Abstract
Objective
Emerging studies highlight the crucial effects of microRNAs on cancer initiation and malignant progression of various tumors. This study focused on the biological effect of miR-377-3p on CUL1 and epithelial–mesenchymal transition (EMT) and Wnt/β-catenin pathways in osteosarcoma (OS).
Methods
We performed quantitative real-time polymerase chain reaction (qRT-PCR) to analyze miR-377-3p and CUL1 expression levels in OS tissues and MG-63 cells. Then, cell counting kit (CCK)-8 and Transwell assay were used to examine the functions of miR-377-3p in OS cell growth and metastasis abilities. Meanwhile, luciferase reporter assay was used to validate CUL1 as direct target of miR-377-3p. qRT-PCR and Western blot were then carried out to detect the impact of miR-377-3p on EMT and Wnt/β-catenin pathways. Tumor xenograft models were established to further examine the effects of miR-377-3p on OS tumorigenesis in vivo.
Results
miR-377-3p downregulation was frequently identified in OS tissues and cells, which was associated with worse prognosis of OS patients. Functional experiments showed miR-377-3p restoration could dramatically repress OS cell growth and migration by regulation of EMT and Wnt/β-catenin pathways. Moreover, luciferase reporter assay revealed that CUL1 acted as a functional target of miR-377-3p. Additionally, the elevated CUL1 expressions in OS tissues also indicated poor prognosis of OS patients. Furthermore, the OS tumor growth was also obviously inhibited by miR-377-3p overexpression in vivo.
Conclusions
Collectively, all the above findings revealed that miR-377-3p exerted anti-OS functions via CUL1 and EMT and Wnt/β-catenin pathways. These results may contribute to the development of clinical OS treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Osteosarcoma (OS) is the most common malignancy affecting children, adolescents, and young adults, accounting for 20% of all bone tumors and approximately 5% of all pediatric tumors [1, 2]. The overall incidence worldwide is 3.4 per million per year [3]. Currently, the most common treatment for osteosarcoma is complete surgical resection combined with chemotherapy. The implementation of neoadjuvant and adjuvant chemotherapy has increased the overall survival of patients diagnosed with non-metastatic osteosarcoma to 70%; however, this has not significantly improved the prognosis of patients with metastasis or recurrence [4]. Current treatment for 5-year survival rate of metastatic or recurrent osteosarcoma patients can be achieved which ranges from 20 to 30% [5]. However, despite multiple attempts to use different chemotherapy regimens over the past 20 years, survival rates have remained essentially unchanged and there have been no successful targeted therapies for osteosarcoma to date [6, 7].
Accumulating evidence indicates that the occurrence, development and prognosis of osteosarcoma are closely related to molecular mechanisms [8]. MicroRNAs (miRNAs) are small non-coding RNAs that can affect mRNA expression by binding to their 3ʹ untranslated regions (3ʹ UTRs) [9]. MiRNAs emerged as post transcriptional regulators in regulating biogenesis and cancer progression, with the potential to be prognostic biomarkers for OS [10, 11]. Many studies have shown that miR-377 may affect the progression of different osteosarcoma by acting as a suppressor gene [12, 13]. In addition, miR-377 was found to have decreased expression in hepatocellular carcinoma, ovarian cancer, suggesting that miR-377 plays an important role in tumorigenesis and progression [14, 15].
Cullin (CUL) 1 is one of the targets of miR-377-3p. CUL1 is an essential component of the SCF E3 ubiquitin ligase complex and plays an important role in tumorigenesis and progression [16]. One key role of cul1 as an oncogene is to promote cell cycle progression [32811853]. Increased CUL1 expression is strongly associated with poor prognosis in osteosarcoma patients [17]. However, the specific mechanisms by which miR-377-3p and CUL1 affect osteosarcoma progression have not been documented.
Therefore, the present study investigated the in vitro and in vivo functions of miR-377-3p in osteosarcoma. In addition, a miRNA–mRNA functional network was also constructed to elucidate the underlying mechanism of osteosarcoma progression.
