Abstract
Purpose
To explore the role of miR-33b in colorectal cancer (CRC) and the correlation between its expression and prognosis.
Methods
The expressions of miR-33b between CRC tissues and normal tissues were measured by real-time PCR. The effects of miR-33b on cell proliferation and cell cycle progression were detected by MTT assay, colony formation assay and flow cytometry. The potential regulations of miR-33b on multiple genes expression were verified by Western blot. Furthermore, the association of miR-33b with CRC clinicopathologic features and prognosis was analyzed by Chi-squared test and Kaplan–Meier tests.
Results
MiR-33b was downregulated in CRC compared with normal colorectal samples and miR-33b inhibited tumor cell growth and induced cell cycle arrest. Western blot assays and correlation analysis showed that miR-33b could regulate multiple growth-related genes. Moreover, the expression of miR-33b was associated with TNM stage and tumor size, and CRC patients with high miR-33b expression had a better prognosis.
Conclusion
Our data suggest that miR-33b functions as a tumor suppressor gene in CRC through regulating cell proliferation and cell cycle.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) is one of the most common human malignant tumors and the fourth leading cause of cancer-related death in China [1, 2]. During the past few decades, the incidence and mortality from CRC are rapidly rising in Asian countries [3]. Thus, there is a growing need for understanding the molecular pathogenesis of CRC to develop novel treatment strategies.
Recently, microRNAs (miRNAs) have been proven to play critical roles in many biological processes and increasing evidence has shown that miRNAs have also emerged as key players in the initiation and progression of a variety of human tumors [4]. MiRNAs are capable of simultaneously targeting different genes and regulating their expression post-transcriptionally. Several miRNAs have been demonstrated to exhibit oncogenic or tumor suppressive role in CRC by directly regulating oncogenes or tumor suppressor genes [5–7].
MiR-33 is a highly conserved miRNA family and two miR-33 genes are present in humans, miR-33a and miR-33b, which are encoded within the intron of the SREBP gene and may cooperate with its host genes to regulate lipid metabolism [8]. A recent work has found that miR-33 could regulate cell proliferation through inhibiting the expression of the cyclin-dependent kinase 6 (CDK6) and cyclin D1 (CCND1) in mice [9]. Besides this, miR-33a has been identified as a tumor suppressor by targeting protooncogene Pim-1 [10], and its antitumor effects have been validated in mouse xenograft tumors model [11]. However, the functional relevance of miR-33b in cancer has not been established.
In this study, aiming to characterize the roles of miR-33b in the pathogenesis of CRC, we compared the expression of miR-33b between CRC tissues and normal tissues. The effects of miR-33b on cell proliferation and cell cycle progression were detected. The potential regulations of miR-33b on multiple gene expression were verified. Furthermore, the association of miR-33b with CRC clinicopathologic features and prognosis was analyzed.
Materials and methods
Clinical samples
A total of 60-paired tumor and adjacent normal tissues were obtained from CRC patients who underwent surgical operations at the Department of General Surgery, Jinshan Hospital in Shanghai, China. Samples were subsequently frozen in liquid nitrogen and evaluated pathologically. The tumor stage was classified according to the TNM classification of World Health Organization [12]. Informed consent was obtained from all patients and the study protocol was approved by the Ethics Committee of Fudan University.
Cell culture
The HT-29, HCT 116 and SW480 CRC cell lines used in this study were obtained from the Shanghai Institute of Cell Biology, Chinese Academy of Science. Cells were cultured in DMEM (Gibco, Rockville, MD) supplemented with 10 % fetal calf serum (FCS) (Life Technologies, Burlington, Canada) and antibiotics (50 units/ml penicillin and 50 mg/ml streptomycin, HyClone, Logan, UT) and incubated in 5 % CO2 at 37 °C.
RNA isolation and real-time quantitative PCR
Total RNA was extracted from tissue samples and cells using miRNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Reverse transcription of miR-33b and mRNA was performed using the One Step PrimeScriptH miRNA cDNA Synthesis Kit (Takara, Tokyo, Japan). Real-time PCR was conducted using the TaqMan human miRNA assay kit for miR-33b (Applied Biosystems; Life Technologies) and the SYBRH Premix Ex TaqTM II (Takara) for mRNA of CDK6, CCND1 and Pim-1 on ABI PRISMH 7900 (Applied Biosystems). The primers for the genes were synthesized by Sangon Biotech (Shanghai, China) and listed in Table 1. The expressions of miR-33b or genes were normalized by RNU6B (U6) or β-actin, respectively, and calculated using the \( 2^{{ - {\Delta \Delta }C_{t} }} \) method [13].
miRNA transfection
Cells were seeded in a 24-well plate at 60 % confluence and kept in an incubator at 37 °C and 5 % CO2 overnight. 50 nM miR-33b mimic or the negative control mimic (Ambion Life Technologies, Austin, TX) were transfected into cells using Lipofectamine 2000 (Invitrogen).
