Abstract
Background
Recent studies have shown that the activation of PI3K/AKT signaling pathway is an essential molecular mechanism participating in trastuzumab resistance in HER2 + GC (gastric cancer). However, how can we effectively inhibit AKT activity associated with drug resistance during trastuzumab treatment? Screening inhibitors against the upstream receptors of PI3K/AKT signaling pathway or interacting proteins of members has become an important way.
Methods
In this study, western blot, qRT-PCR, CCK8, Co-IP and other techniques were used to explore possible mechanisms participating in trastuzumab resistance in vitro. Besides, the xenograft mouse model and GC tissue samples from patients were used to further validate the in-vitro results.
Results
The expression of XB130 adaptor protein was remarkably increased in GC cell lines resistant to trastuzumab, and knockdown of XB130 could reverse the resistance via downregulating p-AKT. In addition, p-SRC (Tyr416) was increased in resistant cells, which could facilitate the binding of XB130 to PI3K p85α. It was also discovered that XB130 could negatively regulate PTEN gene transcription, and thus a positive feedback loop was formed between SRC-XB130-PTEN.
Conclusions
In HER2 + GC, XB130 contributes to trastuzumab resistance by stimulating the PI3K/AKT signaling pathway through binding to PI3K p85α under the mediation of SRC kinase and regulating PTEN gene transcription, and in turn forming a positive feedback loop between SRC-XB130-PTEN.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
GC (gastric cancer) is a prevalent tumor in China, with the 3rd highest incidence and mortality rate as shown by the latest data [1]. As more studies focus on molecular biology, molecular targeted therapy for GC has aroused scholars’ interest, among which human epidermal growth factor receptor 2, abbreviated as HER2, was the first target found to be effective [2]. Trastuzumab, a monoclonal humanized antibody against HER2, can attenuate tumor cell growth via antagonizing delivery of the HER2 signaling pathway [3]. In clinical practice, trastuzumab treatment significantly prolonged the overall survival of patients suffering from HER2 + metastatic GC, however, most patients developed drug resistance within 1 year of effective treatment [4].
XB130, which belongs to the actin filament-related protein family, is a newly identified junctional protein [5]. It contains two PH domains and multiple binding sites for SH2 and SH3 domains, which can activate tumor-related PI3K/AKT and other signals pathway. At the same time, it is also activated by phosphorylation of various tyrosine kinases, such as HER2, EGFR (epidermal growth factor receptor) and SRC [6,7,8]. Therefore, it is widely involved in the differentiation, proliferation, invasion and metastasis of tumor cells [9, 10], which is considered a potential molecular marker for the diagnosis and treatment of tumors [10,11,12].
In our study, it was found that XB130 mediated the development of trastuzumab resistance in GC by binding to PI3K p85α and modulating the PI3K/AKT pathway under the mediation of SRC kinase. Moreover, it was also observed that XB130 formed a positive feedback loop between SRC-XB130-PTEN by regulating PTEN gene transcription and regulated trastuzumab resistance. Taken together, the results indicated that XB130 was a potential biomarker in monitoring the course of HER2 + gastric cancer and a novel target in ameliorating the resistance to trastuzumab.
Materials and methods
Cell lines and cell culture
NCI-N87 and MKN45 cell lines (human gastric cancer cells) were provided by ATCC (American Type Culture Collection). RPMI 1640 (Hyclone) which was added with 10% FBS (Hyclone) and 1% streptomycin-penicillin (Hyclone) was used to culture the cells in an incubator (5% CO2, 37 ℃). Cells were collected for subsequent experiments when logarithmic growth phase was reached.
Establishment of TRNCs (trastuzumab-resistant NCI-N87 cells) and TRMCs (trastuzumab-resistant MKN45 cells)
TRNCs and TRMCs were established based on the procedures mentioned in our previous research [13, 14].
Chemicals and antibodies
The antibodies against the proteins listed below were applied in our research: XB130 (#12,796, CST, USA), PTEN (#9188, CST, USA), PI3K p85α (#4257, CST, USA), AKT (ab38449, Abcam, USA), phosphorylation-AKT (Ser473) (#4060, CST, USA), SRC (ab133283, Abcam, USA), phosphorylation-SRC (Tyr416 and Tyr527) (#59,548, #2105, CST, USA), and GAPDH (#5174, CST, USA). Nuclei or mitochondria were labeled by using 4,6-diamidino-2-phenylindole or MitoTracker Red CMXRos (provided by Invitrogen, Carlsbad, America) staining. Saracatinib, SRC inhibitor, and Trastuzumab were provided by Calbiochem (located in Selleck Chemicals, America) and Roche company, respectively.
