Abstract
Background and aim
Long non-coding RNAs (lncRNAs) have been demonstrated to act as a critical regulator in the processes of tumor biology. In this study, whether lncRNA-ATB is a potential indicator for non-small cell lung cancer (NSCLC) was investigated and its biological function in NSCLC was also determined.
Methods
The expression levels of lncRNA-ATB in NSCLC tissues and cell lines were measured. A549 cell line was explored to investigate the functions of lncRNA-ATB in NSCLC.
Results
Real-time PCR results showed that lncRNA-ATB expression was up-regulated in both in NSCLC tissues and cell lines. High lncRNA-ATB expression in tumor tissue was associated with larger tumor size, lymph node metastasis, and distant metastasis in patients with NSCLC, respectively. In addition, the patients with high expression of lncRNA-ATB presented a lower survival probability. In vitro experiments showed that down-regulation of lncRNA-ATB promoted the cell apoptosis, whereas inhibited the cell viability, cell migration, and cell invasion.
Conclusion
High expression of lncRNA-ATB indicated a poor prognosis and led to the cell proliferation and metastasis in NSCLC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lung cancer is one of the leading causes of cancer death [1], and it is estimated that more than 85% of cases are classified histopathologically as non-small cell lung cancer (NSCLC) [2]. Chemoradiotherapy and operative treatment are the main therapeutic approaches for lung cancer at present [3]. However, the 5-year overall survival of NSCLC is relatively low [4]. To further clarify its molecular mechanisms is significant for developing new therapeutic strategies for NSCLC.
Long non-coding RNAs (lncRNAs) are generally defined as transcribed RNA molecules with a length longer than 200 nucleotides. Emerging evidence has shown that lncRNA is an important regulator in the biological and pathological processes of many human diseases, especially in cancers [5, 6]. The alteration of cellular processes that mediated by lncRNAs includes cell proliferation, cell growth, cell cycle progression, cell apoptosis, cell migration, and cell migration [7]. There are many lncRNAs that function as either tumor suppressors or oncogenes in diverse cancers. Microarray results showed that 571 lncRNAs were differentially expressed between NSCLC tissues and adjacent normal tissues [8]. However, their functions in NSCLC have been demonstrated in only a small part of lncRNAs.
In recent study, researchers have identified that lncRNA activated by transforming growth factor-β (lncRNA-ATB) was closely related to cancer development and progression, such as breast cancer [9], colorectal cancer [10], and pancreatic cancer [11]. The functions of lncRNA-ATB in human diseases have been little known at present, and its role in NSCLC remains unclear. In this study, we investigated whether lncRNA-ATB was a clinical indicator for the prognosis of NSCLC. The role of lncRNA-ATB in tumor biology of NSCLC was also evaluated with in vitro experiments.
Materials and methods
Subject and tissue collection
Eighty-four patients with NSCLC were included in this study. Written informed consent was obtained from all the participants, and this study was approved by the Ethics Committee of Anhui Provincial Hospital. Patients with NSCLC were diagnosed on the basis of the conventional clinical and histopathologic criteria. The clinicopathological characteristics of patients were shown in Table 1. All patients underwent surgery. The cancer tissues and corresponding adjacent non-cancer tissues were collected during surgery and immediately frozen in liquid nitrogen until RNA extraction.
Cell lines and cell culture
Human NSCLC cell lines (A549 and NCI-H1299) and normal human bronchial epithelial cell line (HBE) were purchased from the American Type Culture Collection (ATCC, USA). A549 and NCI-H1299 cells were maintained in Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco, USA) and HBE cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco, USA) that supplemented with 100U/ml penicillin, 100 µg/ml streptomycin, and 10% fetal bovine serum (FBS, Hyclone, USA). The cells were cultured at 37 °C in an atmosphere containing 5% carbon dioxide (CO2), and passaged with the treatment of 0.25% trypsin.
Cell transfection
The interference sequence for lncRNA-ATB (si-ATB) was transfected into A549 cells or NCI-H1299 cells using Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s instructions. The transfection of negative control for siRNA acted as the Mock. The sequences of siRNA against lncRNA-ATB and negative control were as follows: si-ATB: forward TATGGCCTAGATTACCTTTCCATT, reverse TGG AAAGGTAATCTAGGCCATATT; Mock: forward 5′-UUCUCCGAACGUGUCACG-UTT-3′, reverse 5′-ACGUGACACGUUCGGAGAATT-3′. After different times of transfection, cells were harvested to perform the following experiments. The sequences of si-ATB and mock were synthesized by Shanghai GenePharma Co., Ltd (Shanghai, China).
