Abstract
Antibiotic resistance of Staphylococcus aureus has considerably increased among non-clinical or asymptomatic individuals. The formation of biofilms denies antimicrobial access to its targets present on the surface and inside the cell. The present study tested the effect of the combination of flavonoids and antibiotics over the preformed biofilms of S. aureus. The eradication of the preformed biofilms was analyzed using the crystal violet method. It has shown that 2500 µg mL−1 Rutin and 100 µg mL−1 Erythromycin (MIC Concentration) combination efficiently reduced the growth of the cells, which were adhered to the surfaces forming the biofilms. Fluorescence microscopic analysis indicated that the Rutin and Erythromycin (MIC value) combinations could eradicate the preformed biofilm cells more efficiently than other combinations. We found that the flavonoids and antibiotics with MIC concentration show a significant effect over the preformed biofilms cells of S. aureus. In addition, the semi-quantitative real-time PCR analysis for the sRNAs under the treatment of Rutin and Erythromycin combinations showed that few small RNAs expression (SprF, SprG, ArtR, Teg49, Teg41, and RNAIII) are getting downregulated upon the treatment; but again recovers with the incubation time interval increases. Combinations have a significant effect on Teg49 where there is a very faint intensity of the band, but for other small RNAs, there is an irregular pattern on the gel image. It has been concluded that at the initial period of incubation, the combinations have an effect on all the sRNAs but once the incubation increases, the effects have been slowly decreasing. It has been concluded that the combination has been able to reduce the doubling time of S. aureus upon treatment. Whereas, the small RNAs used in the study can be further evaluated for expression profiling through qRT-PCT.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Biofilm causes the physical barrier around the bacteria, which reduces antibiotic penetration into the cell and is one of the attributes of antimicrobial resistance [1]. However, reduced bacterial growth rate, active starvation response, and changes in bacterial gene expression also contribute to biofilm resistance [2,3,4,5] this inherent resistance makes biofilm-associated infections very difficult to overcome [6, 7]. Formation of biofilm remains the major virulence determinant because S. aureus bacteria stays longer in the genitourinary tract when a person gets infected [8]. Other factors that enhance the possibility of biofilm formation in patients are catheters and other prosthetic devices; therefore, it provides the environment for the development of infections in the Urinary tract lining by killing natural barrier molecules [9]. Antibiotic therapy with beta-lactam antibiotics induces biofilm formation in the MSSA even at sub-Minimum Inhibitory Concentration (MIC) levels (Methicillin Susceptible Staphylococcus aureus) [10]. The cells escape from the treatment by deeply burying themselves in the biofilms [11, 12]. Most of the time, data on antimicrobial treatment on planktonic bacteria were used to select the choice of antibiotic therapy, which is not representative of biofilm bacteria.
Usually, the dosage of the antibiotics is defined by their effect on the planktonic cells and how they act against bacteria in the planktonic forms (free forms). Therefore, these antibiotics are used only as inhibitory molecules for the planktonic cells as a chemotherapeutic compound but not the inhibition of biofilm formation or affect the preformed biofilms [13]. The antibiotic’s inhibitory concentration has a positive correlation with the biofilm formation rate [14]. For some antibiotics, 1000-fold more antibiotic concentration is required to kill sessile bacteria than a similar dosage of antibiotics or drugs needed for the killing of planktonic cells of the same microorganisms [15, 16]. The study aimed to determine the drug concentrations required to eradicate in vitro S. aureus biofilm and/or to inhibit biofilm formation using a representative clinical strain of MSSA (Methicillin Susceptible Staphylococcus aureus) isolated from an infection formed on the catheter inserted for the treatment of the bone injury [17].
Small RNA plays a major role by pairing with bases of target mRNA or by interacting with the modulating proteins for both the positive and negative mechanisms of biofilm formation. Regulation of gene expression mediated by sRNAs is more beneficial when compared to proteins during rapid response because it takes a short time for sRNAs to either synthesize or degrade. Various regulatory mechanisms of sRNAs are similar to the regulation of quorum sensing in bacteria. Since the quorum sensing mechanism controls the virulence factor of bacteria, it is considered the major target for finding out new therapeutic methods [18] (Table 1).
In our study, we have examined the effects of different flavonoids in combination with conventional antibiotics against preformed biofilms of S. aureus. Further, the microscopic analysis of the different combinations over the preformed biofilm of S. aureus was studied. In addition, the effect of treatments on small RNAs related to biofilm formation was studied.
