Abstract
Purpose
Nowadays, the oxidative phosphorylation (OXPHOS) correlated with leukemogenesis and treatment response is extensive. Thus, exploration of novel approaches in disrupting OXPHOS in AML is urgently needed.
Materials and methods
Bioinformatical analysis of TCGA AML dataset was performed to identify the molecular signaling of OXPHOS. The OXPHOS level was measured through a Seahorse XFe96 cell metabolic analyzer. Flow cytometry was applied to measure mitochondrial status. Real-time qPCR and western blot were used to analyze the expression of mitochondrial or inflammatory factors. MLL-AF9-induced leukemic mice were conducted to measure the anti-leukemia effect of chidamide.
Results
Here, we reported that AML patients with high OXPHOS level were in a poor prognosis, which was associated with high expression of HDAC1/3 (TCGA). Inhibition of HDAC1/3 by chidamide inhibited cell proliferation and induced apoptotic cell death in AML cells. Intriguingly, chidamide could disrupt mitochondrial OXPHOS as assessed by inducing mitochondrial superoxide and reducing oxygen consumption rate, as well as decreasing mitochondrial ATP production. We also observed that chidamide augmented HK1 expression, while glycolysis inhibitor 2-DG could reduce the elevation of HK1 and improve the sensitivity of AML cells exposed to chidamide. Furthermore, HDAC3 was correlated with hyperinflammatory status, while chidamide could downregulate the inflammatory signaling in AML. Notably, chidamide eradicated leukemic cells in vivo and prolonged the survival time of MLL-AF9-induced AML mice.
Conclusion
Chidamide disrupted mitochondrial OXPHOS, promoted cell apoptosis and reduced inflammation in AML cells. These findings exhibited a novel mechanism that targeting OXPHOS would be a novel strategy for AML treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Acute myeloid leukemia (AML) is the clonal hematological malignancy, mostly attributing to genomic aberration in the leukemia-initiating cells or leukemia stem cells (LSCs), such as gene mutations and chromosomal rearrangements. Nowadays, the metabolic signal associated with leukemogenesis is extensive, and oxidative phosphorylation (OXPHOS) appears to play a vital role in AML progress and treatment response [1]. Numbers of studies have also reported the importance of mitochondrial OXPHOS in LSCs. In primary human AML specimens, STAT3-MYC mediates survival and OXPHOS of LSCs by regulating SLC1A5 [2]. Of note, LSCs are metabolically less active and rely on OXPHOS rather than glycolysis [3], and display a unique mitochondrial morphology [4]. Interestingly, mitochondrial outer membrane protein MTCH2 is essential to the survival of AML cells. MTCH2 inhibition leads to an increase of nuclear pyruvate, as well as pyruvate dehydrogenase to promote histone acetylation, which subsequently improves cell differentiation in AML [5]. Deacetylase SIRT3 reprograms mitochondrial metabolism toward OXPHOS and decreases ROS generation to confer chemoresistance in AML [6]. Hence, the epigenetic regulation is linked to mitochondrial metabolism, highlighting that this process can be acted as potential therapeutic targets in AML.
Since histone acetylation aberration is an important epigenetic event that is mostly modulated by histone deacetylase (HDAC) activity, HDAC inhibition with HDAC inhibitors has been proven to be potent anti-tumor strategies for the treatment of multiple tumor types [7]. Chidamide (CS055 or tucidinostat), a novel oral HDAC inhibitor, is currently applied in multiple clinical trials for the treatment of various solid cancers and hematological malignancy as monotherapy or combination therapy, which inhibits the activity of HDAC1, 2, 3 and 10. Chidamide restrains cell growth and induces apoptotic cell death, especially in lymphoma and leukemia [8]. In the preclinical and clinical models, chidamide displays encouraging therapeutic response in peripheral T-cell lymphoma (PTCL) or T-cell acute lymphoblastic leukemia (T-ALL) by monotherapy or chidamide-based regimens [9, 10]. In AML, chidamide also blocks the cell cycle in G0/G1 phase and inhibits cell growth [11,12,13]. In addition, chidamide induces DNA double-strand break and alters the ratio of anti- and pro-apoptotic proteins [11], and improves the sensitivity to anthracycline drugs by inhibition of the HDAC3-AKT-P21-CDK2 axis [12]. In T-ALL, combination of apatinib and chidamide suppresses mitochondrial respiration and decreases the levels of some rate limiting enzymes in the OXPHOS [10], suggesting that chidamide could exert its anti-leukemia on cellular metabolism.
In this study, we found that high OXPHOS was associated with high expression of HDAC1/3 in AML. Inhibition of HDAC1/3 by chidamide disrupted mitochondrial OXPHOS and reduced inflammatory status of AML, providing a new approach of targeting leukemia in future.