For in vivo experiment, an HT-29 cell line stably expressing miR-33b was established using lentiviral vectors. For lentivirus construction, the precursor sequence for miR-33b or irrelevant sequence (negative control) was inserted into hU6-MCS-PGK-puromycin lentiviral vectors (Hanbio, Shanghai, China). The recombinant lentivirus was produced by co-transfection of 293T cells with plasmids PSPAX2 and PMD2G with LipoFiterTM (Hanbio, Shanghai, China). Lentivirus-containing supernatants were harvested 48 h after transfection and filtered through 0.22-μm cellulose acetate filters (Millipore, USA). Recombinant lentiviruses were concentrated by ultracentrifugation (2 h at 50,000×g).
To establish stable cell lines, HT-29 cells were transducted with lentiviral vector in the presence of 5 μg/ml polybrene. After 24 h, the culture medium was removed and fresh medium added. 72 h after transduction, puromycin was added to the medium at the concentration of 5 mg/ml for stable cell line selection. After antibiotic selection for 3 weeks, corresponding cell lines were obtained and the expression level of miR-33b was determined by real-time PCR.
Cell viability assay
After 24 h of transfection of miR-33b mimic or negative control mimic, the cells were digested, re-seeded in 96-well plates (1.5 × 103 per well) and incubated at 37 °C. At daily intervals (day 1, 2, 3, 4, 5), 10 μl of 5 mg/ml 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT, Sigma-Aldrich, St. Louis, MO) was added into cultured cells and incubated for 4 h. The medium was removed and 100 μl of dimethyl sulfoxide was added at the end of incubation. The absorbance was measured at 570 nm using an ELISA reader (Anthos Labtec Instruments). Each experiment was performed in triplicate and repeated three times.
Colony formation assay
HT-29 cells (5 × 104/well) were plated in a 24-well plate and transfected with miR-33b mimic or negative control mimic. After 24 h, the cells were collected and seeded (1000–1500/well) in a dish for 10 days. Surviving colonies were fixed, stained with 5 % gentian violet (Sigma-Aldrich) and counted. The experiment was carried out in triplicate wells for three times.
In vivo tumorigenicity
HT-29 control cells (control), HT-29 cells (5 × 106) stably transducted with lentiviral-miR-33b or lentiviral-negative control (Lv-NC) were injected subcutaneously into the right flank of each BALB/c nude mice (4 weeks old), respectively (6 mice per group). Tumor size was measured every 3 days and the tumor volume (V) was calculated as 0.5 × ab 2 (a largest diameter, b smallest diameter). Mice were humanely killed on day 30, and then tumors were dissected, photographed and weighed. All experimental procedures were approved by the Animal Ethics Committee of Fudan University.
Cell cycle analysis
After 48 h of transfection of miR-33b mimic or negative control mimic, the cells were harvested, washed with PBS and then fixed with 70 % (v/v) ice-cold ethanol overnight at 4 °C. Cells were resuspended and stained with 50 μg/ml propidium iodide (Sigma-Aldrich) for 30 min at 37 °C in the dark and then analyzed by flow cytometry (FACScan; BD Biosciences, San Diego, CA, USA).
Western blot
Cells were homogenized with 400 ml lysis buffer (50 mM Tris_HCl, pH 8.0, 1 mM EDTA, 1 % Triton X-100, 0.5 % sodium deoxycholate, 0.1 % SDS, 150 mM NaCl, 2× protease inhibitor mix). Cell lysates were centrifuged and the protein-containing supernatant was collected and quantified using the Bradford protein assay. A total of 20 mg protein per sample were separated by SDS polyacrylamide gels electrophoresis and then transferred to polyvinylidene difluoride membranes (Millipore, Bedford, MA). The membranes were probed with rabbit anti-human CDK6, CCND1, Pim-1 and β-actin primary antibody (1:1000 dilution, all from Santa Cruz, Dallas, Tex) overnight at 4 °C. After repeated washing, the membranes were then incubated with horseradish peroxidase-labeled goat anti-rabbit IgG (1:1000, Abcam, Cambridge, MA) at room temperature for 1 h. The bands were visualized using ECL Plus Western blotting detection system (GE Healthcare). Western blot for β-actin was performed as an internal sample loading control.