Materials and Methods
Bacterial Culture, Media and Antimicrobials
All the antibiotics (Erythromycin, Vancomycin, Tetracycline, Rifampicin, and Gentamicin), and flavonoids (Quercetin, Rutin, Morin, and Naringenin) were purchased from Himedia Laboratories Pvt. Ltd., Mumbai, India. Susceptible strain S. aureus (MTCC96) was purchased from Microbial Type Culture Collection and Gene Bank (MTCC), Chandigarh, India. The isolated strain of S. aureus from asymptomatic individuals was used as test organisms [23]. The strain was maintained on the Mannitol Salt Agar (MSA) media. The Tryptic soy broth (TSB) was used to standardize the biofilm formation. Mueller Hinton Broth (MHB) was used to check the effects of combinations of flavonoids and antibiotics against the preformed biofilms of S. aureus.
Estimation of Biofilm Formation of S. aureus
The biofilm formation of bacteria was analyzed using the Congo-red agar (CRA) method [19]. The Congo red agar medium was prepared with BHI broth (37 g L–1, sucrose 50 g L–1), agar 10 g L–1 and Congo red stain 0.8 g L–1. Plates were inoculated with tested microorganisms, incubated for 24–48 h at 37 °C. The change in colour of colonies from pink to dark reddish or black colour indicates the biofilm formation. The efficiency of the biofilm formation of S. aureus was performed using the tissue culture plate method [20, 21]. The method in brief: The overnight culture of S. aureus (6 × 107 CFU/mL) was added to the plate containing TSB medium supplemented with 2% glucose. After incubating for 48 h at 37 °C, the planktonic cells were aspirated out and washed with 1× PBS buffer (pH 7.4) to remove the non-adhered bacteria. The developed biofilm was fixed by incubating at 65 °C for 15 min. The biofilm of adhered bacteria was stained with the addition of a 0.1% (w/v) crystal violet solution. The excess stains were removed by washing them with deionized water. The 33% (v/v) glacial acetic acid was added to dissolve the biofilm, and the absorbance at 570 nm was measured. The efficiency of biofilm formation of bacteria was determined based on the absorbance value: Absorbance < 0.120 is non-adherent and weak biofilm producers; 0.120 < absorbance < 0.240 is moderately adherent and biofilm producers; absorbance > 0.240 strongly adherent and high biofilm producers [22].
Effect of Treatments on Preformed Biofilm
The effect of different combinations on preformed biofilms was evaluated to determine the efficacy of the treatment with previously reported MIC concentration [23]. The S. aureus was diluted to 1:100 in the TSG medium supplemented with 2% glucose, which enhances the biofilm production. Once the absorbance of the culture achieved a 0.1 OD value at 600 nm, it was added to the 24-well plate. After incubating at 37 °C for 48 h, the planktonic cells were removed, and wells were washed thrice with 1X PBS. Further fresh TSB media was added to the wells to study the effects of combinations [24]. The drug combination of flavonoids and antibiotics at MIC concentrations was added to each well with positive control as culture alone, negative control as TSB media alone, and incubated at 37 °C for 12 h. Then, the wells were aspirated and washed thrice with 1 X PBS. After fixing the biofilms by incubating the plate at 65 °C for 15 min, 0.1% (v/v) crystal violet solution was added to each well, incubated for 20 min at room temperature. The wells were washed thrice with 1× PBS to remove the excess stain. After the air dry, 33% glacial acetic acid solution was added to the well to dissolve the fixed biofilm, and intensity was measured at 570 nm. The percentage of biofilm eradication was calculated using the formula descriptive below [25]:
Fluorescence Microscopic Imaging for Biofilm Eradication Assay
The biofilm eradication was measured using microscopic observation [26]. The S. aureus isolated strain was inoculated in TSB growth media supplemented with 2% glucose and cultured over coverslips in 6 well plates. After 48 h of incubation at 37 °C, the plate with coverslips was washed with 1× PBS buffer to remove the planktonic cells and non-adherent cells. After that, the cells were fixed by incubating the plate at 65 °C for 15 min. Then, fresh TSB media was added to the wells and positive control as media + culture, and negative control as media alone was kept for the study. Further, different drug combinations of flavonoids and antibiotics at MIC concentrations, which were previously determined for Planktonic cells [23], were added. The plates were allowed to incubate at 37 °C for 12 h on a shaker. Then the media is removed and again washed with 1× PBS buffer. After the fixation of biofilms, 0.001% (w/v) of acridine orange acidic pH (pH 5.0) was added and incubated for 15 min. The stained coverslips were visualized in a fluorescence microscope Leica DM6 Fluorescent Microscope with Cryostat at an excitation wavelength of 488 nm. Quantitative analysis for the fluorescent imaging was calculated using Image Proplus 10.0 software [27].