Fluorogenic quantitative PCR
Total RNA from tissues and cultured cells was extracted using TRIzol reagent (Invitrogen, USA) according to the manufacturer’s protocol. A total of 1 µg RNA was used to produce first-strand complementary DNA by M-MLV Reverse transcriptase (Invitrogen, USA). The quantitative analysis of lncRNA-ATB expression was performed with Power SYBR® Green PCR Master Mix (Life Technologies, USA) in triplicates by reacting with specific primers for lncRNA-ATB, with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) acting as an internal control. The relative lncRNA-ATB expression level was expressed as fold changes through normalization against GAPDH expression using the 2−△△Ct method. The sequences of specific primers for lncRNA-ATB and GAPDH were as follows: lncRNA-ATB, forward: 5′-CTTCACCAGCACCCAGAGA-3′, reverse: 5′-AAGACAGAAAAACAGTTCCGAGTC-3′; GAPDH: forward: 5′-CAGGAGGCATTGCTGATGAT-3′, reverse: 5′-GAAGGCTGGGGCTCATTT-3′.
MTT assay
The viability of A549 cells was measured by 3-(4,5-dimethyl-2-thiazolyl) -2,5-diphenyl-2-H-tetrazolium bromide (MTT) assay after the transfection of si-ATB. In brief, the cells were seeded in 96-well plates. After transfection, the cells were harvested at 24, 48, 72, and 96 h, respectively, and added 10 µl MTT stock solution (5 mg/ml, Sigma, USA) to incubate for 4 h at 37 °C in an atmosphere containing 5% CO2. The supernatant was then discarded and 100 µl dimethyl sulfoxide (DMSO, Sigma-Aldrich, USA) was added in each well for dissolving crystal. The absorbance was recorded at 492 nm. The cell viability was calculated by following formula:
Flow cytometry for cell cycle analysis
At the 48 h after the transfection of si-ATB, the cell cycle was measured and analyzed. In brief, the cells in logarithmic growth phase were harvested and treated with trypsin. After washing with cold phosphate balanced solution (PBS) containing ethylene diamine tetraacetic acid (EDTA), the cells were fixed in 75% ethanol for 24 h at 4 °C, centrifuged the cells at 12,000 rpm for 3 min, and washed and re-suspended in PBS-EDTA twice. Then, the cells were incubated with 50 µl RNase Staining Buffer (10 mg/ml) for 20 min and subsequently incubated with 50 µl propidium iodide (PI, 500 ng/µl) for 30 min in the dark. The DNA content was measured by BD FACSCalibur Flow Cytometer, and the proportions of cells in G0/G1, S, and G2/M phases were analyzed using the CellQuest Pro software.
Flow cytometry for cell apoptosis analysis
At the 48 h after the transfection of si-ATB, the cell apoptosis was measured by Annexin V-FITC/PI Apoptosis Detection Kit (Sigma-Aldrich, USA) following the manufacturer’s protocol. In brief, a volume of 5 µl Annexin V-FITC and 5 µl PI was added to incubate for 10 min in the dark at room temperature. Then, the samples were analyzed by BD FACSCalibur Flow Cytometer (BD Biosciences, USA).
Scratch-wound assay for cell migration analysis
At the 48 h after the transfection of si-ATB, the cell migration was determined by scratch-wound assay. In brief, when the cells grew to 80% confluence, the medium was removed. A vertical scratch wound was made using 10 µl pipette tip. The cells were then washed with PBS thrice for removing the scratched cells, and incubated with serum-free RPMI 1640 medium for 24 h. The representative photographs were captured and the scratch area was analyzed using the Image software.
Transwell assay for cell invasion analysis
At the 48 h after the transfection of si-ATB, the cell invasion was determined by Transwell assay. In brief, A549 cells were seeded on upper chambers pre-coated with matrigel and added 200 μl of serum-free medium. In the lower chambers, 800 μl RPMI 1640 medium containing 10% FBS was added. Then, the transwell plates were incubated at 37 °C in an atmosphere containing 5% CO2 for 48 h. After removing the cells in the upper chamber, the cells in the lower chamber were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. Six fields of vision were randomly selected, and the cells were counted to calculate the mean percentage of cell invasion.
Statistical analysis
Statistical analysis was conducted with software SPSS 18.0. The difference of means between two groups was analyzed using a Chi square test or a Student’s t test appropriately. Survival analysis was performed using the Kaplan–Meier method, and the log-rank test was used to test the statistical difference. P value less than 0.05 was considered to be statistically significant.