Materials and methods
Bioinformatics analysis
The Cancer Genome Atlas AML dataset (173 patients in all and 162 patients with full survival data) was downloaded from XENA (https://xenabrowser.net/hub/). Gene set enrichment analysis (GSEA) was performed on Gene Ontology and Hallmark using clusterProfiler package [14]. Immune infiltration was analyzed by the xCell algorithm [15] and ssGSEA. Glycolysis score and OXPHOS score were assessed based on gene set variation analysis (GSVA). ChIP sequence analysis of GSE111293 from Gene Expression Omnibus (GEO) was performed by Intergrative Genomics Viewer (IGV).
Cell lines and reagents
HL-60 and MOLM-13 were cultured in RPMI-1640 (Gibco, Grand Island, NY, USA) with 10% fetal bovine serum (Gibco) in an incubator at 37℃ with 5% CO2. Chidamide (Chipscreen Bioscience Ltd., Shenzhen, China) was dissolved in dimethyl sulfoxide at a concentration of 50 mM.
Cell viability assay
Leukemic cells were cultured in 96-well plates and treated with various doses of chidamide alone or combination for indicated time. The cell viability was detected by Cell Counting Kit-8 (CCK-8, APExBIO, Houston, TX, USA). After different treatment, 10 μL CCK-8 solution was added to each well for incubation for another 4 h, and then cells were detected using a microplate reader by the optical density (OD) at 450 nm.
Colony forming unit (CFU) assay
HL-60 and MOLM-13 cells were cultured in methylcellulose (R&D Systems, Minneapolis, MN, USA) and treated with chidamide. The CFU were counted and measured on the indicated day.
Cell death detection
HL-60 and MOLM-13 cells were incubated with chidamide alone or combination with other drugs, and cell apoptotic death was measured using Annexin V-FITC and propidium iodide (PI) staining (Vazyme Biotech, China). Briefly, cells were collected, and resuspended in binding buffer before being incubated with Annexin V-FITC and PI for 15 min. Cell apoptosis was analyzed using BD FACSCanto instrument. SYTOX staining (Thermo Fisher Scientific, Waltham, MA, USA) was performed as manufacturer’s protocols and cells were finally subjected to flow cytometry detection.
Analysis of mitotracker and mitosox
After treated with or without chidamide for 48 h, AML cells were collected and washed with PBS, and then incubated with 200 nM MitoTracker™ Deep Red FM (Invitrogen Life Technologies, Carlsbad, CA, USA) in RPMI-1640 for 30 min at 37℃, or incubated with 5 μM MitoSOX™ Red (Invitrogen) in RPMI-1640 for 15 min. After incubation, cells were washed and resuspended with PBS, following with measurement by flow cytometry.
ATP content measurement
ATP content in AML cells with or without treatment of chidamide was detected using the Enhanced ATP Assay Kit (Beyotime Biotech, China) according to the manufacturer’s protocols. Briefly, cells were lysed in the ATP detection lysis buffer and then centrifuged at 12,000 g for 5 min at 4 ℃. The supernatant was collected for luminescence detection using TECAN Infinite 200 PRO and the result was normalized by protein content.
Real-time qPCR assays
Cellular RNA was extracted from AML cells by TRIzol reagent (Invitrogen), and the cDNA was attained by a Reverse Transcription System (Thermo Fisher Scientific). qPCR was performed with qPCR Mix (Vazyme) following the instructions of the manufacturer. Primer sequences of qPCR are shown in Table S1, and ACTB was used as an internal standard.
Western blot analysis
Cells were collected, and lysed with ice-cold RIPA buffer (Cell Signaling Technology, Danvers, MA, USA). After centrifugation at 12,000 rpm for 15 min at 4 ℃, the protein lysates was quantified by a bicinchoninic acid (BCA) assay and separated by SDS-PAGE. The lysates was then transferred onto the nitrocellulose membranes (Merk Millipore, Billerica, MA, USA). After blocked with 5% bovine serum albumin, the membranes were incubated with antibodies at 4 ℃ overnight. Primary antibodies were used as following: HK1, LDHA, PFKP, p-AKT, AKT, acetyl-Histone H3 Lys9 (H3K9ac), acetyl-Histone H3 Lys27 (H3K27ac), p-IκBα, IκBα, NF-κB (p65), GAPDH, β-actin, and Histone Deacetylase Antibody Sampler Kit (Cell Signaling Technology). The membranes were then washed, and exposed to the corresponding secondary antibodies for 1 h. Finally, immunoreactive bands were monitored by chemiluminescent imaging system.
Mitochondrial oxygen consumption rate measurement
To analyze the mitochondrial function, oxygen consumption rate (OCR) was determined according to the instruction of the Seahorse XF Cell Mito Stress Test Kit (Agilent Technologies, Santa Clara, CA, USA). Briefly, the sensor cartridge in Seahorse XF Calibrant was hydrated in a non-CO2 incubator at 37℃ overnight. AML cells were adhered to XFe96 cell culture microplates using Cell-Tak (BD Biosciences, San Jose, CA, USA) and incubated at 37 ℃ for 1 h before analysis. Oligomycin (1 μM), carbonyl cyanide-4- (trifluoromethoxy) phenylhydrazone (FCCP, 1 µM), Rotenone (0.5 µM) and Antimycin A (0.5 µM) were added sequentially at the indicated time points.