Materials and methods
Osteosarcoma patients
This study was approved by the Research Ethics Committee of Nanxishan hospital of Guangxi Zhuang Autonomous Region (Guangxi, China). All participants were aware of the content of this study and signed an informed consent. We collected 10 osteosarcoma tissue samples during the surgical procedure. Normal bone samples were obtained from 10 participants who succumbed to mortality in traffic accidents. The characteristics of the patients are shown in Table S1.
Cell culture
The human OS cell lines (MG-63), human embryonic kidney cell line (HEK293T) and immortalized human osteoblast cells hFOB (hFOB1.19) were purchased from Shanghai Institutes for Biological Sciences Cell Resource Center (Shanghai, China). Dulbecco's modified Eagle's medium (DMEM; Hyclone, MA, USA) supplemented with 10% fetal bovine serum and 1% antibiotic and antimycotic solution. Cells were put into the incubator for culture at 37 °C and 5% CO2.
Construction and transfection of miR-377-3p
Human miR-377-3p mimics (5′-UACAG UACUGUGAUAACUGAACAGUUAUCACAGUACUGU AUU-3′) and the scramble mimics were synthesized in GenePharma (Shanghai, China). About the transfection, 80 nm miR-377-3p mimics and scramble mimics were transfected into MG-63 using Lipofectamine® 2000 (Thermo, MA, USA) according to the manufacturer's instructions. These cells were then incubated for 48 h.
Luciferase reporter assays
For the luciferase reporter plasmids, the full-length 3′-UTR of CUL1, and 3′-UTR mutant for the miR-377-3p-binding site 3′-UTRs of the gene were cloned individually into pMIR-REPORT luciferase reporter vectors (Ambion, TX, USA). Then, vectors were co-transfected with miR-377-3p mimics into HEK293T through Lipofectamine 2000 reagent. The cells were collected and measured luciferase activities after 24 h using Dual-Luciferase reporter assay kit (Promega, WI, USA).
Quantitative real-time polymerase chain reaction
The total RNA from tissue samples and cultured cells was extracted using Trizol kit (Thermo). Total RNA was reverse transcribed to cDNA utilizing a PrimeScript™ RT reagent Kit (Invitrogen, USA) or miRNA 1st Strand cDNA Synthesis Kit (Vazyme, Nanjing, China). Quantitative real-time polymerase chain reaction (qRT-PCR) was performed using the SYBR Green qPCR Mix (Invitrogen). The results from qRT-PCR were normalized to GAPDH or U6. The primers listed below: miR-377-3p, 5′-TGGTGTCGTGGAGTCG-3′ and 5′-ACACTCCAGCTGGGATCACACAAAGGCA-3′; CUL1, 5′- GCCGTGTCGCACGCAG-3′ and 5′- GTCCCGCCAAGGTCTAGCTG-3′; GAPDH, 5′-CTTTGGTATCGTGGAAGGACTC-3′ and 5′-GTAGAGGCAGGGATGATGTTCT-3′. U6, 5′-CTCGCTTCGGCAGCACA-3′ and 5′-AACGCTTCACGAATTTGCGT-3′. The genes relative expression was calculated using the 2−ΔΔCt method. All reactions were carried out in triplicate.
Western blot analysis
Cells were thoroughly mixed with RIPA cell lysis buffer (Beyotime, Shanghai, China) on ice. Proteins were quantified, then separated using 10% SDS/polyacrylamide gels and transferred onto PVDF membranes (Millipore, USA). Membranes were blocked using 5% fat-free milk at room temperature. The membranes were incubated with antibodies against GAPDH, Vimentin, Twist, N-cadherin, Snail, β-catenin, Wnt3a, and cyclin D1 (Abcam, MA, USA) overnight at 4 °C. The membranes were then incubated with the following secondary antibodies. Immunoreactivity was detected using an ECL Kit (Pierce). The results were analyzed using Image LabTM Software (Bio-Rad Laboratories). Protein bands were detected using an ECL Kit (Beyotime).
Cell Counting Kit (CCK)-8
MG-63 cells transduced with miR-377-3p mimics and scramble mimics were seeded in 96-well plate (2.5 × 103/well), cells were cultured for 96-h at 37 °C in a 5% CO2 incubator. Then, 10 µl CCK-8 reagent was added to each well and incubated for 2 h. The absorbance of each well was detected at a wavelength of 450 nm.