Myr-AKT plasmids
Active AKT expression plasmids were created by cloning the cDNA into pCAGGS-IRESEGFPpA vectors. The cDNA encoded Myr-AKT (myristoylated-human AKT) without the PH domain.
Transient transfection
XB130 shRNA (small hairpin RNA), XB130-OE (Over Expressed), control vectors, pGL-base-PTEN, PTEN shRNA, PTEN-wt, PTEN-mut and NC (negative control) were generated by GenePharma (Shanghai, China). The cells were then transiently transfected with the recombinant plasmids mentioned above by Lipofectamine™ 3000 reagent (provided by Invitrogen, CA, America) following the instructions, when cell confluence reached 50–60%. Into a 6-well plate the cells were seeded (3 × 105 cells in each well) containing the culture medium added with 10% FBS for twenty-four hours prior to drug treatment.
Establishing stable cell lines after transfection
Lentiviral vectors (10 µl) added with control vectors and XB130 shRNA were used for transfection. Puromycin at a minimum lethal dose (0.5 mg/ml) was used to screen the cells which were stably transfected at 48 h post infection. The transfection efficiency was confirmed by western blot.
Cell viability assay
Into a 96-well plate cells were inoculated (3 × 103 cells in each well) and added with trastuzumab at certain concentrations. After 72 h incubation, we determined cell viability by the CCK8 reagent following the protocol. The optical density was detected by using the spectrophotometer (Thermo Electron Corporation, MA, USA) at a wavelength of 450 nm. The experiments were conducted in triplicate.
Quantitative real-time PCR
Total cellular RNAs were obtained using the Trizol reagent (Invitrogen, Carlsbad, CA, USA). RNA was eluted with DEPC water, and the concentration was quantified using a NanoDrop system (NanoDrop; Thermo Fisher Scientific). Then, 1.5 µg RNA was reversed into cDNA in a total reaction system of 10 µl using cDNA reverse transcriptase kit (Applied Biosystems) and qRT-PCR was conducted in triplicates using 10 μl of SYBR Master Mixture (Applied Biosystems). The following thermal cycling conditions were used: initial denaturation at 95 °C for 3 min; 40 cycles of denaturation at 95 °C for 30 s, annealing at 58 °C for 30 s and extension at 72 °C for 45 s; Finally, it was extended at 72 °C for 10 min. (The primers are listed in Supplementary Table 1). GAPDH was used as the internal control to normalize the raw data. The 2−∆∆Cq method [15] was used for data analysis, and the results are presented as the fold changes in the relative mRNA expression.
Total tissue RNAs were isolated from the tumor samples by Trizol according to the manufacturer's instructions.
Western blot analysis
The total cellular protein was collected by using RIPA protein lysate (Sigma-Aldrich). BCA protein assay kit (Thermo Scientific) was used to determine protein concentrations. SDS-PAGE gels (10% gels) were used to separate the proteins, followed by transferring onto PVDF (polyvinylidene fluoride) membranes (provided by Millipore). After being blocked by 5% nonfat milk (Sangon Biotech) in TBS added with 0.05% Tween-20, primary antibodies (XB130; #12,796, CST; 1:1000; PTEN; #9188, CST; 1:1000; PI3K p85α; #4257, CST; 1:1000; AKT; ab38449, Abcam; 1:1000; phosphorylation-AKT (Ser473); #4060, CST; 1:1000; SRC; ab133283, Abcam; 1:1000; phosphorylation-SRC (Tyr416 and Tyr527); #59,548, #2105, CST; 1:1000; GAPDH; #5174, CST; 1:1000;) were incubated with the membranes at 4 °C overnight. Then, the membranes were incubated with HRP-linked Goat anti-Rabbit secondary antibodies (1:6000; Boster) for 1 h. Protein bands were observed using commercial ECL kit (provided by Beyotime, China).
Co-IP (co-immunoprecipitation)
Co-IP assays were conducted using the Pierce Co-Immunoprecipitation Kit (Z-CHIP, MILLIPORE). Each 1.5 ml EP tube containing cells was added with pre-cooled equal volume IP cell lysis buffer, which were then lysed on ice for 10 min and on a mixer at 4 °C for 30 min. Then, the lysate was centrifuged at 13,000 g at 4 °C for 10 min, and the supernatant (50 µl) was collected as Input and another 50 µl was collected into a new 1.5 ml EP tube for Co-IP experiment. A proper amount of Anti-Flag magnetic beads (M8823, Sigma-Aldrich) were rinsed with PBST for 3 times and then with PBS for 1 time. The magnetic beads were added into the supernatant and incubated at 4 °C on a mixer overnight. On the next day, the supernatant was placed on a magnetic separator, and the magnetic beads were rinsed with lysis buffer for 3 times, and then with Flag elution buffer for 15 min. The magnetic separator separated the magnetic beads from the liquid, and the liquid was sucked into a new Ep tube (1.5 ml) to obtain the eluent. The IP product was added with loading buffer, mixed, boiled in boiling water for 10 min, and then centrifuged at 13,000 g for 3 min at 4 °C for western blot. Rabbit IgG was applied as internal control.