In vivo experiment
Bone marrow cells from 5-FU-treated C57BL/6 J mice were infected with MLL-AF9 retrovirus with IL-3, IL-6, and SCF (Peprotech, Rocky Hill, NJ, USA). After receiving with X-ray irradiation, the recipient mice were transplanted with bone marrow cells transfected with MLL-AF9 to develop leukemia. The bone morrow cells from the leukemic mice were later collected for analysis or applied for second transplantation treated with or without chidamide (25 mg/kg/d) orally each day for 14 days. The animal study was conducted with the approval of Institutional Animal Care and Use Committee.
Statistical analysis
Data were analyzed using GraphPad Prism 8.0 software (GraphPad Software Inc., La Jolla, CA, USA). The result was considered significant if p value was less than 0.05 (*p < 0.05, **p < 0.01, ***p < 0.001).
Results
High OXPHOS is correlated high expression of HDAC1/3 in AML
To investigate the relation of OXPHOS and AML progression, a survival analysis was applied using AML data from TCGA. We divided AML patients into two groups based on OXPHOS score and found that AML patient with high OXPHOS displayed a short survival (Fig. 1A). GSVA showed OXPHOS, as well as apoptosis, reactive oxygen species, glycolysis and fatty acid metabolism pathway were enriched in high OXPHOS group (Fig. 1B). Additionally, NF-κB and HDACs signaling were also upregulated when OXPHOS was increased by performing GSEA (Fig. 1C). To further investigate the function of HDAC family in AML OXPHOS, we analyzed the expression of HDAC1-10 and found that Class I of HDACs, such as HDAC1 and HDAC3, was upregulated in high OXPHOS group by heatmap analysis (Fig. 1D). Quantitative expression analysis further confirmed that HDAC1 and HDAC3 were significantly high expressed in high OXPHOS AML patients (Fig. 1E, F).
OXPHOS is correlated to HDACs expression in AML. A AML dataset from TCGA was divided into two groups based on OXPHOS score (high and low) and a survival analysis was performed. B High OXPHOS group and low OXPHOS group of AML patients from TCGA was subjected to GSVA to illustrate the difference of pathway enrichment. C GSEA was used to analyze the signaling of high OXPHOS group and low OXPHOS group. D Differently expressed genes were analyzed between high OXPHOS group and low OXPHOS group patients and the heatmap showed the different gene expressed pattern of HDAC family. The expression of HDAC1 (E) and HDAC 3 (F) in high OXPHOS group (red) or low OXPHOS group (blue) was displayed
We then divided the AML patients into groups according to HDAC1 or HDAC3 expression level. Figure 2A, B shows that both patients with high expression of HDAC1 or HDAC3 were associated with OXPHOS, reactive oxygen species, and fatty acid metabolism pathway. GSEA revealed that gluconeogenesis, glucose metabolism and PD-1 signaling were decreased in low expression of HDAC1 or HDAC3 group (Fig. 2C, D). Glycolysis score and OXPHOS score were greatly increased in high HDAC3 group, while high HDAC1 group just showed upregulation of OXPHOS score (Fig. 2E, F). These data suggested that both HDAC1 and HDAC3 played important roles in AML OXPHOS.
AML patients with high HDAC1 or HDAC3 expression display high OXPHOS. AML patients were divided into two groups based on HDAC1 (A) or HDAC3 (B) expression and GSVA was performed to analyze the difference of pathway enrichment. GSEA was applied to analyze the signaling in low HDAC1 group and high HDAC1 group (C), and in low HDAC3 group and high HDAC3 group (D). AML patients were divided into groups based on the expression of HDAC1 (E) and HDAC3 (F), and the glycolysis score and OXPHOS score in each group were displayed
Chidamide inhibits HDACs and suppresses cell growth in AML
To investigate the alternation of OXPHOS by inhibition of HDACs, we applied chidamide, an HDACs inhibitor, to measure the relationship of HDACs, OXPHOS and leukemic cell growth. Western blot showed that chidamide could inhibit HDACs expression, especially HDAC3 in HL-60 and MOLM-13 cells, and greatly induce H3K9ac and H3K27ac expression. Meanwhile, LDHA and PFKP, as well as phosphorylated AKT, were reduced, while HK1 was slightly increased (Fig. 3A). Then, we tested the proliferation of HL-60 and MOLM-13 cells after chidamide treatment at various concentrations by CCK-8 assay. The results showed that chidamide suppressed cell viability in dose- and time-dependent manners (Fig. 3B, C). Both after exposure of 36 h, 1 μM of chidamide could significantly reduce cell viability. Similarly, a carboxyfluorescein succinimidyl ester (CFSE) assay confirmed that chidamide reduced cell growth in a concentration-dependent manner for 48 h treatment (Fig. 3D, E), indicative of chidamide could inhibit a short-term proliferation. To continue to measure the long-term proliferative suppression, HL-60 and MOLM-13 cells were cultured in the methylcellulose and treated with chidamide for 10 days. Figure 3F shows that chidamide inhibited the high proliferated cells in a long-term culturing. Chidamide decreased the CFU numbers as well as the diameters at a low concentration of 0.15 μM (Fig. 3F). Thus, chidamide could greatly inhibit cell growth in AML cells.