Results
Upregulation of lncRNA-ATB expression in NSCLC
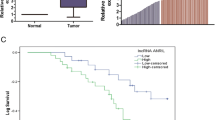
The relative expression of lncRNA-ATB in tissues and cells of NSCLC was determined. As shown in Fig. 1a, the expression level of lncRNA-ATB in cancer tissues was significantly up-regulated than that in corresponding non-cancer tissues. The up-regulation of lncRNA-ATB level was also observed in NSCLC cell lines A549 and NCI-H1299 than that in normal HBE cell line (Fig. 1b).
lncRNA-ATB expression in NSCLC tissues and cell lines. a lncRNA-ATB expression in cancer and corresponding non-cancer tissues. b lncRNA-ATB expression in NSCLC cell lines A549 and NCI-H1299 and normal human bronchial epithelial cell line (HBE). *P < 0.05, vs. non-cancer or HBE cell line. **P < 0.01, vs. non-cancer or HBE cell line
lncRNA-ATB expression associates with clinical parameters of tumor aggressiveness in patients with NSSLC
The clinicopathological characteristics of 84 patients with NSCLC were shown in Table 1. The distribution of lncRNA ATB expression was analyzed according to different clinical factors. The results showed that high lncRNA ATB expression was associated with larger tumor size, lymph node metastasis, and distant metastasis, whereas the statistical correlations between lncRNA ATB expression level and other clinical factors including age, gender, TNM stages, and smoking history were not observed.
High lncRNA-ATB expression was associated with poor prognosis in patients with NSCLC
We then investigated the relationship between lncRNA-ATB expression and overall survival (OS) in NSCLC patients. The lncRNA-ATB expression in all patients was measured. The average relative expression of lncRNA-ATB was 2.28, which was considered as the standard for evaluating the high or low lncRNA-ATB expression. As shown in Fig. 2, the patients with high expression of lncRNA-ATB presented a lower survival probability. The overall 3-year accumulative survival rates of patients with high lncRNA expression and low lncRNA expression were 32.6% and 68.4%, respectively.
Down-regulation of lncRNA-ATB promoted the cell apoptosis of A549 cells
To further investigate the effect of lncRNA-ATB on cell function of NSCLC, A549 cells were transfected with si-ATB for inhibiting lncRNA-ATB expression. si-ATB effectively downregulated the expression of lncRNA-ATB (Fig. 3a). Down-regulation of lncRNA-ATB led to the decrease of cell viability in A549 cells (Fig. 3b). The results also showed that si-ATB contributed to the cell-cycle arrest in the G0/G1 phase (Fig. 3c) and the promotion of cell apoptosis (Fig. 3d).
Effects of lncRNA-ATB down-regulation on cell viability, cell cycle, and apoptosis of A549 cells. a The transfection efficiency was confirmed by determining the expression of lncRNA-ATB after transfection of si-ATB (Mock acting as control). b Cell viability, c cell cycle, and d cell apoptosis were evaluated after transfection. **P < 0.01, vs. Mock
Down-regulation of lncRNA-ATB inhibited cell migration and invasion of A549 cells
The roles of lncRNA-ATB in cell migration and invasion of NSCLC were then evaluated. As shown in Fig. 4, transfection of si-ATB induced the decease of migration and invasion in A549 cells. The same effects were also observed in NCI-H1299 cells (supplementary Figs. 1 and 2).
Discussion
The discovery of non-coding RNA (ncRNA) opens the new understanding for various cellular and pathophysiologic processes. LncRNA is one of the important types of ncRNAs [12]. Although the biological functions of lncRNAs in many human diseases have been partly identified [13, 14], more details of the discovered lncRNAs should be discussed and many other un-known lncRNAs should be found. In recent years, mounting studies have focused on the relationship between lncRNAs and tumorigenesis [15]. In this study, we found that lncRNA-ATB expression was up-regulated in NSCLC, and high expression of lncRNA-ATB indicated a poor prognosis of NSCLC. In vitro experiments demonstrated that the lncRNA-ATB down-regulation led to the promotion of cell apoptosis and the inhibition of cell migration and invasion in A549 cells. These findings suggested that lncRNA-ATB might be a new biomarker for indicating NSCLC.