Apoptosis assay
Cells were washed with PBS followed by incubation with Annexin V PE-A and PC5.5-A using the Annexin V Apoptosis Detection Kit (BD Biosciences, NJ, USA) for 15–20 min at room temperature. Cell apoptosis was then analyzed by flow cytometry (FACSCalibur) at 480 nm. Data were analyzed using FlowJo version 10 (FlowJo LLC, Ashland, OR, USA).
Transwell
The cells were seeded into the upper well of a Millicell hanging cell culture insert of a Transwell chamber (8-μm pore size, Millipore), for the migration assays after serum-free culture. The culture medium with serum was added into the lower chamber and subsequently incubated for 24 h. The cells migrating to or invading the bottom of the membrane were fixed with methanol and stained with crystal violet.
Animal experiments
To establish OS murine xenograft model, the 6-week-old BALB/c nude mice were used. MG6-3 cells (7 × 106 cells) transfected with miR-377-3p mimics or scramble mimics were subcutaneously injected into the armpit of the nude mice (10 mice each group). After injection for 7 days, tumor growth was measured every 3 days, and these mice were euthanized after 19 days. The tumor growth inhibition rate was then calculated. The length (L) and width (W) of tumors were measured and assessed using the formula: (L × W2)/2. All experimental procedures were approved by the Ethics Committee of Animal Research of Nanxishan hospital of Guangxi Zhuang Autonomous Region.
Statistical analysis
Statistical analyses were performed through SPSS 20.0. Data are presented as the mean ± standard deviation (SD). Statistical analyses were performed with Student's t test and the statistical significance level was set at α = 0.05 (two-side). Values of P < 0.05 were regarded as statistically significant.
Results
Expression of miR-377-3p and CUL1 in osteosarcoma tissues and cell lines
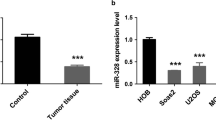
First, we examined the expression of miR-377-3p and CUL1 using qRT-PCR in 10 osteosarcoma and 10 control samples. The results suggested that the level of miR-377-3p was lower in osteosarcoma tissues than in controls, whereas CUL1 showed increased expression in osteosarcoma tissues (Fig. 1a). In addition, we also found that the expression level of miR-377-3p was decreased, while the expression of CUL1 was clearly increased in the osteosarcoma cell line MG-63, compared with osteoblast hFOB1.19 (Fig. 1b).
miR-377-3p targets binding to CUL1
We predicted the binding sites of miR-377-3p and CUL1 in the TargetScan online website (Fig. 2a). We suggested that miR-377-3p may be involved in the regulation of CUL1 protein expression. miR-377-3p was predicted to interact with 8-mer seed match with the CUL1-3′ UTR from position 417–422. To confirm whether miR-377-3p directly targets CUL1, a wild-type (WT) CUL1 3′-UTR luciferase reporter vector and a miR-377-3p homologous sequence mutant CUL1 3′-UTR luciferase reporter vector were constructed. The results showed that the luciferase activity of CUL1-WT cells with miR-377-3p mimics was significantly decreased (Fig. 2b). There was no significant change in luciferase reporter activity in the mutant CUL1 3′-UTR group. It suggested that the expression regulation of cul1 was regulated by miR-377-3p targeting.
The miR-377-3p could regulate CUL1 expression by targeting CUL1. a Though TargetScan online prediction, the sequence of the miR-377-3p-binding site in the 3′-UTR of CUL1 was shown at the upper site. Mutated residues were shown at the lower site. b Luciferase activity of the wild-type 3′-UTR reporters and the mutant 3′-UTR reporters in 293 T cells treated with control mimics or miR-377-3p mimics was shown. *P < 0.05
MiR-377-3p mimics inhibit proliferation and migration of MG-63 cells
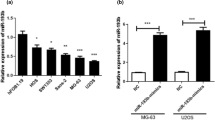
To explore the effect of miR-377-3p on osteosarcoma, miR-377-3p mimics were transfected into MG-63 cells to overexpress miR-377-3p (Figure S1). CUL1 expression in MG-63 cells decreased significantly after transfection with miR-377-3p mimic detected by qRT-PCR (Fig. 3a). Higher expression of miR-377-3p resulted in significantly lower proliferation of MG-63 cells compared with cells transfected with scramble mimics which detected using CCK-8 kit (Fig. 3b). In contrast, the level of apoptosis was clearly elevated with high expression of miR-377-3p (Fig. 3c). Cell migration was slightly decreased in cells treated with miR-377-3p mimics compared with cells of the scramble mimics group (Fig. 3d).