Chidamide inhibits HDACs and suppresses cell growth in AML. A AML cells were treated with different doses of chidamide for 48 h and subjected to western blot for the expression of HDACs and metabolic proteins. AML cells were treated chidamide for 24 h (B) or 36 h (C), and CCK-8 assay was applied to detect cell viability. HL-60 (D) and MOLM-13 (E) cells were exposed to 1 μM and 2 μM chidamide for 48 h and subjected to CFSE staining measurement. F AML cells were treated with 0.15 μM chidamide for 10 days, and the numbers and diameters of CFU were measured. Data were presented as mean ± SD, *p < 0.05, **p < 0.01, ***p < 0.001
Next, Annexin V-PI staining assay was used to test whether chidamide could induce apoptotic cell death. We found that chidamide increased apoptosis in a dose-dependent manner both in HL-60 and MOLM-13 cells after 48 h treatment (Fig. 4A, B). Similarly, more cellular death was observed by SYTOX staining assay after cells were exposed to 1 μM and 2 μM chidamide (Fig. 4C, D), indicating that chidamide could promote apoptosis in AML cells.
Chidamide induces apoptosis in AML. HL-60 (A) and MOLM-13 (B) cells were exposed to 1 μM and 2 μM chidamide for 48 h and subjected to Annexin V-PI staining measurement. HL-60 (C) and MOLM-13 (D) cells were exposed to 1 μM and 2 μM chidamide for 48 h and the cells were then stained with SYTOX and detected by flow cytometry. Data were presented as mean ± SD, *p < 0.05, **p < 0.01, ***p < 0.001
Chidamide disrupts OXPHOS in AML
Since HDAC inhibitors could change the enrichments of histone acetylation at the promoter region of target genes and regulate gene expression, we then continued to analyze ChIP sequence data from GSE111293, of which U937 cells were treated with HDAC1/HDAC3 inhibitor Entinostat and pan HDAC inhibitor Panobinostat, and then subjected to ChIP sequence using antibody against H3K9ac and H3K27ac. The results showed that Entinostat (E) and Panobinostat (P) treatment decreased histone H3 acetylation enrichment at the promoters of mitochondrial metabolism related genes, including ATP5A and UQCRC2 (Fig. 5A, B). Similarly, in HL-60 and MOLM-13 cells, chidamide also reduced the expression of ATP5A and UQCRC2 (Fig. 5C), further suggesting that the mitochondrial metabolism were regulated by HDAC inhibitors.
ChIP sequence analysis of mitochondrial-related genes after HDAC inhibitor treatment. ChIP sequence analysis of U937 cells from GSE111293 for H3K9ac and H3K27ac on the promoters of ATP5A (A) and UQCRC2 (B) after treatment with HDAC inhibitors. C qPCR was used to detect the expression of ATP5A and UQCRC2 in HL-60 and MOLM-13 cells. **p < 0.01, ***p < 0.001
To address the role of OXPHOS in AML, we detected the change of metabolism after chidamide treatment. AML cells cultured with chidamide had exhibited higher production of mitochondrial superoxide than control cells as assessed by MitoSOX staining (Fig. 6A, B). MitoTracker staining also confirmed the mitochondrial balance was disrupted in HL-60 and MOLM-13 cells (Fig. 6C, D). Additionally, we measured mitochondrial respiration using the XFe96 cell metabolic analyzer. Culture with chidamide significantly reduced mitochondrial respiration in AML cells, as evidenced by a decrease in the levels of basal respiration, maximal respiration and ATP production (Fig. 6E–H). Since maximal respiration reflects the cell ability to respond to energy demands, our result indicated that AML cells treated with chidamide could decrease metabolic demand (Fig. 6G, H). Furthermore, ATP measurement was also confirmed by luminescence detection. Figure 6I, J reveals that chidamide effectively reduced ATP content in HL-60 and MOLM-13 cells. Hence, chidamide could disrupt OXPHOS in AML.
Chidamide disrupts mitochondrial OXPHOS in AML. HL-60 (A) and MOLM-13 (B) were exposed to 0.5 μM and 1 μM chidamide for 48 h and MitoSOX staining was detected. HL-60 (C) and MOLM-13 (D) were exposed to 0.5 μM and 1 μM chidamide for 48 h and subjected to MitoTracker measurement. AML cells were cultured with chidamide for 48 h, and exposed to Oligomycin (1 μM), FCCP (1 µM) and Rotenone and Antimycin A (Rot/AA, 0.5 µM), and the OCR was analyzed in HL-60 (E) and MOLM-13 (F). ATP production, basal respiration and maximal respiratory capacity in HL-60 (G) and MOLM-13 (H) treated with chidamide were shown. Chidamide treated HL-60 (I) and MOLM-13 (J) cells were applied to measure ATP production by luminescence detection. Data were presented as mean ± SD, *p < 0.05, **p < 0.01, ***p < 0.001
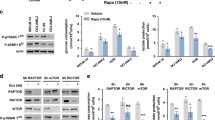
Importantly, we observed that glycolysis inhibitor 2-Deoxy-D-glucose (2-DG) could enhance the apoptotic effect of chidamide and lead to growth blockage (Fig. 7A–C) in AML cells. However, FCCP, Oligomycin, or Metformin could not improve cell sensitivity (Figure S1). Furthermore, chidamide augmented HK1 expression, while 2-DG reduced the elevation of HK1 (Fig. 7D). Thus, 2-DG-inhibited glycolysis might promote OXPHOS in AML cells, which facilitated AML cells to be more sensitive to OXPHOS inhibitor to achieve therapeutic benefit.