Dual-luciferase reporter assay
Plasmid pGL-base-PTEN was transiently transfected into TRNCs XB130 shRNA or NC cells. After 48 h, the luciferase activity was measured according to the instructions of luciferase reporter gene kit. pRL-CMV(Renilla) was used as the internal reference for co-transfection.
Nude mice cancer xenograft model
We strictly abided by the Guidelines for the Care and Use of Laboratory Animals (NIH publication no. 80–23, revised in 1996) and the institutional ethical guidelines for animal experimentation in all animal experiments. The animals were randomly grouped, and investigators knew nothing about grouping details throughout the research. The same athymic nude BALB/C mice (female, 5–6 weeks) were given subcutaneous injection of 5 × 106 NCI-N87, TRNCs, TRNCs shXB130, or stable control cells. When the xenograft’s volume was ~ 50 mm3, trastuzumab (20 mg/kg) was injected into the mice through the tail vein 2 times per week for 6 weeks (n = 5). The size of the tumor was calculated by using W (width) and L (length) measured by the calipers. The formula was (L × W2) × 0.5. This research obtained approval from the Animal Care and Ethics Committee, Southern Medical University.
IHC (immunohistochemistry)
Immunohistochemistry assays were conducted as depicted in the previous article [13]. Dako Envision System (provided by Dako, Glostrup, Denmark) was used to conduct IHC staining.
Patient data
Tissue samples were collected from 12 patients who underwent PD (progressive disease) of HER2 + gastric cancer after trastuzumab treatment between April 2019 and October 2020 in the department of oncology, Nanfang hospital. The age of patients (n = 12; female, n = 5; male, n = 7) was between 39 and 67. Tumor-node-metastasis (TNM) was staged according to pathological reports and imaging examinations. The study was approved by the ethics committee of the Nanfang hospital. And the patients provided oral informed consent for future research when collecting tissues.
Statistical analysis
Difference was compared by one-way ANOVA or Student's t-test (two-tailed) using SPSS 17.0 (bought from SPSS Inc., America) and GraphPad Prism (from GraphPad Software, Inc., America). The data were presented as mean ± SD (standard deviation). P < 0.05 indicated significant difference.
Results
XB130 mediated the resistance to trastuzumab of HER2 + GC cells in vitro
According to our previous research, NCI-N87 and MKN45 expressing a high level of HER2 were used to establish TRNCs and TRMCs through exposing the cells to increasing concentrations of trastuzumab [13, 14]. As shown by the curve of concentration effects, TRNCs and TRMCs were less sensitive to the treatment of trastuzumab in comparison with NCI-N87 and MKN45, with a resistance index RI of 9.3 and 6.2 (RI = IC50 of resistant cells / IC50 of parent cells), respectively (Fig. 1a). However, in comparison with the parent cells, XB130 expression levels were remarkably increased in the resistant cells (Fig. 1b, c). In the presence of trastuzumab, XB130 and p-AKT protein expressions were greatly down-regulated in NCI-N87 and MKN45, while those in the resistant cells remained significantly elevated (Fig. 1d). Afterwards, to detect if XB130 was a modulating factor of the resistance of HER2 + gastric cancer to trastuzumab, XB130 was silenced by transfecting shRNA into TRNCs and TRMCs (Fig. 1e). It was found that p-AKT protein expression was remarkably decreased (Fig. 1e), and trastuzumab sensitivity was significantly up-regulated (Fig. 1f).
XB130 mediated the resistance to trastuzumab of HER2 + GC cells in vitro. a NCI-N87 cells, MKN45 cells, TRNCs and TRMCs after 72 h trastuzumab treatment indicated by CCK8 assay. b XB130 levels in NCI-N87 cells, MKN45 cells, TRNCs and TRMCs determined by qRT-PCR. c p-AKT, AKT and XB130 expressions in NCI-N87 cells, MKN45 cells, TRNCs and TRMCs determined by western blot. d AKT, p-AKT and XB130 expression in NCI-N87 cells, MKN45 cells, TRNCs and TRMCs after trastuzumab (20 µg/ml) treatment for 0, 24, 48 h determined by western blot. e Expressions of XB130, AKT, p-AKT in TRNCs and TRMCs after transfection of control or XB130 shRNA determined by western blot. f TRNCs and TRMCs after they were transfected with XB130 shRNA or control followed by trastuzumab treatment for 72 h, as shown by CCK8 assay. Error bars indicated SD (***p < 0.001)
To figure out the function of XB130 in trastuzumab resistance in HER2 + gastric cancer cells, we transfected colonies of ectopic XB130-OE and XB130 shRNA and the controls into NCI-N87 and MKN45 cells. It was observed that the increased XB130 remarkably decreased cell viability, while the opposite result was observed in the low expression group (Fig. S1). The results suggested that XB130 could result in the resistance of HER2 + gastric cancer cells to trastuzumab.
