastric cancer cells. Artif Cells Nanomed Biotechnol. 2018;46(sup2):607–15.
Jiang L, Zhou B, Fu D, Cheng B. lncRNA TUG1 promotes the development of oral squamous cell carcinoma by regulating the MAPK signaling pathway by sponging miR-593-3p. Cell Cycle. 2022;21(17):1856–66.
Huang Q, Peng L, Sun Y, Huang J, Han T, Li Y, et al. miR-593-3p promotes proliferation and invasion in prostate cancer cells by targeting ADIPOR1. Onco Targets Ther. 2021;14:3729–37.
Gao Z, Ye X, Bordeaux A, Hettich S, Lin S, Han F, et al. miR-26b regulates cell proliferation and apoptosis of CD117+CD44+ ovarian cancer stem cells by targeting PTEN. Eur J Histochem. 2021.
Nero C, Ciccarone F, Pietragalla A, Scambia G. PTEN and gynecological cancers. Cancers (Basel). 2019;11(10):1458.
Gong J, Xu X, Zhang X, Zhou Y. Circular RNA-9119 suppresses in ovarian cancer cell viability via targeting the microRNA-21-5p-PTEN-Akt pathway. Aging (Albany NY). 2020;12(14):14314–28.
Acknowledgements
This study was funded by Nantong Science and Technology Plan Project (JC2021005, MS22022005); Jiangsu Province Maternal and Children Health Research Project (F202139); Nantong Health Commission Research Project (MB2021017); Scientific Research and Innovation Team Project of Kangda College of Nanjing Medical University (KD2022KYCXTD010).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Second Affiliated Hospital of Nantong University.
Patient consent for publication
Written informed consent was obtained from all patients for the publication of data.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Han, Y., Zheng, Y., You, J. et al. Hsa_circ_0001535 inhibits the proliferation and migration of ovarian cancer by sponging miR-593-3p, upregulating PTEN expression. Clin Transl Oncol 25, 2901–2910 (2023). https://doi.org/10.1007/s12094-023-03152-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-023-03152-2