2-DG enhances cell sensitivity to chidamide. A AML cells were treated chidamide (1 μM) or/and 2-DG (HL-60: 2.5 mM and 5 mM; MOLM-13: 5 mM and 10 mM) for 24 h, and CCK-8 assay was applied to detect cell viability. HL-60 (B) and MOLM-13 (C) cells were exposed to chidamide (1 μM) or/and 2-DG (HL-60: 2.5 mM; MOLM-13: 5 mM) for 48 h and subjected to apoptosis detection and western blot (D). Data were presented as mean ± SD, *p < 0.05, ***p < 0.001
Chidamide suppresses inflammation in AML
Moreover, ChIP sequence analysis from GSE111293 also showed that Entinostat and Panobinostat treatment reduced histone H3 acetylation enrichment at the promoter of IκB (Fig. 8A). Consistently, IκB/NF-κB signaling was decreased in low HDAC3 patients with low expression of a set of genes related to IκB/NF-κB signaling, including some important inflammatory factors, such as TLRs and S100A (Fig. 8B, C). Thus, chidamide was used to inhibit HDAC3 to test the inflammatory status. We showed that chidamide inhibited the expression p-IκBα and NF-κB (p65) (Fig. 8D), which was consistently with the result in Fig. 8A, B. Meanwhile, inflammatory cytokines, such as IFI44, ISG15 and CXCL10 was also reduced after low dose of chidamide treatment (Fig. 8E), which might lead to reduce the inflammatory state and reactivate immune cells in the microenvironment.
Chidamide relieves cell inflammatory status. A ChIP sequence analysis from GSE111293 for H3K9ac and H3K27ac on the promoter of IκB after treatment with HDAC inhibitors. B GO-GSEA was applied to analyze the inflammatory signaling in AML patients from TCGA. C The heatmap of inflammatory factors in NF-κB signaling between HDAC3 high and low groups was displayed. D HL-60 and MOLM-13 were exposed to different doses of chidamide for 48 h and subjected to western blot. E Cells were treated with 0.5 μM chidamide for 48 h and qPCR was performed. Data were presented as mean ± SD, *p < 0.05, **p < 0.01, ***p < 0.001
Since OXPHOS signature is correlated to inflammation or immunity [16], and chidamide could decrease the production of inflammatory cytokines, we continued to investigate how HDACs influence immunity in AML. We found that patients with high HDAC3 expression displayed low enrichment of CD8 T cells and NK cells (Figure S2A) by performing ssGSEA using TCGA data. Unsupervised clustering revealed comparable patterns of checkpoint proteins between high and low HDAC3 groups. However, activated NK cells showed effective infiltration in low HDAC3 group (Figure S2B). Although the relationship between the immune cells in HDAC3 high and low groups was comparable (Figure S2C), significant abundance of immune infiltration in CD8 T cells and NK cells were also observed in low HDAC3 group compared with the high group (Figure S2D). These data suggested that immune infiltration, especially CD8 T and NK cell, might be regulated by HDAC3 in AML patients, implying the HDAC inhibitors might improve the status of AML microenvironment.
Chidamide eradicates leukemic cells in vivo
Notably, the bone marrow cells from MLL-AF9-induced mice (Fig. 9A, B) were used for CCK-8, CFU assay, GFP detection and Annexin V-7AAD staining (Fig. 9C–F). The MLL-AF9 leukemic cells exposure to chidamide led to reduction of cell viability, as well as CFU numbers and diameters (Fig. 9C, D). Chidamide also reduced GFP expression of leukemic cells and increased cell apoptosis (Fig. 9E, F). Similarly, chidamide decreased the expression of UQCRC2 and ATP5A in murine leukemic cells (Fig. 9G). After second transplantation of MLL-AF9 leukemic cells, the mice were treated intragastrically with or without chidamide. A prolonged life span was observed after treatment with chidamide (Fig. 9H). Flow cytometric analysis showed decreased c-Kit+Gr-1− leukemic cells of the peripheral blood in chidamide-treated mice (Fig. 9I). Therefore, chidamide could eradicate leukemic cells in vivo.