MiR-377-3p overexpression affected the proliferation, apoptosis, and migration of cells. a The mRNA levels of CUL1 after transfection of miR-377-3p mimic. b MG-63 cell growth curves were determined with CCK-8 assay. Transfection of miR-377-3p mimics significantly inhibited cell proliferation. *P < 0.05. OD optical density. c Flow cytometry results showed that miR-377-3p mimics increased the apoptosis rate of MG-63 cells. The apoptosis rate of MG-63 cells after transfection with miR-377-3p mimics was 39.70%, the apoptosis rate in which transfection with scramble mimics was 7.57%. D. miR‐613 mimics decreased MG-63 migration detected by Transwell
MiR-377-3p inhibits EMT and Wnt/β-catenin signaling pathway in MG-63
Vimentin, Twist, N-cadherin, and Snail are markers of EMT [18], and β-catenin, Wnt3a, and cyclin D1 are three key components in the Wnt/β-catenin signaling pathway [19]. In KEGG website (https://www.kegg.jp/entry/K03347), we identified that CUL1 participates in Wnt signaling pathway. To investigate the effect of miR-377-3p on EMT and the Wnt/β-catenin signaling pathway, we performed qRT-PCR and western blot assays on cells transfected with miR-377-3p mimics (Fig. 4a, b). The results showed that the EMT and Wnt/β-catenin signaling pathways may be suppressed in MG-63 cells with high expression of miR-377-3p. These results suggested that miR-377 may target CUL1 to suppress EMT and Wnt/β-catenin signaling pathways in MG-63 cells.
The inhibition of signal pathway in cells transfected with miR-377-3p mimics. a The mRNAs of key markers in EMT and Wnt/β-catenin signaling pathway were detected by qRT-PCR. *P < 0.05. b The EMT and Wnt/β-catenin signaling pathway was inhibited in MG-63 cells transfected with miR-377-3p mimics detected by western blot. *P < 0.05
Tumor-suppressive effect of miR-377-3p
We further investigated the effect of miR-377-3p on the growth of MG-63 cells in nude mice in vivo. Tumor growth was slow in nude mice injected with MG-63 cells transfected with miR-377-3p mimics (Fig. 5a, b). Compared with the control group, nude mice overexpressing miR-377-3p had significantly smaller tumors. The body weight of nude mice overexpressing miR-377-3p was higher than that of control mice (Fig. 5c). On the 19th day, we calculated that the tumor growth inhibition rate was 78.64%. Using qRT-PCR assay, we found that the level of CUL1 in the miR-377-3p overexpression group was significantly lower than that in the control group in tumor tissue (Fig. 5d).
A mouse osteosarcoma model showed that miR-377-3p has a tumor suppressive role. a Effects on tumor growth rate were assessed in each mouse on the day 0. miR-377-3p mimics groups are shown in red, control groups in blue. *P < 0.05. b Tumor size was observed after cells were injected for 7 days. c The body weight was assessed in each mouse on the day 0. miR-377-3p mimics groups are shown in red, control groups in blue. *P < 0.05. **P < 0.01. D. The levels of CUL1 mRNA in the mice were determined by qRT-PCR. ***P < 0.001
Discussion
Although surgery combined with systemic chemotherapy is the gold standard for the treatment of osteosarcoma, the prognosis is poor due to the resistance to chemotherapy [20]. Therefore, it is urgent to find a new target for the treatment of osteosarcoma in order to achieve more effective treatment for osteosarcoma. In the present study, we designed a series of experiments to explore the role of miR-377-3p in osteosarcoma. Our experimental results showed that miR-377-3p was down-expressed in osteosarcoma samples and osteosarcoma cell lines compared with normal controls. Overexpression of miR-377-3p in osteosarcoma cells inhibited cell proliferation and migration. In vivo experiments also confirmed that miR-377-3p overexpression inhibited the growth of tumors. These results suggest that miR-377-3p plays an inhibitory role in osteosarcoma progression.