Up-regulation of lncRNA-ATB was firstly proposed in hepatocellular carcinoma samples to contribute to promote both the early and late steps of cancer metastasis [16]. Evidences also showed that lncRNA-ATB participated in other cancers, including colorectal cancer [10], pancreatic cancer [17], renal cell carcinoma [18], breast cancer [9], and glioma [19]. Yuan et al. firstly showed that lncRNA-ATB, which is activated by TGF-β, binds, and stimulates autocrine interleukin (IL) -11 production for triggering signal transducer and activator of transcription 3 (STAT3) signaling in tumor cells to promote colonization [16]. Li et al. demonstrated that lncRNA-ATB could increase zinc finger E-box-binding homeobox (ZEB) 1 and ZEB2 expression through competitively binding and sequestering microRNA-200 s (miR-200 s), thereby inducing epithelial-mesenchymal transition (EMT) and promoting tumorigenesis [20]. In this study, we also observed that lncRNA-ATB expression was up-regulated in NSCLC tissues and cells, which has not been reported. When NSCLC cell line A549 transfected with si-ATB for inhibiting lncRNA-ATB expression, the results showed that si-ATB significantly reduced the cell viability, cell migration, and cell invasion, whereas si-ATB also markedly elevated the cell apoptosis. However, we did not evaluate the role of lncRNA-ATB in the tumor growth in vivo experiments. Except for lncRNA-ATB, many other lncRNAs, such as HOX transcript antisense RNA (HOTAIR) [21], metastasis-associated lung adenocarcinoma transcript 1 (MALAT-1) [22], urothelial cancer associated 1 (UCA1) [23], antisense non-coding RNA in the INK4 locus (ANRIL) [24], and so on, have been demonstrated to influence the tumor biology of NSCLC.
The role of lncRNA in NSCLC as a clinical biomarker has also been demonstrated. For example, Nie et al. showed that microvascular invasion in hepatocellular carcinoma (MVIH) indicated a poor prognosis for NSCLC [25]. Cancer susceptibility candidate 2 (CASC2) is involved in the development and progression of NSCLC and its low expression indicates a poor prognosis of NSCLC [26]. In our study, we investigated the expression of lncRNA-ATB and its relationship with clinicopathological characteristics of NSCLC. The results showed that high lncRNA ATB expression was associated with larger tumor size, lymph node metastasis, and distant metastasis. In addition, we then determined the relationship between lncRNA-ATB expression and OS in NSCLC patients. It showed that the patients with high expression of lncRNA-ATB presented a lower survival probability, and the overall 3-year accumulative survival rates of patients with high lncRNA-ATB expression were significantly lower than that of patients with low lncRNA-ATB expression. The data indicated that high lncRNA-ATB expression in NSCLC was associated with survival time of patients, and the interference of lncRNA-ATB might be a useful target for the treatment of NSCLC.
In conclusion, our study demonstrated that high expression of lncRNA-ATB indicated a poor prognosis and functioned as a promoter in cell proliferation and metastasis in NSCLC. These findings are important for better understanding the pathophysiology of NSCLC. More information about lncRNA-ATB in tumor biology of NSCLC and other cancers should be investigated.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. doi:10.3322/caac.21254.
Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer. 2014;14(8):535–46. doi:10.1038/nrc3775.
Tarasevych S, Lauwers P, Vandaele F, van Meerbeeck JP. Novel treatment options in stage I non-small-cell lung cancer. Expert Rev Anticancer Ther. 2014;14(9):1007–20. doi:10.1586/14737140.2014.929500.
Society AC. Cancer facts & figures 2016. Atlanta: American Cancer Society; 2016.
Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861–74.
Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10(1):1.
Zhang H, Chen Z, Wang X, Huang Z, He Z, Chen Y. Long non-coding RNA: a new player in cancer. J Hematol Oncol. 2013;6(1):1.
Li P, Li J, Yang R, Zhang F, Wang H, Chu H, et al. Study on expression of lncRNA RGMB-AS1 and repulsive guidance molecule b in non-small cell lung cancer. Diagn Pathol. 2015;10:63. doi:10.1186/s13000-015-0297-x.
Shi SJ, Wang LJ, Yu B, Li YH, Jin Y, Bai XZ. LncRNA-ATB promotes trastuzumab resistance and invasion-metastasis cascade in breast cancer. Oncotarget. 2015;6(13):11652–63. doi:10.18632/oncotarget.3457.
Iguchi T, Uchi R, Nambara S, Saito T, Komatsu H, Hirata H, et al. A long noncoding RNA, lncRNA-ATB, is involved in the progression and prognosis of colorectal cancer. Anticancer Res. 2015;35(3):1385–8.
Qu S, Yang X, Song W, Sun W, Li X, Wang J, et al. Downregulation of lncRNA-ATB correlates with clinical progression and unfavorable prognosis in pancreatic cancer. Tumour Biol. 2016;37(3):3933–8. doi:10.1007/s13277-015-4252-y.