Chidamide eradicates murine leukemic cells in vivo. A The chart of procedure for collecting leukemic cells in mice. B Murine bone marrow cells transfected with the MLL-AF9 were transplanted to the recipient mice to develop leukemia. Then the leukemic cells were collected for PCR to detect MLL-AF9 fusion gene expression (n = 3). CCK-8 (C) and CFU (D) assays were performed after chidamide treatment. The GFP expression (E), apoptosis (F), and UQCRC2 and ATP5A mRNA expression (G) were displayed. (H) The survival of mice treated with or without chidamide. (I) Peripheral blood cells were collected to detect the ratio of c-Kit+Gr-1− after treatment with chidamide for 14 days. *p < 0.05, **p < 0.01, ***p < 0.001
Discussion
OXPHOS, as well as other catabolic pathways, such as glycolysis, glutamine metabolism, and fatty acid metabolism, is crucial for the cellular energy metabolism and survival. OXPHOS-associated metabolic rewiring is extensive in leukemogenesis, leukemia maintenance and treatment resistance [1]. An increase in OXPHOS could help AML cells to survive in stressed condition and evade apoptosis. Pharmacological targeting of mitochondrial OXPHOS metabolism is therefore an attractive area in AML therapy [17].
Aberrant expression and recruitment of HDACs play critical roles in leukemogenesis and disease progression. HDACs are the key epigenomic modulators in the nucleus that forms multiprotein complexes to deacetylate histones, controlling the transcription of numbers of genes which are required for leukemia initiation and development. Alternation of the expression or activity of HDACs are observed in solid tumors as well as leukemia [18]. For example, HDAC3 is reported to be critical to control DNA damage as well as maintaining genome stability, while deletion of HDAC3 greatly reduces DNA repair capacity [19, 20]. Chemotherapy upregulates HDAC3 expression in leukemic cells, while inhibition of HDAC3 decreases drug resistance through repressing the activation of AKT [20]. Consistently, upregulation of HDAC3-AKT-P21-CDK2 signal is observed in anthracycline-resistant AML cells [12]. In t(8;21) AML cells, HDAC3 binds to RUNX1 to collaborate with RUNX1, contributing to repressing AML1-ETO-dependent transcription [21]. HDAC3 inhibition by RGFP966 increases apoptosis and attenuates ontogenetic protein expression, including β-catenin, MYC and WT1 [22]. Importantly, HDAC3 translocates to mitochondria to drive inflammation in an IL-1β-dependent manner by deacetylating and inactivating the FAO enzyme HADHA [23], suggesting that HDAC3 couples mitochondria through configuring cell metabolism. In this study, we showed that both HDAC1 and HDAC3 were strongly correlated to AML OXPHOS (Figs. 1, 2). Chidamide effectively inhibited HDAC3 expression and reduced mitochondrial OXPHOS, giving evidence that HDAC3 was a target to intervene OXPHOS in AML (Figs. 3, 5, 6).
The metabolism of tumor cell has been identified as a hallmark, and tumor cells switch between glycolysis and OXPHOS to adapt their energy requirement and to satisfy the needs of proliferation. Recent studies report that in numbers of tumor types, cells switch to mitochondrial OXPHOS for maintaining a high proliferative rate [24]. AML cells also heavily rely on OXPHOS [3], while inhibition of OXPHOS could efficiently reduce cell proliferation [17, 25]. Cells acquire metabolic remodeling by regulating metabolic enzymes by translation or post-translation modification. Acetylation/deacetylation is considering as a major regulatory metabolic mechanism and alters in tumors [26]. The role of HDAC3 in controlling DNA damage and gene expression is well reported. However, we provided new data that HDAC3 could regulate mitochondrial OXPHOS in AML cells by reducing OCR and ATP production (Fig. 6).
In addition to supplying the power for the cell survival, mitochondria are also regarded as the powerhouses of immunity [16]. In colorectal adenocarcinoma, OXPHOS-related prognostic signature is associated with immune infiltration. Higher gene expression of immune checkpoint and tumor mutation burden exist positively with higher OXPHOS risk score [27]. Similarly, in hepatocellular carcinoma, the five-gene OXPHOS-related signature is associated with immune cell infiltration as well as tumor stemness [28]. In AML, EVT-701, an electron transfer chain complex I inhibitor, reduces the OXPHOS and PD-L1 expression in cytarabine-resistant leukemic cells to improve the anti-tumor response [29]. Thus, OXPHOS-mediated immune response can be used for prognostic prediction for tumor patients. In this study, we also found that HDAC3 expression predicted CD8 T and NK cell infiltration and inflammatory status, while chidamide could reduce inflammatory signaling (Fig. 8, S2), revealing its novel role in regulating metabolism-controlled inflammation and immune infiltration.
Chidamide has been studied as a monotherapy in several cancer types, which is currently used as a second-line therapy in T-cell lymphoma treatment [30]. Currently, chidamide is mostly used in combination with chemotherapy, immunotherapy or targeted therapy in numbers of clinical trials in hematological malignancies [8]. Daunorubicin, idarubicin, and cytarabine were DNA-damaging agents that widely used in AML, and low dose of chidamide could improve the therapeutic effect [31]. A Phase I/II study shows the strategy of chidamide, decitabine, cytarabine, aclarubicin, and granulocyte colony-stimulating factor combination in patients with relapsed or refractory AML achieves favorable benefit [32]. Chidamide also enhances the anti-leukemic activity of venetoclax, a selective Bcl-2 inhibitor, by increasing DNA damage and promoting apoptotic cell death [11, 13]. In this study, we found that chidamide restrained leukemic cells by reduction of cell proliferation and induction of apoptosis (Figs. 3, 4, 9). Furthermore, inhibition of glycolysis by 2-DG could improve the sensitivity of AML cells to chidamide. Mechanically, 2-DG reduced the expression of HK1 increased by chidamide (Fig. 7), indicating that switching the AML cells to OXPHOS by inhibition glycolysis could improve the efficacy of HDAC inhibitors.