However, studies on the genes regulated by miR-377-3p targets are currently lacking. A previous study showed that miR-377-3p targets to regulate CUL4A expression and is involved in ovarian cancer metastasis [15]. There are also studies confirming that miR-377-3p can target DNA methyltransferase 1 to induce senescence in human dermal fibroblasts [21]. However, no study has yet determined whether miR-377-3p targets CUL1.
Cullin ubiquitin ligases affect a variety of important cellular functions such as cell cycle progression, signaling, and DNA repair [22]. Among them, one key role of CUL1 as an oncogene is to promote cell cycle progression [23]. CUL1 is highly expressed in both lung adenocarcinoma and colorectal cancer and plays a tumor promoting role [24, 25]. In this study, through sequence complementation analysis and dual luciferase assay, we identified that CUL1 was an inhibitory target of miR-377-3p and its expression was regulated by miR-377-3p. Upon miR-377-3p overexpression, a decrease in CUL1 levels inhibits the proliferation and promotes the apoptosis of osteosarcoma cells.
In addition, to better understand the mechanism by which miR-377-3p regulates the phenotype of osteosarcoma cells, we examined the expression of epithelial–mesenchymal transition (EMT) and Wnt signaling pathway-related proteins. The results showed that transfection of miR-377-3pmimics in the osteosarcoma cell line MG-63 significantly decreased the expression of Vimentin, Twist, N-cadherin, Snail, β-catenin, Wnt3a, and cyclin D1. The downregulation of these proteins probably decreased the activity of the pathway.
EMT is considered an important factor in tumor invasion and metastasis. EMT is a dynamic multistep process that includes loss of intercellular adhesion, disruption of the cancer cell basement membrane and extracellular matrix, remodeling of the cytoskeleton and enhancement of cell motility [26]. In addition to driving tumor initiation and metastasis, EMT generates chemoresistance that helps tumor cells evade destruction by the immune system [27]. This makes it difficult to treat cancer clinically. EMT is characterized by decreased expression of epithelial markers such as E-cadherin and γ-Catenin and increased expression of mesenchymal markers such as N-cadherin, vimentin, snail, and twist [28]. Translocation of β-Catenin from the cytoplasm to the nucleus regulates the activation of these proteins, thereby playing a key role in the EMT process [29]. Β-Catenin is considered a key protein in Wnt signaling and an executor of the regulatory function of its downstream targets [30]. As miRNAs influence the Wnt/β-catenin signaling pathway, more and more attention has been paid to the interaction between miRNAs and EMT [31, 32]. The role of miRNAs as tumor suppressors in the regulation of EMT may facilitate the search for potential therapeutic targets.
Our experimental results suggest that miR-377-3p suppresses the proliferation of osteosarcoma cells and tumor growth in vivo. Therefore, we speculated that miR-377-3p could negatively regulate EMT and Wnt/β-catenin signaling pathways to inhibit the progression of osteosarcoma.
Our study also has certain limitations. First, the clinical patient's sample size is too small, which may affect the interpretation of the results. Second, there is a lack of findings that CUL1 directly regulates the EMT and Wnt pathways.
Conclusion
In conclusion, this study suggests that miR-377-3p is a tumor suppressor gene in OS cancer, which promotes cancer cell apoptosis and inhibits tumor growth, inhibiting EMT and Wnt/β-catenin signaling pathways. These results provide a possible molecular mechanism for OS cancer initiation and progression. Overexpression of miR-377-3p inhibited the expression of CUL1 and may serve as a novel therapeutic strategy for OS.
References
Moore DD, Luu HH. Osteosarcoma. Cancer Treat Res. 2014;162:65–92.
Tang N, Song WX, Luo J, Haydon RC, He TC. Osteosarcoma development and stem cell differentiation. Clin Orthop Relat Res. 2008;466:2114–30.