Zlotorynski E. Non-coding RNA: X chromosome inactivation unravelled. Nat Rev Genet. 2015;16(6):315.
Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339(2):159–66. doi:10.1016/j.canlet.2013.06.013.
Lalevee S, Feil R. Long noncoding RNAs in human disease: emerging mechanisms and therapeutic strategies. Epigenomics. 2015;7(6):877–9. doi:10.2217/epi.15.55.
Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–61. doi:10.1038/nm.3981.
Yuan J-H, Yang F, Wang F, Ma J-Z, Guo Y-J, Tao Q-F, et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25(5):666–81.
Qu S, Yang X, Song W, Sun W, Li X, Wang J, et al. Downregulation of lncRNA-ATB correlates with clinical progression and unfavorable prognosis in pancreatic cancer. Tumor Biol. 2016;37(3):3933–8.
Xiong J, Liu Y, Jiang L, Zeng Y, Tang W. High expression of long non-coding RNA lncRNA-ATB is correlated with metastases and promotes cell migration and invasion in renal cell carcinoma. Jpn J Clin Oncol. 2016;46(4):378–84.
Ma C-C, Xiong Z, Zhu G-N, Wang C, Zong G, Wang H-L, et al. Long non-coding RNA ATB promotes glioma malignancy by negatively regulating miR-200a. J Exp Clin Cancer Res. 2016;35(1):1.
Li W, Kang Y. A new Lnc in metastasis: long noncoding RNA mediates the prometastatic functions of TGF-β. Cancer Cell. 2014;25(5):557–9.
Zhou C, Ye L, Jiang C, Bai J, Chi Y, Zhang H. Long noncoding RNA HOTAIR, a hypoxia-inducible factor-1α activated driver of malignancy, enhances hypoxic cancer cell proliferation, migration, and invasion in non-small cell lung cancer. Tumor Biol. 2015;36(12):9179–88.
Schmidt LH, Gorlich D, Spieker T, Rohde C, Schuler M, Mohr M, et al. Prognostic impact of Bcl-2 depends on tumor histology and expression of MALAT-1 lncRNA in non-small-cell lung cancer. J Thorac Oncol. 2014;9(9):1294–304. doi:10.1097/JTO.0000000000000243.
Nie W, Ge HJ, Yang XQ, Sun X, Huang H, Tao X, et al. LncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett. 2016;371(1):99–106. doi:10.1016/j.canlet.2015.11.024.
Lin L, Gu ZT, Chen WH, Cao KJ. Increased expression of the long non-coding RNA ANRIL promotes lung cancer cell metastasis and correlates with poor prognosis. Diagn Pathol. 2015;10:14. doi:10.1186/s13000-015-0247-7.
Nie FQ, Zhu Q, Xu TP, Zou YF, Xie M, Sun M, et al. Long non-coding RNA MVIH indicates a poor prognosis for non-small cell lung cancer and promotes cell proliferation and invasion. Tumour Biol. 2014;35(8):7587–94. doi:10.1007/s13277-014-2009-7.
He XZ, Liu ZL, Su J, Yang JS, Yin DD, Han L, et al. Low expression of long noncoding RNA CASC2 indicates a poor prognosis and regulates cell proliferation in non-small cell lung cancer. Tumor Biol. 2016;31(7):1–8. doi:10.1007/s13277-016-4787-6.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Informed consent
Written informed consents were obtained from all the participants, and this study was approved by the Ethics Committee of Anhui Provincial Hospital.
Conflict of interest
All authors declare that there is no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.

12094_2016_1572_MOESM1_ESM.jpg
Effects of lncRNA-ATB down-regulation on cell viability of NCI-H1299 cells. a Transfection efficiency was confirmed by determining the expression of lncRNA-ATB after transfection of si-ATB (Mock acting as control). b Cell viability was evaluated after transfection. ** P<0.01, vs. Mock (JPEG 551 kb)

12094_2016_1572_MOESM2_ESM.jpg
Effects of lncRNA-ATB down-regulation on cell migration and invasion of NCI-H1299 cells. NCI-H1299 cells were transfected with si-ATB (Mock acting as control). a Cell migration and b cell invasion were evaluated after transfection. ** P<0.01, vs. Mock 2 (JPEG 330 kb)
Rights and permissions
About this article
Cite this article
Ke, L., Xu, SB., Wang, J. et al. High expression of long non-coding RNA ATB indicates a poor prognosis and regulates cell proliferation and metastasis in non-small cell lung cancer. Clin Transl Oncol 19, 599–605 (2017). https://doi.org/10.1007/s12094-016-1572-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-016-1572-3