Therefore, chidamide could act as an OXPHOS inhibitor to disrupt AML mitochondrial OXPHOS and eradicate leukemic cells. This study demonstrated that targeting OXPHOS will be a new strategy by reducing cell metabolism and improving immune status in AML treatment.
Data availability
The data presented in this study are available on request from the corresponding author.
References
de Beauchamp L, Himonas E, Helgason GV. Mitochondrial metabolism as a potential therapeutic target in myeloid leukaemia. Leukemia. 2022;36(1):1–12.
Amaya ML, Inguva A, Pei S, Jones C, Krug A, Ye H, et al. The STAT3-MYC axis promotes survival of leukemia stem cells by regulating SLC1A5 and oxidative phosphorylation. Blood. 2022;139(4):584–96.
Lagadinou ED, Sach A, Callahan K, Rossi RM, Neering SJ, Minhajuddin M, et al. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell. 2013;12(3):329–41.
Pei S, Minhajuddin M, Adane B, Khan N, Stevens BM, Mack SC, et al. AMPK/FIS1-mediated mitophagy Is required for self-renewal of human AML stem cells. Cell Stem Cell. 2018;23(1):86-100.e6.
Khan DH, Mullokandov M, Wu Y, Voisin V, Gronda M, Hurren R, et al. Mitochondrial carrier homolog 2 is necessary for AML survival. Blood. 2020;136(1):81–92.
Ma J, Liu B, Yu D, Zuo Y, Cai R, Yang J, et al. SIRT3 deacetylase activity confers chemoresistance in AML via regulation of mitochondrial oxidative phosphorylation. Br J Haematol. 2019;187(1):49–64.
Cheng Y, He C, Wang M, Ma X, Mo F, Yang S, et al. Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials. Signal Transduct Target Ther. 2019;4:62.
Sun Y, Hong JH, Ning Z, Pan D, Fu X, Lu X, et al. Therapeutic potential of tucidinostat, a subtype-selective HDAC inhibitor, in cancer treatment. Front Pharmacol. 2022;13: 932914.
Gui L, Cao J, Ji D, Zhang H, Fan Q, Zhu J, et al. Chidamide combined with cyclophosphamide, doxorubicin, vincristine and prednisone in previously untreated patients with peripheral T-cell lymphoma. Chin J Cancer Res. 2021;33(5):616–26.
Zhong M, Lin F, Jiang Y, Pan G, Tan J, Zhou H, et al. Therapeutic interaction of Apatinib and Chidamide in T-Cell acute lymphoblastic Leukemia through interference with mitochondria associated biogenesis and intrinsic apoptosis. J Pers Med. 2021;11(10):977.
Chen K, Yang Q, Zha J, Deng M, Zhou Y, Fu G, et al. Preclinical evaluation of a regimen combining chidamide and ABT-199 in acute myeloid leukemia. Cell Death Dis. 2020;11(9):778.
Wang H, Liu YC, Zhu CY, Yan F, Wang MZ, Chen XS, et al. Chidamide increases the sensitivity of refractory or relapsed acute myeloid leukemia cells to anthracyclines via regulation of the HDAC3 -AKT-P21-CDK2 signaling pathway. J Exp Clin Cancer Res. 2020;39(1):278.
Li G, Li D, Yuan F, Cheng C, Chen L, Wei X. Synergistic effect of chidamide and venetoclax on apoptosis in acute myeloid leukemia cells and its mechanism. Ann Transl Med. 2021;9(20):1575.
Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–7.
Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18(1):220.
Mills EL, Kelly B, O’Neill LAJ. Mitochondria are the powerhouses of immunity. Nat Immunol. 2017;18(5):488–98.
Molina JR, Sun Y, Protopopova M, Gera S, Bandi M, Bristow C, et al. An inhibitor of oxidative phosphorylation exploits cancer vulnerability. Nat Med. 2018;24(7):1036–46.
Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6(1):38–51.
Bhaskara S, Knutson SK, Jiang G, Chandrasekharan MB, Wilson AJ, Zheng S, et al. Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell. 2010;18(5):436–47.
Long J, Fang WY, Chang L, Gao WH, Shen Y, Jia MY, et al. Targeting HDAC3, a new partner protein of AKT in the reversal of chemoresistance in acute myeloid leukemia via DNA damage response. Leukemia. 2017;31(12):2761–70.