Analysis of Biofilm Formation Related to Small RNA Expression Under Antimicrobial Treatment
Staphylococcus aureus cultures was grown in brain heart Infusion at 37 °C, which is the suitable media for the. The cells were allowed to grow for 6 h, and then the combination was given to the cells and incubated for three different time intervals 12 h, 18 h and 24 h respectively. The cells were harvested and the RNA was isolated with slight modifications [28]. The quality and quantity of RNA were evaluated using a bio-photometer (Thermo Scientific, USA) with a ratio of A230/280 ratio. The RNA was stored at − 80 °C for further use. The small RNAs were taken from the Staphylococcus regulatory RNA database (SRD database) for the NCTC8325 strain.
The first strand of the cDNA was synthesized using isolated RNA under different drug treatments. PCR amplification of cDNA was performed under the following conditions: the mixture was incubated at 25 °C for 5 min. The 20 µL cDNA synthesis reaction was performed at 42 °C for one hour. The enzyme was inactivated at 80 °C for 5 min. The synthesized cDNA was stored at − 20 °C. The specific small RNA (sRNAs) primers were used for second-strand synthesis (Table 2). The SYBR Green qPCRs were performed using the following parameters: 3 min at 95 °C; 35 cycles of 95 °C for 20 s and 58 °C for 30 s; and 30 s at 72 °C. A melting curve analysis was added to ensure the specificity of the PCR product. Semiquantitative analyses were done for all the sRNAs after the RT-PCR. Further, the qRT-PCR expression of the two small RNA RNAIII and ArtR in S. aureus was measured using 16 s rRNA as an endogenous control. The expression level was calculated using the (2 − ΔΔCt) method [29].
Statistical analysis
Data from the experiments are presented as the mean ± SEM. The level of statistical analysis was performed by ANOVA and the P values < 0.01 were considered significant. The data for the qRT-PCR are mean ± S.D. for triple-independent experiments (** = P < 0.05). The statistical analysis was performed using GraphPad Prism software.
Results
Evaluation of Biofilm Formation by S. aureus
The crystal violet method was used to evaluate the biofilm formed by S. aureus. The absorbance values at 570 nm indicate the level of biofilm formed in the microtiter plate. The biofilm formation was optimized in a microtiter plate to obtain the appropriate biofilm production to study biofilm eradication. During normal conditions, the formation of biofilm, when estimated using crystal violet, the absorbance was not sufficient to perform the study. After optimizing the media by supplementing the 2% glucose, there was a significant increase in the production of biofilms. Initially, we evaluated the biofilm formation under normal conditions with 1% glucose to find the efficiency of the biofilm formation. After 48 h of incubation, an inadequate amount of biofilm was formed (Absorbance at 570 nm: sensitive strain: 0.175 and isolated strain: 0.370), which was insufficient to perform the biofilm eradication assay. The increase in carbon source (glucose) from 1% (w/v) to 2% (w/v) enhances the biofilm formation (sensitive strain 96—0.370, isolated strain—0.760) within 48 h as the bacterial growth increased (Fig. 1). Glucose acts as a carbon source and metabolite, which has various effects on the growth of bacteria and the involvement of biofilm formation. It has been reported that in an adhesion-dependent manner, the glucose-induced formation of the biofilm of S. aureus was regulated by accessory protein GbaAB in a polysaccharide intercellular [30].
Effect of Flavonoids in Combinations with Antibiotics on Preformed Biofilms
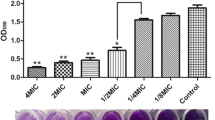
We have performed a biofilm eradication assay to find the effect of the treatment on the preformed biofilms of S. aureus. The antibiotics and flavonoids were used in combination to test the biofilm eradication. The crystal violet assay was used to calculate the percentage of eradication of preformed biofilms. The combinatory treatments showed better biofilm eradication effects on preformed bacterial biofilms. The MIC concentrations, which were reported in the previous research, have been used for the preformed biofilm eradication assay [23]. Different combinations and their concentrations were given in Table 3. For susceptible strain, the higher percentage of biofilm eradication was found in the following combinations: 125 μg mL−1 Rutin + 6.25 μg mL−1 Erythromycin—86%, 250 μg mL−1 Quercetin + 6.25 μg mL−1 Vancomycin—83%. Similarly, for isolated strain, the combinations with high biofilm eradication were 2500 μg mL−1 Rutin + 100 μg mL−1 Erythromycin—68%, 2500 μg mL−1 Rutin + 25 μg mL−1 Vancomycin—68% (Fig. 2).