Mirabello L, Troisi RJ, Savage SA. International osteosarcoma incidence patterns in children and adolescents, middle ages and elderly persons. Int J Cancer. 2009;125:229–34.
Lilienthal I, Herold N. Targeting molecular mechanisms underlying treatment efficacy and resistance in osteosarcoma: a review of current and future strategies. Int J Mol Sci. 2020;21:6885.
Bajpai J, Chandrasekharan A, Simha V, Talreja V, Karpe A, Pandey N, Singh A, Rekhi B, Vora T, Ghosh J, Banavali S, Gupta S. Outcomes in treatment-naive patients with metastatic extremity osteosarcoma treated with OGS-12, a novel non-high-dose methotrexate-based, dose-dense combination chemotherapy, in a tertiary care cancer center. J Glob Oncol. 2018;4:1–10.
Zhang Y, Yang J, Zhao N, Wang C, Kamar S, Zhou Y, He Z, Yang J, Sun B, Shi X, Han L, Yang Z. Progress in the chemotherapeutic treatment of osteosarcoma. Oncol Lett. 2018;16:6228–37.
Casali PG, Bielack S, Abecassis N, Aro HT, Bauer S, Biagini R, Bonvalot S, Boukovinas I, Bovee J, Brennan B, Brodowicz T, Broto JM, Brugieres L, et al. Bone sarcomas: ESMO-PaedCan-EURACAN Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2018;29:iv79–95.
Wang S, Gao H, Zuo J, Gao Z. Cyclooxygenase-2 expression correlates with development, progression, metastasis, and prognosis of osteosarcoma: a meta-analysis and trial sequential analysis. FEBS Open Bio. 2019;9:226–40.
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97.
Ji Q, Xu X, Song Q, Xu Y, Tai Y, Goodman SB, Bi W, Xu M, Jiao S, Maloney WJ, Wang Y. miR-223-3p inhibits human osteosarcoma metastasis and progression by directly targeting CDH6. Mol Ther. 2018;26:1299–312.
Czarnecka AM, Synoradzki K, Firlej W, Bartnik E, Sobczuk P, Fiedorowicz M, Grieb P, Rutkowski P. Molecular biology of osteosarcoma. Cancers (Basel). 2020;12:2130.
Xia P, Gu R, Zhang W, Shao L, Li F, Wu C, Sun Y. MicroRNA-377 exerts a potent suppressive role in osteosarcoma through the involvement of the histone acetyltransferase 1-mediated Wnt axis. J Cell Physiol. 2019;234:22787–98.
Huang YF, Lu L, Shen HL, Lu XX. LncRNA SNHG4 promotes osteosarcoma proliferation and migration by sponging miR-377–3p. Mol Genet Genomic Med. 2020;8:e1349.
Bu N, Dong Z, Zhang L, Zhu W, Wei F, Zheng S. CircPVT1 regulates cell proliferation, apoptosis and glycolysis in hepatocellular carcinoma via miR-377/TRIM23 axis. Cancer Manag Res. 2020;12:12945–56.
Yu R, Cai L, Chi Y, Ding X, Wu X. miR377 targets CUL4A and regulates metastatic capability in ovarian cancer. Int J Mol Med. 2018;41:3147–56.
Huang YF, Zhang Z, Zhang M, Chen YS, Song J, Hou PF, Yong HM, Zheng JN, Bai J. CUL1 promotes breast cancer metastasis through regulating EZH2-induced the autocrine expression of the cytokines CXCL8 and IL11. Cell Death Dis. 2018;10:2.
Cheng Q, Yin G. Cullin-1 regulates MG63 cell proliferation and metastasis and is a novel prognostic marker of osteosarcoma. Int J Biol Markers. 2017;32:e202–9.
Davis FM, Azimi I, Faville RA, Peters AA, Jalink K, Putney JW Jr, Goodhill GJ, Thompson EW, Roberts-Thomson SJ, Monteith GR. Induction of epithelial-mesenchymal transition (EMT) in breast cancer cells is calcium signal dependent. Oncogene. 2014;33:2307–16.
Savagner P. The epithelial-mesenchymal transition (EMT) phenomenon. Ann Oncol. 2010;21(Suppl 7):89–92.