Guo C, Li J, Steinauer N, Wong M, Wu B, Dickson A, et al. Histone deacetylase 3 preferentially binds and collaborates with the transcription factor RUNX1 to repress AML1-ETO-dependent transcription in t(8;21) AML. J Biol Chem. 2020;295(13):4212–23.
Beyer M, Romanski A, Mustafa AM, Pons M, Buchler I, Vogel A, et al. HDAC3 Activity is essential for human leukemic cell growth and the expression of beta-catenin, MYC, and WT1. Cancers (Basel). 2019;11(10):1436.
Chi Z, Chen S, Xu T, Zhen W, Yu W, Jiang D, et al. Histone deacetylase 3 couples mitochondria to drive IL-1beta-dependent inflammation by configuring fatty acid oxidation. Mol Cell. 2020;80(1):43-58.e7.
Weinberg SE, Chandel NS. Targeting mitochondria metabolism for cancer therapy. Nat Chem Biol. 2015;11(1):9–15.
Sriskanthadevan S, Jeyaraju DV, Chung TE, Prabha S, Xu W, Skrtic M, et al. AML cells have low spare reserve capacity in their respiratory chain that renders them susceptible to oxidative metabolic stress. Blood. 2015;125(13):2120–30.
Xiong Y, Guan KL. Mechanistic insights into the regulation of metabolic enzymes by acetylation. J Cell Biol. 2012;198(2):155–64.
Wang C, Cui G, Wang D, Wang M, Chen Q, Wang Y, et al. Crosstalk of oxidative phosphorylation-related subtypes, establishment of a prognostic signature and immune infiltration characteristics in colorectal adenocarcinoma. Cancers (Basel). 2022;14(18):4503.
Chen W, Yang Z, Chen Y. A novel oxidative phosphorylation-associated gene signature for prognosis prediction in patients with hepatocellular carcinoma. Dis Markers. 2022;2022:3594901.
Luna-Yolba R, Marmoiton J, Gigo V, Marechal X, Boet E, Sahal A, et al. Disrupting mitochondrial electron transfer chain complex i decreases immune checkpoints in murine and human acute myeloid leukemic cells. Cancers (Basel). 2021;13(14):3499.
Wang J, Fang Y, Ma S, Su N, Zhang Y, Huang H, et al. Comparison of chidamide-contained treatment modalities versus chemotherapy in the second-line treatment for relapsed or refractory peripheral T-cell lymphoma. Leuk Res. 2021;111: 106705.
Li Y, Wang Y, Zhou Y, Li J, Chen K, Zhang L, et al. Cooperative effect of chidamide and chemotherapeutic drugs induce apoptosis by DNA damage accumulation and repair defects in acute myeloid leukemia stem and progenitor cells. Clin Epigenetics. 2017;9:83.
Wang L, Luo J, Chen G, Fang M, Wei X, Li Y, et al. Chidamide, decitabine, cytarabine, aclarubicin, and granulocyte colony-stimulating factor (CDCAG) in patients with relapsed/refractory acute myeloid leukemia: a single-arm, phase 1/2 study. Clin Epigenetics. 2020;12(1):132.
Acknowledgements
We thank Dr. Shou-Jie Wang from Center for Precision Medicine, SYSU Platform of Metabolomics for the assistance with Seahorse XF Cell Mito Stress experiment.
Funding
This study was supported by Talent Training Project of The Third Affiliated Hospital of Sun Yat-sen University.
Author information
Authors and Affiliations
Contributions
JYW and ZJL: conceived and designed the experiments. JDW and JQX: performed the experiments. JDW analyzed the data and wrote the article. All authors revised the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Informed consent
Not applicable.
Ethical approval and Research involving human participants and/or animals
The animal study was conducted with the approval of Institutional Animal Care and Use Committee.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
12094_2023_3079_MOESM1_ESM.tif
Supplementary file1 Effects of different metabolic inhibitors with chidamide. AML cells were treated chidamide (1 μM) or/and FCCP (1 µM/2 µM) (A, E), Oligomycin (1 µM/2 µM) (B, F), Rotenone (0.5 µM/1 µM) (C, G) or Metfoemin (5 mM/10 mM) (D, H) for 24 h and CCK-8 assay was applied to detect cell viability. Data were presented as mean ± SD, *p<0.05, **p<0.01, ***p<0.001 (TIF 9851 KB)
12094_2023_3079_MOESM2_ESM.tif
Supplementary file2 Relation between HDAC3 and immune cell infiltration is performed. (A) Immune cell enrichment was performed in HDAC3 high and low AML patients by ssGSEA. (B) Unsupervised clustering of immune checkpoint genes and infiltrating cells were analyzed in HDAC3 high and low groups. (C) Correlation between each infiltrating cell signature was assessed by spearman analysis. (D) Infiltrating cell infiltration abundance analysis in HDAC3 high and low groups was shown (TIF 32323 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, JD., Xu, JQ., Long, ZJ. et al. Disruption of mitochondrial oxidative phosphorylation by chidamide eradicates leukemic cells in AML. Clin Transl Oncol 25, 1805–1820 (2023). https://doi.org/10.1007/s12094-023-03079-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-023-03079-8