Microscopic Determination of Biofilm Eradication Assay
Micro-morphological study of preformed biofilm eradication was performed using a fluorescence microscope stained with acridine orange dye to differentiate the log and lag phases of cells. Active cells or live cells are appeared in green colour after the treatment with the Acridine Orange dye (acidic pH), whereas red fluorescence has been observed when it has bounds with ssDNA or RNA and it is termed as Phagocytosed cells [27]. Growth has been inhibited, and the cells are in the log phase. A fluorescence microscope (Fig. 3) evaluated the analysis of the biofilm architecture in the presence of different combinations at MIC concentration. Fluorescence microscope results showed a wrinkled or less compact biofilm architecture whereas non-treated cells showed highly compact biofilm architecture. In addition, a reduction in the cell number reflects the eradication activity of the combinations of flavonoids and antibiotics against preformed bacterial biofilms. Whereas few treatments showed medium compactness of the biofilm architecture, which indicates that the combinations were not able to eradicate the preformed biofilms much efficiently. In addition, the relative fluorescence intensity determination of the cells showed a clear difference between the treatment and control samples (non-treatment) (Fig. 4). In most cases, eradicating the preformed biofilms in S. aureus is considered as the most challenging task. Because of the attachment of the bacteria on the surfaces, produces various virulence factors and adhesins, and forms a complex architecture that encourages the resistivity pattern in the bacteria [31, 32]. Previous studies have found several anti-biofilm agents which were highly active in inhibiting biofilm yet were unable to disperse the pre-existing mature one [33, 34].
Graphical representation of the corrected total cell fluorescent (CTCF) and Integrated Density for the different treatment combinations in biofilm eradication assay. A Control, B Quercetin + Erythromycin, C Naringenin + Erythromycin, D Rutin + Vancomycin, E Quercetin + Vancomycin, F Morin + Vancomycin, G Rutin + Erythromycin, H Morin + Erythromycin, I Naringenin + Vancomycin
Semi-quantitative PCR for Small RNA (sRNA)
The Rutin and erythromycin combinations showed a better effect in reducing the preformed biofilms with the lowest FIC values among the treatment of Rutin and Erythromycin combinations. The virulence-related small RNA for biofilm formation was analyzed under the treatment of this antimicrobial combination. The non-clinical isolated strains of S. aureus were grown under the different combinations of MIC concentrations of Rutin and erythromycin at different time intervals of 12 h, 18 h, and 24 h. The RNA was isolated from the treated samples and control samples. The isolated RNA was used to perform reverse transcriptase PCR for selected virulence-related small RNAs, including Teg49, ArtR, Teg41, sprX, SprG1, SprF1, SprC, and RNAIII. The differential expression of this small RNA under different treatments was evaluated.
Using the intensity of the bands and the Image J analysis software, we have determined the relative expression of the small RNAs. The differentially expressed small RNA expression level under different conditions of time intervals for Rutin and Erythromycin combinations were analyzed. From the relative expression, it has been observed that most of the sRNAs are downregulated after the treatment in the initial phase. Among all the sRNAs, only ArtR and Teg41 have been upregulated more in comparison to the control gene (16sr RNA), but other sRNAs are downregulated. Even the RNAIII, which is considered the standard sRNA being expressed in every condition during virulence production, was downregulated when it is treated in the initial phase (Fig. 5). Whereas some sRNAs such as srpG (12 h), sprF (18 h), and Teg49 (12 h, 18 h and 24 h) do not show any expression, which means that these sRNAs are downregulated completely when they are treated in the combinations (Fig. 4). From this data, we can state that these combinations are having a significant effect in downregulating the small RNA. However, the effect starts reducing in some of the sRNAs when the incubation time increase, with this we can infer that the combination can act as a bacteriostatic agent rather than a bactericidal agent. In addition, the other reason for the upregulation of sRNAs upon the increase of time interval is that under stress conditions, the production of virulence factors reduces the effect of the combinations.