XB130 mediated trastuzumab resistance through binding with PI3K p85α induced by SRC kinase
It was confirmed that XB130 mediated the resistance of HER2 + gastric cancer cells to trastuzumab, and we sought to define the molecular mechanisms. XB130 can regulate the PI3K/AKT signaling pathway by binding to the regulatory subunit p85α of PI3K [16]. SRC, as a member in the tyrosine kinases that mediate the activation of XB130, plays a key part in promoting the binding of XB130 to PI3K p85α [6,7,8]. As expected, the p-SRC (Tyr416) protein level was remarkably higher in trastuzumab-resistant cells in comparison with parent cells (Fig. 2a). Meanwhile, the Co-IP assay suggested that the binding of XB130 to PI3K p85α was dramatically enhanced in the cells resistant to trastuzumab (Fig. 2b). To further confirm the effect of SRC, TRNCs and TRMCs were treated with SRC inhibitor saracatinib. The results demonstrated that the binding level of XB130 to PI3K p85α was dramatically decreased (Fig. 2c). To ensure the regulatory pathway “XB130-PI3K/AKT-trastuzumab resistance”, Myr-AKT (a constitutively activator of AKT) and XB130 shRNA were used in the co-transfection of TRNCs and TRMCs. The expression of p-AKT was reduced when the XB130 expression was suppressed by shRNA, but Myr-AKT could reverse this effect (Fig. 2d). Moreover, Myr-AKT restored the sensitivity to trastuzumab (Fig. 2e). These results implied that XB130 bound to PI3K p85α under the mediation of SRC kinase and caused the activation of PI3K/AKT pathway, which in turn induced the resistance of HER2 + gastric cancer cells to trastuzumab.
XB130 mediated trastuzumab resistance through binding to PI3K p85α induced by SRC kinase. a p-SRC (Tyr416), p-SRC (Tyr527), SRC in NCI-N87 cells, MKN45 cells, TRNCs and TRMCs determined by western blot. b The binding level of XB130 to p58α in NCI-N87 cells, MKN45 cells, TRNCs and TRMCs detected by Co-IP assay. c The binding level between XB130 and p58α in TRNCs and TRMCs with or without SRC inhibitor saracatinib detected by Co-IP assay. d The expression of XB130, AKT, p-AKT after transfection of XB130 shRNA with or without Myr-Akt into TRNCs and TRMCs prior to 72 h trastuzumab (20 µg/ml) treatment. e TRNCs and TRMCs post transfection of XB130 shRNA with or without Myr-Akt prior to 72 h trastuzumab treatment, as shown by CCK8 assay. Error bars indicated SD
XB130 mediated trastuzumab resistance by forming a positive feedback loop between SRC-XB130-PTEN through regulating PTEN gene transcription
The PI3K/AKT signaling pathway can be continuously activated due to PTEN (phosphatase and tensin homologue) deficiency, which induces the development of trastuzumab resistance in GC [17, 18]. It was observed that the PTEN expression level was significantly lower in trastuzumab-resistant GC cells (Fig. 3a, b), while PTEN was remarkedly increased when resistant cells were treated with saracatinib (Fig. 3c). In addition, we found that XB130 might bind to the GAGCAA site of the PTEN promoter sequence through bioinformatics (Fig. 3d, Fig. S2). Moreover, luciferase assay results suggested that XB130 negatively regulated PTEN gene transcription in TRNCs (Fig. 3e). Next, we transfected TRNCs and TRMCs with PTEN-wt or PTEN-mut which resulted in significant down-regulation of p-SRC and p-AKT and increased sensitivity to trastuzumab in the PTEN-wt group, while there was no significant change in the PTEN-mut group (Fig. 3f, g). Also, when XB130 shRNA and PTEN shRNA were transiently co-transfected into the resistant cells, it was observed that PTEN shRNA enhanced the activity of SRC kinase and the trastuzumab resistance recovered (Fig. 3h, i). These results confirmed that there was a positive feedback effect between SRC-XB130-PTEN, and XB130 participated in the modulation of resistance of GC to trastuzumab via regulating PTEN gene transcription.