Statistical analysis
Data from at least three independent experiments were expressed as mean values ± standard error. Results of miR-33b level between paired samples were analyzed by the Wilcoxon matched pairs test. Student’s t test was used to analyze the difference between two groups. The Pearson’s Chi-squared test was used to analyze the association of miR-33b expression and clinicopathologic parameters. Kaplan–Meier tests were used for survival analysis. All statistical analyses were performed using SPSS 16.0 software (SPSS, Chicago, IL). P < 0.05 was considered to be statistically significant.
Results
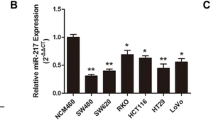
MiR-33b is downregulated in primary CRC and colon cancer cell lines
Total RNA was extracted from 60 pairs of CRC tissues and adjacent normal tissues, and the expression of miR-33b was detected using real-time PCR. As shown in Fig. 1a, the expression level of miR-33b was significantly lower in tumors than in matched non-tumorous tissues. In addition, we identified miR-33b expression in three CRC cell lines and the results showed that miR-33b level was reduced in CRC cell lines, especially in HT-29 cells, compared with normal colorectal tissues (Fig. 1b).
miR-33b was downregulated in primary colorectal cancer tissues and colon cancer cell lines. a The expression of miR-33b in primary colorectal cancer tissues and adjacent normal tissues was determined by real-time PCR (n = 60, normalized with U6). b The expression of miR-33b in CRC cell lines and six randomly selected normal colorectal tissues by real-time PCR
MiR-33b inhibits colon cancer cell growth in vitro and in vivo through inducing cell cycle arrest at G1 phase
To explore the effect of miR-33b on CRC cell proliferation, miR-33b mimic was transfected into HT-29 cells. After transfection, significantly enhanced expression of miR-33b was observed (Fig. 2a). MTT assay and colony formation assay showed that overexpression of miR-33b significantly inhibited the cell proliferation and formed colony of HT-29 cells (Figs. 2, 3). Furthermore, HT-29 cells line stably expressing miR-33b was established and xenograft tumor growth assay was performed to confirm the growth inhibitory effect of miR-33b on CRC cells in vivo. The results showed that miR-33b overexpression significantly suppressed tumor growth and lowered the tumor volume (Fig. 2d, e).
miR-33b inhibited the colon cancer cell growth in vitro and in vivo. HT-29 cells were transfected with 50 nM of miR-33b mimic or negative control mimic (miR-NC). a Expression of miR-33b was detected by real-time PCR. b Cell growth were detected by MTT assay. c Representative images of colony formation assay and quantitative analysis of colony numbers (n = 3). d HT-29 cells transfected with or without miR-33b-overexpression lentivirus or control lentivirus were injected into the nude mice. Tumor size was measured and tumor growth curve was drawn. e Representative images of the tumors were shown. *p < 0.05, compared with control
A recent study revealed that miR-33 could regulate cell cycle progression in mice [9]. Therefore, we investigated whether the anti-proliferative effect of miR-33b in HT-29 cells correlated with cell cycle arrest. As shown in Fig. 3, the number of CRC cells in the G1 phase was significantly increased after transfection with miR-33b, compared to the control (p < 0.05).
MiR-33b inhibits multiple gene expression in colon cancer cell
Analysis of potential targets by TargetScan showed that miR-33b may have same binding sites as miR-33a in multiple genes 3′-UTR, including CDK6, CCND1 and Pim-1 (Supplementary Figure 1). Then we tested whether miR-33b regulates the protein levels of these three genes in CRC cells. Western blot showed that transfection of miR-33b resulted in significant reduction of CDK6, CCND1 and Pim-1 protein expression (Fig. 4a). Moreover, the expression of miR-33b and mRNA of the three genes exhibited significant inverse correlations in 60 primary CRC tissues.