RT-PCR confirmation of differential expression in Staphylococcus aureus for differentially expressed small RNAs under combinatorial treatment in different time intervals. The relative levels of sRNA expression in comparison with the wild type are plotted in a bar graph with relative expression levels
The expression of SprF reduced as the increase in treatment time in comparison with untreated samples, while the increase in RNAIII expression was observed with increased treatment time (Fig. 6). The treatment reduces the expression of Teg41 and it completely suppressed the expression of Teg49. No change in the expression of the reference gene was observed (16sRNA). The results indicated the treatment-driven expression change in the virulence-related small RNA. The expression pattern of sprG differs from wild type to different time intervals, where the intensity of bands showed that there was a mild expression in 18 h. For 12 h, there was no expression from the band intensity. For ArtR, the expression pattern bright band for wild type shows a good expression, and then the faint expression of 12 h indicated that there might be a little expression of sRNA under the treatments. Similarly, for 18 h and 24 h, moderate expression was observed (Fig. 7).
Further, qRT-PCR analysis was performed for two specific small RNAs (RNAIII and ArtR) which are specifically involved in the regulation of virulence genes regulation and regulated by the agrA promotor. Through relative fold change, it has been found that under 12 h of treatment RNAIII has downregulated with 1.89-fold change, in 18 h it has upregulated to 4.1-fold change, and in 24 h, it has upregulated to 8.15-fold change with respect to wild type of S. aureus. Since in ArtR sRNA, wild type has prominent expression, with respect to fold change; for 12 h under treatment, it has downregulated to 11.56-fold change, but it is upregulating in 18 h of treatment with 0.8-fold change with 2.4-fold change after 24 h of treatment (Fig. 8). The reason for the inconsistency expression of these sRNAs is due to the production of virulence factors, which indirectly suppress the effects of the treatment combinations.
Expression levels of the two sRNA in three different time intervals of treatment relative to those in the wild type were determined by quantitative RT-PCR. 16s rRNA was used as a housekeeping gene. Expression levels were normalized to 16s rRNA level. The data are mean ± S.D. for triple-independent experiments (** = P < 0.05)
Discussions
To determine the antibiotic susceptibility pattern of the microorganisms, MIC has been used effectively and is considered the gold standard [35]. If the determined MIC concentration states that the drug is ineffective in the in-vitro model, then it is not considered for the clinical trials since it will not have any significant effect on the patients [36]. In another case, if the drug is having an effect in the in-vitro model, it cannot ensure that it will have a significant effect in the in-vivo model [37,38,39]. In research laboratories, the effect of the drug is reported only through MIC determination in planktonic cells, which may not be as effective in infected patients. The biofilm formation pattern in the microorganisms protects them from the effect of antibiotics.
Biofilms have overexpressed efflux pumps, a multilayer of cells through which permeability is an issue and have anaerobic conditions in the inner layers of cells. These conditions in addition to quorum sensing are reasons for higher resistance. Biofilm is produced through quorum sensing means by releasing the exo-polysaccharides. EPS helps in the formation of a connection between cells and thereby forming a network-like structure. In sub-MIC, does bacteria cannot be killed. Biofilm is formed by bacteria attaching to a specific surface enclosed in extra polymeric substances the antibiotics or antimicrobials are usually difficult to reach the bacteria, and thus the efficacy of antibiotics is reduced. In addition, there is a quorum sensing effect among the colonies when biofilm forms on the surface, which can also increase bacterial resistance [40]. The effect of the treatment over preformed biofilms was tested with a biofilm eradication assay. The result showed that Rutin and Erythromycin had better activity in eradicating the preformed biofilms. The fluorescence microscopic imaging also confirmed that Rutin and Erythromycin have a better effect on preformed biofilm eradication. For the first time, it has been reported that there is an increase in the antimicrobial resistance pattern of S. aureus among the skin of asymptomatic individuals.