Song M, Wang Y, Zhang Z, Wang S. PSMC2 is up-regulated in osteosarcoma and regulates osteosarcoma cell proliferation, apoptosis and migration. Oncotarget. 2017;8:933–53.
Xie HF, Liu YZ, Du R, Wang B, Chen MT, Zhang YY, Deng ZL, Li J. miR-377 induces senescence in human skin fibroblasts by targeting DNA methyltransferase 1. Cell Death Dis. 2017;8:e2663.
Jang SM, Redon CE, Aladjem MI. Chromatin-bound cullin-ring ligases: regulatory roles in DNA replication and potential targeting for cancer therapy. Front Mol Biosci. 2018;5:19.
Sweeney MA, Iakova P, Maneix L, Shih FY, Cho HE, Sahin E, Catic A. The ubiquitin ligase Cullin-1 associates with chromatin and regulates transcription of specific c-MYC target genes. Sci Rep. 2020;10:13942.
Liu J, Su S, He H, Wang H, Zhang D. Effects of Cullin1 on the Biological Characteristics of Lung AdenocarcinomaA549 and H1395 Cells. Zhongguo Fei Ai Za Zhi. 2021;24:69–77.
Deng J, Chen W, Du Y, Wang W, Zhang G, Tang Y, Qian Z, Xu P, Cao Z, Zhou Y. Synergistic efficacy of Cullin1 and MMP-2 expressions in diagnosis and prognosis of colorectal cancer. Cancer Biomark. 2017;19:57–64.
Chen HT, Liu H, Mao MJ, Tan Y, Mo XQ, Meng XJ, Cao MT, Zhong CY, Liu Y, Shan H, Jiang GM. Crosstalk between autophagy and epithelial-mesenchymal transition and its application in cancer therapy. Mol Cancer. 2019;18:101.
Lavin DP, Tiwari VK. Unresolved complexity in the gene regulatory network underlying EMT. Front Oncol. 2020;10:554.
Lei Y, Chen L, Zhang G, Shan A, Ye C, Liang B, Sun J, Liao X, Zhu C, Chen Y, Wang J, Zhang E, Deng L. MicroRNAs target the Wnt/betacatenin signaling pathway to regulate epithelialmesenchymal transition in cancer (Review). Oncol Rep. 2020;44:1299–313.
Guo Q, Qin W. DKK3 blocked translocation of beta-catenin/EMT induced by hypoxia and improved gemcitabine therapeutic effect in pancreatic cancer Bxpc-3 cell. J Cell Mol Med. 2015;19:2832–41.
Cai J, Guan H, Fang L, Yang Y, Zhu X, Yuan J, Wu J, Li M. MicroRNA-374a activates Wnt/beta-catenin signaling to promote breast cancer metastasis. J Clin Invest. 2013;123:566–79.
Peng L, Liu Z, Xiao J, Tu Y, Wan Z, Xiong H, Li Y, Xiao W. MicroRNA-148a suppresses epithelial-mesenchymal transition and invasion of pancreatic cancer cells by targeting Wnt10b and inhibiting the Wnt/beta-catenin signaling pathway. Oncol Rep. 2017;38:301–8.
Jin Y, Wang J, Han J, Luo D, Sun Z. MiR-122 inhibits epithelial-mesenchymal transition in hepatocellular carcinoma by targeting Snail1 and Snail2 and suppressing WNT/beta-cadherin signaling pathway. Exp Cell Res. 2017;360:210–7.
Author information
Authors and Affiliations
Contributions
KL, LL and QL are responsible for the conception or design of the work. QO, LJ and GW contribute the acquisition, analysis, or interpretation of data for the work. All authors finally approved the manuscript version to be published.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The study was approved by Ethical Committee of Nanxishan Hospital of Guangxi Zhuang Autonomous Region and conducted in accordance with the ethical standards.
Informed consent
Subjects signed the informed consent.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liang, K., Liao, L., Liu, Q. et al. microRNA-377-3p inhibits osteosarcoma progression by targeting CUL1 and regulating Wnt/β-catenin signaling pathway. Clin Transl Oncol 23, 2350–2357 (2021). https://doi.org/10.1007/s12094-021-02633-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-021-02633-6