miR-33b inhibits multiple gene expression. a HT-29 cells were transfected with miR-33b mimic or negative control mimic (miR-NC). The protein expressions of CDK6, CCND1 and Pim-1 were detected by Western blot. b Correlation analysis between expressions of miR-33b and mRNA of the three genes in 60 primary CRC tissues. The expressions of miR-33b and genes were normalized by RNU6B (U6) or β-actin, respectively
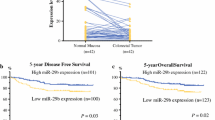
High miR-33b expression is associated with better survival
We then examined the association of miR-33b expression with clinical outcome. The patients were divided into two groups based on the relative expression level of miR-33b in paired tumor tissues and normal tissues of the same patient. Patients with lower level of miR-33b in tumor than normal tissues were assigned to the low expression group (n = 37) and the rest were assigned to the high expression group (n = 23). The association of the expression of miR-33b with clinicopathological factors was analyzed (Table 2). The reduced expression of miR-33b was significantly related to TNM stage (p < 0.05) and tumor size (p < 0.01). However, no significant differences were observed regarding age, gender, tumor differentiation, lymph node metastasis and depth of wall invasion. Kaplan–Meier survival analysis showed that patients with high miR-33b expression had better survival than those with low miR-33b expression (Fig. 5, p < 0.05).
Discussion
In this study, we explored the potential function of miR-33b in CRC and found its tumor suppressor activity. Our results indicated that miR-33b was downregulated in tumors compared with normal colorectal samples and that miR-33b inhibited tumor cell growth through regulating multiple genes. Further analysis showed that low expression of miR-33b was associated with TNM stage, tumor size and worse prognosis.
There are two members in the miR-33 family in humans, miR-33a and miR-33b, which are located within intronic sequences of SREBP-2 (chromosome 22) and SREBP-1 (chromosome 17), respectively. It has been shown that miR-33a/b regulates similar cellular processes to their host genes, such as cholesterol and fatty acid metabolism, by targeting the relevant genes [8, 14]. Recent works indicated that miR-33a could regulate cell cycle progression and suggested its role as a tumor suppressor [10]. However, the function of miR-33b in cancer remains unclear.
Here, we evaluated the expression of miR-33b in CRC and found that miR-33b was downregulated in CRC tissue and cells compared with the normal tissues. In addition to CRC, downregulation of miR-33b has been reported in gastric cancer [15]. We next characterized the function and potential mechanisms of miR-33b in regulating the biological behavior of CRC cells. Our results showed that the restoration of miR-33b in the colon cancer cell lines HT-29 significantly inhibited cell proliferation in vitro as evidenced by MTT and colony formation assays. Overexpression of miR-33b also repressed the growth of the xenograft tumor in vivo. Furthermore, cell cycle analysis evaluated by flow cytometry revealed a marked increase of cells in G0/G1, which could contribute to inhibition of cell proliferation. These data suggested that miR-33b played a similar role as a tumor suppressor to miR-33a. Since miR-33b and miR-33a have similar sequences and bind sites, we therefore validated whether miR-33b could regulate the genes which have been confirmed as targets of miR-33 or miR-33a. Western blot assay showed that restoration of miR-33b reduced the expression of CDK6, CCND1 and Pim-1 in HT-29 cells. Negative correlations were also observed between miR-33b expression and mRNA expression of these genes in the CRC tissue. Both CDK6 [16] and CCND1 [17] are critical mediators of cell cycle progression and are demonstrated as targets of miR-33a [9]. Pim-1 is another confirmed target gene of miR-33a and functions as a proto-oncogene [10, 18]. These results suggested that miR-33b played comparable roles with miR-33a by regulating multiple genes.
Moreover, to investigate the clinical value of miR-33b, we analyzed the relationship of miR-33b expression and clinicopathological features. The expression of miR-33b was considered to be associated with TNM stage and tumor size. In the Kaplan–Meier survival analysis, there is a significantly better survival rate in patients with high miR-33b expression than those with low expression. These findings suggested the potential implications of miR-33b as a prognostic marker and therapeutic target.
On the other hand, like many intronic miRNAs, miR-33b is co-transcribed with its host gene. Emerging evidences indicated that miR-33a/b act in concert with the SREBP host genes to control cholesterol and lipid homoeostasis [8, 14], implying that they could be potential therapeutic targets for metabolic syndrome [19, 20]. However, our finding suggested that the risk of oncogenesis and tumor development should be considered when using miR-33 inhibitor as a therapeutic strategy. These results also hint that there may be an association between impaired lipid metabolism and the risk of cancer. More recently, the association was observed in epidemiologic studies [21–23], but the mechanisms of this link and whether miR-33 is involved deserve further investigation.