There is no previous report available on combinatorial treatments and their effect on small RNA expression. However, some reports have studied the effect of Linezolid alone on the small RNA-regulated virulence factor in S. aureus [41]. Another study has reported that the regulation of sRNA has been associated with secretions of extracellular vesicles in S. aureus. Several sRNA and tRNA molecules with different regulatory functions have been reported to have associations with extracellular vesicles. Therefore, targeting these small RNAs could provide us insight into developing or finding a novel anti-virulence therapy to overcome the resistance pattern among S. aureus infections [42]. We are reporting for the first time that the combination of Rutin and Erythromycin can downregulate the expression of virulence regulating small RNAs. The combinations against preformed biofilms have not been tested earlier and the study related to small RNA expression profiling. Since the individual effect of flavonoids and antibiotics against planktonic cells of S. aureus has already been reported in our earlier paper [23], we have used the Minimum Inhibitory Concentrations of that article to determine the effect of MICs combinations of the flavonoids and antibiotics against the preformed biofilms of the S. aureus. It has been earlier reported that antibiotics alone are not able to completely reduce or inhibit the formation of the biofilm. In our study, we have directly evaluated the effect of the combinations of antibiotics and flavonoids for the eradication of preformed biofilms in S. aureus. In addition, the combinations are significant in eradicating the preformed biofilms in the well plate. Quercetin is the most abundant flavanol present in the plant species, Morin is the isomeric form of Quercetin and it has been reported to have antibacterial activity. Rutin is the glycosidic form of isomeric Quercetin. However, the reports for the antibacterial efficiency of these flavonoids are variables, particularly regarding the MICs concentrations. Because of the methods used for inter and intra-assay differences in determining susceptibility testing or may be due to the difference in the genetic variation in the bacterial strains used in the assay [43, 44].
The semi-quantitative reverse transcriptase PCR analysis for the differential expression of virulence-related small RNA under the drug treatments. The small RNA Teg41 does not show any expressions for three different time intervals under treatments. Similarly, sprG1 and sprF1 do not show any differential expression for 12 h and 18 h respectively. The RNAIII showed prominent expression under the treatments and its expression pattern gradually increases from 12 to 24 h. Other small RNAs show a similar expression pattern to RNAIII. Still, many questions related to the physiological roles of small RNAs have been unanswered. With the help of transcriptomic profiling, we can reveal the role of small RNAs and the effect of individual or combinations of drugs during the infection process in S. aureus, and how the biofilm eradication process takes place.
There are certain limitations of this study because, upon the treatment with combinations at different time intervals; there are certain small RNAs that have been expressed with the increasing time intervals. This states that even though the combinations have been able to kill the planktonic cells efficiently; and be able to reduce the formation of preformed biofilms when sRNAs expression was analyzed it has been found that there are different expression patterns with the increasing time interval. This indicates that further study need to be carried out to know the actual mechanism behind the regulation of small RNA expression under the stress condition or under the treatment conditions using transcriptomic profiling. This will help the researcher to understand and develop the drug targeting the sRNAs-mediated virulence factor production in a more efficient manner.
References
Suci PA, Mittelman MW, Yu FP, Geesey GG (1994) Investigation of ciprofloxacin penetration into Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 38:2125–2133. https://doi.org/10.1128/AAC.38.9.2125
Evans DJ, Allison DG, Brown MR, Gilbert P (1990) Effect of growth rate on resistance of gram-negative biofilms to cetrimide. J Antimicrob Chemother 26:473–478. https://doi.org/10.1093/jac/26.4.473