XB130 mediated PTEN transcription and formed a positive feedback loop between SRC-XB130-PTEN. a PTEN levels in NCI-N87 cells, MKN45 cells, TRNCs and TRMCs detected by qRT-PCR. b PTEN in NCI-N87 cells, MKN45 cells, TRNCs and TRMCs determined by western blot. c PTEN in TRNCs and TRMCs after treated with or without SRC inhibitor saracatinib determined by western blot. d Bioinformatics was used to predict whether XB130 binds to the promoter region of PTEN. It was found XB130 may bind to the GAGCAA site of the PTEN promoter sequence. e After 48 h transfection of XB130 shRNA/NC and pGL-base-PTEN into TRNCs, we determined the luciferase activity using luciferase reporter assay. f The expression of PTEN, XB130, p-SRC(Tyr416), SRC, p-AKT, AKT of TRNCs and TRMCs after transfection with PTEN-wt or PTEN-mut detected by western blot. g CCK8 analysis of TRNCs and TRMCs transfected with PTEN-wt or PTEN-mut after 72 h trastuzumab treatment. h The expression of XB130, PTEN, p-SRC(Tyr416), SRC, p-AKT, AKT of TRNCs and TRMCs after transfection of XB130 shRNA with or without PTEN shRNA/NC detected by western blot. i CCK8 assay showed that TRNCs and TRMCs transfected of XB130 shRNA with or without PTEN shRNA/NC after 72 h trastuzumab treatment. Error bars indicated SD (**p < 0.01, ***p < 0.001)
XB130 mediated resistance of HER2 + gastric cancer to trastuzumab in vivo
To further verify the involvement of XB130 in regulating trastuzumab resistance in GC in vivo, we constructed tumor-bearing nude mouse models of NCI-N87, TRNCs, TRNCs shXB130, or stable control cells. It was demonstrated the tumor volumes were larger in TRNCs group in comparison with NCI-N87 group (Fig. 4a). XB130 and p-SRC levels were significantly increased and PTEN significantly down-regulated in tumor tissues under the treatment with trastuzumab, whereas the results were contrary in TRNCs shXB130 group (Fig. 4a, b).
XB130 mediated the resistance of HER2 + gastric cancer to trastuzumab in vivo. a Subcutaneous xenograft assay of NCI-N87, TRNCs, TRNCs shXB130, or stable control cells in nude mice after subcutaneously injected with trastuzumab for certain days. The tumor volume was calculated (n = 5 per group). b Representative immunohistochemistry images of tumor sections from different groups (n = 5 per group). Scale bar: 50 μm. Error bars indicated SD (**p < 0.01)
XB130 mediated the resistance to trastuzumab in patients with HER2 + gastric cancer
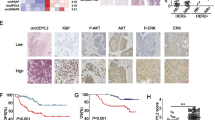
We evaluated 12 pair of tissue samples collected from the cases with PD of HER2 + gastric cancer after trastuzumab was prescribed according to RECIST 1.1 (response evaluation criteria in solid tumors version 1.1). It was shown that XB130 (Fig. 5a, b) and p-SRC (Fig. 5b) were upregulated and PTEN (Fig. 5b) was downregulated in patients with PD. These results indicated that XB130 might be correlated with the resistance of cases with HER2 + gastric cancer to trastuzumab.
XB130 mediated resistance of cases with HER2 + gastric cancer to trastuzumab. a XB130 mRNA levels in tissue samples derived from patients suffering from progressive disease of HER2 + GC pre or post trastuzumab treatment (n = 12 in each group). b Representative immunohistochemistry images of tumor sections from cases with HER2 + gastric cancer pre or post treatment with trastuzumab (n = 12 per group). Scale bar: 50 μm. c A schematic diagram showing XB130 based signaling circuit of resistance to trastuzumab in HER2 + GC: XB130 adaptor protein regulated trastuzumab resistance by stimulating PI3K/AKT signaling pathway in HER2 + GC. XB130 bound with PI3K p85α, which was induced by SRC kinase. XB130 formed a positive feedback loop between SRC-XB130-PTEN by regulating PTEN gene transcription. Error bars indicated SD (**p < 0.01)
A pattern diagram indicating the role of XB130 in HER2 + gastric cancer cells was depicted (Fig. 5c). The molecular mechanisms of trastuzumab resistance might be that XB130 activates the PI3K/AKT signaling pathway through binding to PI3K p85α medicated by SRC kinase and forming a positive feedback loop between SRC-XB130-PTEN by regulating PTEN gene transcription.