In conclusion, our study demonstrated that miR-33b was a tumor suppressor in CRC. MiR-33b was frequently underexpressed in CRC and restoration of miR-33b inhibited cell growth both in vitro and in vivo. The underlying mechanisms by which miR-33b functions in CRC may involve inhibition of cell cycle progression and multiple gene expression. Furthermore, miR-33b was identified to be associated with TNM stage, tumor size and overall survival, implying that miR-33b can be a potential prognostic marker.
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90.
Yang L, Parkin DM, Li L, Chen Y. Time trends in cancer mortality in China: 1987–1999. Int J Cancer. 2003;106(5):771–83.
Sung JJY, Lau JYW, Goh KL, Leung WK. Increasing incidence of colorectal cancer in Asia: implications for screening. Lancet Oncol. 2005;6(11):871–6.
Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–66.
Zhang L, Dong Y, Zhu N, Tsoi H, Zhao Z, Wu CW, et al. microRNA-139-5p exerts tumor suppressor function by targeting NOTCH1 in colorectal cancer. Mol Cancer. 2014;13:124.
Rokavec M, Öner MG, Li H, Jackstadt R, Jiang L, Lodygin D, et al. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J Clin Investig. 2014;124(4):1853–67.
Xiang KM, Li XR. MiR-133b acts as a tumor suppressor and negatively regulates TBPL1 in colorectal cancer cells. Asian Pac J Cancer Prev. 2014;15(8):3767–72.
Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, et al. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328(5985):1566–9.
Cirera-Salinas D, Pauta M, Allen RM, Salerno AG, Ramírez CM, Chamorro-Jorganes A, et al. Mir-33 regulates cell proliferation and cell cycle progression. Cell Cycle. 2012;11(5):922–33.
Thomas M, Lange-Grunweller K, Weirauch U, Gutsch D, Aigner A, Grunweller A, et al. The proto-oncogene Pim-1 is a target of miR-33a. Oncogene. 2012;31(7):918–28.
Ibrahim AF, Weirauch U, Thomas M, Grünweller A, Hartmann RK, Aigner A. MicroRNA replacement therapy for miR-145 and miR-33a is efficacious in a model of colon carcinoma. Cancer Res. 2011;71(15):5214–24.
Sobin LH, Gospodarowicz MK, Wittekind Ch. TNM classification of malignant tumors. 7th ed. Oxford: Wiley-Blackwell; 2009.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8.
Davalos A, Goedeke L, Smibert P, Ramirez CM, Warrier NP, Andreo U, et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci USA. 2011;108(22):9232–7.
Miyachi K, Sawada Y, Shida Y, Sugawara A, Hisatomi H. Lipogenic gene expression profile in patients with gastric cancer. Mol Clin Oncol. 2013;1(5):825–7.
Rader J, Russell MR, Hart LS, Nakazawa MS, Belcastro LT, Martinez D, et al. Dual CDK4/CDK6 inhibition induces cell-cycle arrest and senescence in neuroblastoma. Clin Cancer Res. 2013;19(22):6173–82.
Schafer KA. The cell cycle: a review. Vet Pathol. 1998;35(6):461–78 (Online).
Nawijn MC, Alendar A, Berns A. For better or for worse: the role of Pim oncogenes in tumorigenesis. Nat Rev Cancer. 2011;11(1):23–34.
Rayner KJ, Esau CC, Hussain FN, McDaniel AL, Marshall SM, van Gils JM, et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 2011;478(7369):404–7.
Ono K, Horie T, Nishino T, Baba O, Kuwabara Y, Yokode M, et al. MicroRNA-33a/b in lipid metabolism—novel “thrifty” models. Circ J. 2015;79(2):278–84.
Giovannucci E. Metabolic syndrome, hyperinsulinemia, and colon cancer: a review. Am J Clin Nutr. 2007;86(3):836S–42S.
Zhao W, Guan J, Horswell R, Li W, Wang Y, Wu X, et al. HDL cholesterol and cancer risk among patients with type 2 diabetes. Diabetes Care. 2014;37(12):3196–203.
Strickler HD, Wylie-Rosett J, Rohan T, Hoover DR, Smoller S, Burk RD, et al. The relation of type 2 diabetes and cancer. Diabetes Technol Ther. 2001;3(2):263–74.
Author information
Authors and Affiliations
Corresponding author
Additional information
W. Liao and C. Gu contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liao, W., Gu, C., Huang, A. et al. MicroRNA-33b inhibits tumor cell growth and is associated with prognosis in colorectal cancer patients. Clin Transl Oncol 18, 449–456 (2016). https://doi.org/10.1007/s12094-015-1388-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-015-1388-6