Mah TF, Pitts B, Pellock B, Walker GC, Stewart PS, O’Toole GA (2003) A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426:306–310. https://doi.org/10.1038/nature02122
Zhang L, Mah TF (2008) Involvement of a novel efflux system in biofilm-specific resistance to antibiotics. J Bacteriol 190:4447–4452. https://doi.org/10.1128/JB.01655-07
Nguyen D, Joshi-Datar A, Lepine F et al (2011) Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334:982–986. https://doi.org/10.1126/science.1211037
Stewart PS, Costerton JW (2001) Antibiotic resistance of bacteria in biofilms. Lancet 358:135–138. https://doi.org/10.1016/s0140-6736(01)05321-1
Donlan RM, Costerton JW (2002) Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev 15:167–193. https://doi.org/10.1128/CMR.15.2.167-193.2002
Costerton JW, Stewart PS, Greenberg EP (1999) Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. https://doi.org/10.1126/science.284.5418.1318
Delcaru C, Alexandru I, Podgoreanu P, Grosu M, Stavropoulos E, Chifiriuc MC, Lazar V (2016) Microbial biofilms in urinary tract infections and prostatitis: etiology, pathogenicity, and combating strategies. Pathogens 5:65. https://doi.org/10.3390/pathogens5040065
Dibah S, Arzanlou M, Jannati E, Shapouri R (2014) Prevalence and antimicrobial resistance pattern of methicillin-resistant Staphylococcus aureus (MRSA) strains isolated from clinical specimens in Ardabil, Iran. Iran J Microbiol 6:163–168
Ng M, Epstein SB, Callahan MT et al (2013) Induction of MRSA biofilm by low-dose β-lactam antibiotics: specificity, prevalence and dose-response effects. Dose-Response 12:152–161. https://doi.org/10.2203/dose-response.13-021
Singh R, Ray P, Das A, Sharma M (2010) Penetration of antibiotics through Staphylococcus aureus and Staphylococcus epidermidis biofilms. J Antimicrob Chemother 65:1955–1958. https://doi.org/10.1093/jac/dkq257
Jorgensen JH, Ferraro MJ (2009) Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin Infect Dis 49:1749–1755. https://doi.org/10.1086/647952
Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O (2010) Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents 35:322–332. https://doi.org/10.1016/j.ijantimicag.2009.12.011
Nickel JC, Ruseska I, Wright JB, Costerton JW (1985) Tobramycin resistance of Pseudomonas aeruginosa cells growing as a biofilm on urinary catheter material. Antimicrob Agents Chemother 27:619–624. https://doi.org/10.1128/AAC.27.4.619
Aslam S (2008) Effect of antibacterials on biofilms. Am J Infect Control? 36:S175.e9–11. https://doi.org/10.1016/j.ajic.2008.10.002
Neopane P, Nepal HP, Shrestha R et al (2018) In vitro biofilm formation by Staphylococcus aureus isolated from wounds of hospital-admitted patients and their association with antimicrobial resistance. Int J Gen Med 11:25–32. https://doi.org/10.2147/ijgm.s153268
Svenningsen SL (2018) Small RNA-based regulation of bacterial quorum sensing and biofilm formation. Microbiol Spectrum. https://doi.org/10.1128/microbiolspec.rwr-0017-2018
Freeman DJ, Falkiner FR, Keane CT (1989) New method for detecting slime production by coagulase-negative Staphylococci. J Clin Pathol 42:872–874. https://doi.org/10.1136/jcp.42.8.872
Christensen GD, Simpson WA et al (1985) Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of Staphylococci to medical devices. J Clin Microbiol 22:996–1006. https://doi.org/10.1128/jcm.22.6.996-1006.1985
Mathur T, Singhal S, Khan S, Upadhyay DJ, Fatma T, Rattan A (2006) Detection of biofilm formation among the clinical isolates of Staphylococci: an evaluation of three different screening methods. Indian J Med Microbiol 24:25–29. https://doi.org/10.4103/0255-0857.19890
Singh AK, Prakash P, Achra A et al (2017) Standardization and classification of in vitro biofilm formation by clinical Isolates of Staphylococcus aureus. J Glob Infect Diseases 9:93–101. https://doi.org/10.4103/jgid.jgid_91_16
Mishra R, Rathinasabapathi P (2020) Antibiotics susceptibility profile and synergistic effects of flavonoids with antibiotics against resistant Staphylococcus aureus from asymptomatic individuals. J Pure Appl Microbiol 14:2669–2676. https://doi.org/10.22207/JPAM.14.4.44
Khan F, Lee J-W, Pham DTN et al (2019) Streptomycin mediated biofilm inhibition and suppression of virulence properties in Pseudomonas aeruginosa PAO1. Appl Microbiol Biotechnol 104:799–816. https://doi.org/10.1007/s00253-019-10190-w
Cruz CD, Shah S, Tammela P (2018) Defining conditions for biofilm inhibition and eradication assays for Gram-positive clinical reference strains. BMC Microbiol. https://doi.org/10.1186/s12866-018-1321-6