Discussion
At present, trastuzumab provides a novel direction for treating GC. However, drug resistance is still the most common and biggest obstacle in clinical application of trastuzumab, and it is also the most urgent problem to improve the survival of cases with advanced gastric cancer. Thus, it is of vital importance to explore the mechanism and countermeasures of trastuzumab resistance and to find predictive markers indicating the resistance to trastuzumab. Recent studies have suggested that activated PI3K/AKT signaling pathway is a main molecular mechanism related to the resistance to trastuzumab in HER2 + GC [5, 6, 19, 20]. In our study, we aimed to find the inhibitors of PI3K/AKT signaling pathway upstream receptor or interacting proteins of members.
XB130, also known as PI3KAP (phosphatidylinositol 3-kinase-associated protein), is a novel adaptor protein, which belongs to the family of actin filament-related protein. The full length of human XB130 gene is 375 bp, encoding a 130 kDa protein containing 818 amino acids, which is highly expressed in thyroid and spleen, but rarely expressed in other organs [5]. As a typical adaptor protein, XB130 structurally contains two PH domains and multiple binding sites of SH2 and SH3 domains, which can activate many signal pathways such as tumor-related PI3K/AKT, etc. At the same time, XB130 is also activated by many tyrosine kinases such as HER2, EGFR and SRC. In recent years, we have intensively investigated the biological role and molecular mechanisms of XB130 in GC. It was reported for the first time that XB130 was an independent risk factor related to the prognosis of GC patients [21]. Silenced XB130 with down-regulated p-AKT expression by shRNA could lead to decreased invasion, proliferation, and migration ability of GC cells, indicating that XB130 was a potential therapeutic target for GC [22]. In this study, we demonstrated that XB130 and p-AKT expression levels in trastuzumab-resistant GC cells were higher compared with parent cells. After XB130 was silenced by shRNA, p-AKT expression was remarkably decreased and the sensitivity to trastuzumab was markedly enhanced in the cells resistant to trastuzumab. These results indicated that XB130 participated in modulating the resistance of HER2 + gastric cancer to trastuzumab via the activation of AKT.
XB130 regulates PI3K/AKT signaling pathway via binding to the upstream member PI3K regulatory subunit p85α. Tyrosine kinase-mediated phosphorylation of the N-terminal YxxM motif of XB130 protein is the key for XB130 binding to p85α subunit [16]. It was reported that XB130 regulated the expression and phosphorylation levels of PI3K/AKT signaling pathway and its downstream target genes through binding to PI3K p85α and ultimately regulated tumor cell survival and proliferation, in which SRC kinases played a key part in promoting the binding of XB130 to PI3K p85α [6,7,8]. SRC, the first identified proto-oncogene, belongs to the non-receptor tyrosine kinase family and regulates tumor cell survival, differentiation, invasion, metastasis and drug resistance by interacting with multiple tumor-related signaling pathways [23,24,25,26,27]. Increased research has shown SRC kinase activation is related to the resistance of gastric cancer to trastuzumab, but the molecular mechanism of its regulation is not clear [25,26,27]. Our results indicated that the expression of p-SRC (Tyr416) protein was significantly elevated and facilitated the binding of XB130 to PI3K p85α in trastuzumab-resistant cells. In addition, it was also found that XB130 regulated the development of trastuzumab resistance by activating AKT through binding to PI3K p85α under the mediation of SRC kinase in GC.
PTEN gene is the first oncogene with dual specificity for phosphatase activity identified to date [28,29,30]. Accumulating research has shown that PTEN is highly correlated with the tumor development of various cancers [31,32,33,34,35]. The main substrate of PTEN is PIP3, which is maintained at a low level by dephosphorizing PIP3, thereby inhibiting the activated PI3K/AKT signaling pathway [36, 37]. It was reported that PTEN deficiency attenuated PI3K/AKT signaling pathway activation and induced resistance of gastric cancer to trastuzumab [17, 18]. In addition, the results of Zhang S et al. confirmed that PTEN directly and specifically dephosphorylated pTyr416 of SRC through its protein phosphatase activity, thereby inhibiting SRC kinase activation [25]. Our results showed that PTEN levels were remarkably reduced in trastuzumab-resistant cells and XB130 negatively regulated the transcription of PTEN gene. Moreover, it was also found that PTEN inhibited the activation of SRC kinase which in turn formed a positive feedback loop between SRC-XB130-PTEN and finally induced trastuzumab resistance through the activation of PI3K/AKT signaling pathway.
In conclusion, this research demonstrated that XB130 could activate PI3K/AKT signaling pathway through binding with PI3K p85α and regulating PTEN gene transcription, which finally mediated the development of trastuzumab resistance in GC. Our findings revealed the molecular mechanism of XB130 regulating trastuzumab resistance in GC at three levels: cellular, mouse and clinical samples. Thus, we obtained reliable evidence of XB130 as a molecular marker of trastuzumab resistance and laid a molecular foundation for the study of new targets for reversing the resistance of HER2 + gastric cancer to trastuzumab.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- Co-IP:
-
Co-immunoprecipitation
- EGFR:
-
Epidermal growth factor receptor
- GC:
-
Gastric cancer
- HER2:
-
Human epidermal growth factor receptor 2
- IHC:
-
Immunohistochemistry
- NC:
-
Negative control
- OE:
-
Over expressed
- PD:
-
Progressive disease
- PI3KAP:
-
Phosphatidylinositol 3-kinase-associated protein
- PTEN:
-
Phosphatase and tensin homologue
- shRNA:
-
Small hairpin RNA
- TRNCs:
-
Trastuzumab-resistant NCI-N87 cells
- TRMCs:
-
Trastuzumab-resistant MKN45 cells
References
Chen WQ, Li H, Sun KX, Zheng RS, Zhang SW, Zeng HM, et al. Cancer incidence and mortality in China, 2014. Chin J Cancer Res. 2018;30(1):1–12.
Selim JH, Shaheen S, Sheu WC, Hsueh CT. Targeted and novel therapy in advanced gastric cancer. Exp Hematol Oncol. 2019;11(8):25.
Hudis CA. Trastuzumab–mechanism of action and use in clinical practice. N Engl J Med. 2007;357(1):39–51.
Kelly CM, Janjigian YY. The genomics and therapeutics of HER2-positive gastric cancer-from trastuzumab and beyond. J Gastrointest Oncol. 2016;7(5):750–62.
Xu J, Bai XH, Lodyga M, Han B, Xiao H, Keshavjee S, et al. XB130, a novel adaptor protein for signal transduction. J Biol Chem. 2007;282(22):16401–12.
Bai XH, Cho HR, Moodley S, Liu M. XB130-a novel adaptor protein: gene, function, and roles in tumorigenesis. Scientifica. 2014;2014:1–9.
Zhang R, Zhang J, Wu Q, Meng F, Liu C. XB130: a novel adaptor protein in cancer signal transduction. Biomed Rep. 2016;4(3):300–6.
Shiozaki A, Shen-Tu G, Bai X, Iitaka D, De Falco V, Santoro M, et al. XB130 mediates cancer cell proliferation and survival through multiple signaling events downstream of Akt. PLoS ONE. 2012;7(8): e43646.
Cho HR, Wang Y, Bai X, Xiang YY, Lu C, Post A, et al. XB130 deficiency enhances carcinogen-induced skin tumorigenesis. Carcinogenesis. 2019;40(11):1363–75.
Shen J, Jin C, Liu Y, Rao H, Liu J, Li J. XB130 enhances invasion and migration of human colorectal cancer cells by promoting epithelial-mesenchymal transition. Mol Med Rep. 2017;16(4):5592–8.
Zhang J, Jiang X, Zhang J. Prognostic significance of XB130 expression in surgically resected pancreatic ductal adenocarcinoma. World J Surg Oncol. 2014;1(12):49.
Chen B, Liao M, Wei Q, Liu F, Zeng Q, Wang W, et al. XB130 is overexpressed in prostate cancer and involved in cell growth and invasion. Oncotarget. 2016;7(37):59377–87.
Zuo Q, Liu J, Zhang J, Wu M, Guo L, Liao W. Development of trastuzumab-resistant human gastric carcinoma cell lines and mechanisms of drug resistance. Sci Rep. 2015;25(5):11634.
Liu J, Pan C, Guo L, Wu M, Guo J, Peng S, et al. A new mechanism of trastuzumab resistance in gastric cancer: MACC1 promotes the Warburg effect via activation of the PI3K/AKT signaling pathway. J Hematol Oncol. 2016;9(1):76.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–8.
Lodyga M, De Falco V, Bai XH, Kapus A, Melillo RM, Santoro M, et al. XB130, a tissue-specific adaptor protein that couples the RET/PTC oncogenic kinase to PI 3-kinase pathway. Oncogene. 2009;28(7):937–49.
Kim C, Lee CK, Chon HJ, Kim JH, Park HS, Heo SJ, et al. PTEN loss and level of HER2 amplification is associated with trastuzumab resistance and prognosis in HER2-positive gastric cancer. Oncotarget. 2017;8(69):113494–501.
Deguchi Y, Okabe H, Oshima N, Hisamori S, Minamiguchi S, Muto M, et al. PTEN loss is associated with a poor response to trastuzumab in HER2-overexpressing gastroesophageal adenocarcinoma. Gastric Cancer. 2017;20(3):416–27.