Neeraja M, Lakshmi V, Padmasri C, Padmaja K (2017) Utility of Acridine Orange staining for detection of bacteria from positive blood cultures. J Microbiol Methods 139:215–217. https://doi.org/10.1016/j.mimet.2017.06.014
Kannan S, Solomon A, Krishnamoorthy G, Marudhamuthu M (2021) Liposome encapsulated surfactant abetted copper nanoparticles alleviate biofilm mediated virulence in pathogenic Pseudomonas aeruginosa and MRSA. Sci Rep. https://doi.org/10.1038/s41598-020-79976-7
Oh ET, So JS (2003) A rapid method for RNA preparation from Gram-positive bacteria. J Microbiol Methods 52:395–398. https://doi.org/10.1016/S0167-7012(02)00218-X
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
You Y, Xue T, Cao L, Zhao L, Sun H, Sun B (2014) Staphylococcus aureus glucose-induced biofilm accessory proteins, GbaAB, influence biofilm formation in a PIA-dependent manner. Int J Med Microbiol 304:603–612. https://doi.org/10.1016/j.ijmm.2014.04.003
Das T, Sehar S, Koop L et al (2014) Influence of calcium in extracellular DNA mediated bacterial aggregation and biofilm formation. PLoS ONE 9:e91935. https://doi.org/10.1371/journal.pone.0091935
Sheraton MV, Yam JKH, Tan CH et al (2018) Mesoscopic energy minimization drives Pseudomonas aeruginosa biofilm morphologies and consequent stratification of antibiotic activity based on cell metabolism. Antimicrob Agents Chemother 62:e02544-e2617. https://doi.org/10.1128/AAC.02544-17
Zhao X, Zhao F, Wang J, Zhong N (2017) Biofilm formation and control strategies of foodborne pathogens: food safety perspectives. RSC Adv 7:36670–36683. https://doi.org/10.1039/c7ra02497e
Otto M (2013) Staphylococcal infections: mechanisms of biofilm maturation and detachment as critical determinants of pathogenicity. Annu Rev Med 64:175–188. https://doi.org/10.1146/annurev-med-042711-140023
Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM (1995) Microbial biofilms. Annu Rev Microbiol 49:711–745. https://doi.org/10.1146/annurev.mi.49.100195.003431
Potera C (1999) Forging a link between biofilms and disease. Science 283:1837–1839. https://doi.org/10.1126/science.283.5409.1837
Pratt LA, Kolter R (1999) Genetic analyses of bacterial biofilm formation. Curr Opin Microbiol 2:598–603. https://doi.org/10.1016/s1369-5274(99)00028-4
Mendoza-Olazarán S, Morfín-Otero R et al (2015) Antibiotic susceptibility of biofilm cells and molecular characterisation of Staphylococcus hominis isolates from blood. PLoS ONE 10:e0144684. https://doi.org/10.1371/journal.pone.0144684
Algburi A, Comito N, Kashtanov D, Dicks LMT, Chikindas ML (2017) Control of biofilm formation: antibiotics and beyond. Appl Environ Microbiol 83:e02508-e2516. https://doi.org/10.1128/AEM.02508-16
Singh S, Datta S, Narayanan KB, Rajnish KN (2021) Bacterial exo-polysaccharides in biofilms: role in antimicrobial resistance and treatments. J Genet Eng Biotechnol 19:140. https://doi.org/10.1186/s43141-021-00242-y
Gao W, Guérillot R, Lin YH et al (2020) Comparative transcriptomic and functional assessments of linezolid-responsive small RNA genes in Staphylococcus aureus. msystems. 5:e00665-e719. https://doi.org/10.1128/mSystems.00665-19
Joshi B, Singh B, Nadeem A, Askarian F, Wai SN, Johannessen M, Hegstad K (2021) Transcriptome profiling of Staphylococcus aureus associated extracellular vesicles reveals presence of small RNA-Cargo. Front Mol Biosci 7:566207. https://doi.org/10.3389/fmolb.2020.566207
Anand David AV, Arulmoli R, Parasuraman S (2016) Overviews of biological importance of quercetin: a bioactive flavonoid. Pharmacogn Rev 10:84–89. https://doi.org/10.4103/0973-7847.194044
Roy R, Tiwari M, Donelli G, Tiwari V (2018) Strategies for combating bacterial biofilms: a focus on anti-biofilm agents and their mechanisms of action. Virulence 9:522–554. https://doi.org/10.1080/21505594.2017.1313372
Menard G, Silard C, Suriray M et al (2022) Thirty years of sRNA-mediated regulation in Staphylococcus aureus: from initial discoveries to in vivo biological implications. Int J Mol Sci 23:7346. https://doi.org/10.3390/ijms23137346
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mishra, R.A.K., Muthukaliannan, G.K. & Rathinasabapathi, P. Effects of Flavonoids and Antibiotics Combination on Preformed Biofilms and Small RNA of Staphylococcus aureus. Indian J Microbiol 63, 307–316 (2023). https://doi.org/10.1007/s12088-023-01086-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12088-023-01086-5