Díaz-Serrano A, Angulo B, Dominguez C, Pazo-Cid R, Salud A, Jiménez-Fonseca P, et al. Genomic profiling of HER2-positive gastric cancer: PI3K/Akt/mTOR pathway as predictor of outcomes in HER2-positive advanced gastric cancer treated with Trastuzumab. Oncologist. 2018;23(9):1092–102.
Noorolyai S, Shajari N, Baghbani E, Sadreddini S, Baradaran B. The relation between PI3K/AKT signalling pathway and cancer. Gene. 2019;25(698):120–8.
Shi M, Huang W, Lin L, Zheng D, Zuo Q, Wang L, et al. Silencing of XB130 is associated with both the prognosis and chemosensitivity of gastric cancer. PLoS ONE. 2012;7(8): e41660.
Shi M, Zheng D, Sun L, Wang L, Lin L, Wu Y, et al. XB130 promotes proliferation and invasion of gastric cancer cells. J Transl Med. 2014;4(12):1.
Espada J, Martín-Pérez J. An update on Src family of nonreceptor tyrosine kinases biology. Int Rev Cell Mol Biol. 2017;331:83–122.
Roskoski R. Src protein-tyrosine kinase structure, mechanism, and small molecule inhibitors. Pharmacol Res. 2015;94:9–25.
Zhang S, Huang WC, Li P, Guo H, Poh SB, Brady SW, et al. Combating trastuzumab resistance by targeting SRC, a common node downstream of multiple resistance pathways. Nat Med. 2011;17(4):461–9.
Han S, Meng Y, Tong Q, Li G, Zhang X, Chen Y, et al. The ErbB2-targeting antibody trastuzumab and the small-molecule SRC inhibitor saracatinib synergistically inhibit ErbB2-overexpressing gastric cancer. MAbs. 2014;6(2):403–8.
Jin MH, Nam AR, Park JE, Bang JH, Bang YJ, Oh DY. Resistance mechanism against trastuzumab in HER2-positive cancer cells and its negation by Src inhibition. Mol Cancer Ther. 2017;16(6):1145–54.
Chen CY, Chen J, He L, Stiles BL. PTEN: tumor suppressor and metabolic regulator. Front Endocrinol (Lausanne). 2018;9(9):338.
Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275(5308):1943–7.
Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23 3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15(4):356–62.
Leslie NR, Foti M. Non-genomic loss of PTEN function in cancer: not in my genes. Trends Pharmacol Sci. 2011;32(3):131–40.
Silva A, Yunes JA, Cardoso BA, Martins LR, Jotta PY, Abecasis M, et al. PTEN posttranslational inactivation and hyperactivation of the PI3K/Akt pathway sustain primary T cell leukemia viability. J Clin Invest. 2008;118(11):3762–74.
Trotman LC, Wang X, Alimonti A, Chen Z, Teruya-Feldstein J, Yang H, et al. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell. 2007;128(1):141–56.
Wang X, Trotman LC, Koppie T, Alimonti A, Chen Z, Gao Z, et al. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell. 2007;128(1):129–39.
Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147(2):344–57.
Haddadi N, Lin Y, Travis G, Simpson AM, Nassif NT, McGowan EM. PTEN/PTENP1: Regulating the regulator of RTK-dependent PI3K/Akt signaling, new targets for cancer therapy. Mol Cancer. 2018;17(1):37.
Hawkins PT, Anderson KE, Davidson K, Stephens LR. Signalling through Class I PI3Ks in mammalian cells. Biochem Soc Trans. 2006;34(Pt 5):647–62.
Acknowledgements
Not applicable.
Funding
This work was supported by Natural Science Foundation of Guangdong Province (2018A030313558) and National Natural Science Foundation of China (81872009).
Author information
Authors and Affiliations
Contributions
SNY, BBW and JQL conceived this study, conducted experiments and analyzed data; ZYH, XXZ, SLC and ZQW performed experiments and curated the data; SNY, BBW, JQL, XYZ and QZ wrote, reviewed and revised papers; QZ supervised the project, developed the research concept and received funding.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval and consent to participate
The study was approved by Ethical Committee of Nanfang Hospital and conducted in accordance with the ethical standards.
Consent for publication
All authors have read and approved the final paper.
Human and animal rights
The study was approved by Ethical Committee of Nanfang Hospital and conducted in accordance with the ethical standards.
Informed consent
Informed consent was obtained from the patient.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, S., Wang, B., Liao, J. et al. Molecular mechanism of XB130 adaptor protein mediates trastuzumab resistance in gastric cancer. Clin Transl Oncol 25, 685–695 (2023). https://doi.org/10.1007/s12094-022-02974-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-022-02974-w