Abstract
Previous studies have demonstrated that the C-terminal of E1A binding proteins (CtBPs) influences tumorigenesis by participating in cell signal transduction in various human malignancies. However, the detailed expression patterns of CtBP isoforms in human osteosarcoma (OS) and the molecular mechanisms of CtBP involvement in tumor cell phenotypes requires further investigation. In the present study, the expression patterns of CtBP2 in OS cells and tissues were explored by immunohistochemistry. Fetal osteoblast cells were transfected with a eukaryotic expression plasmid to overexpress CtBP2, and the endogenous CtBP2 in OS cells was silenced via a short hairpin RNA. These transfections were validated and the phosphorylation levels of the JAK1/Stat3 signaling pathway were explored via western blotting. Furthermore, the malignant phenotype of OS cells was evaluated via a Cell Counting Kit-8 assay, cell colony formation assay, cell migration assay and scratch wound healing assay. The results revealed that the expression of CtBP2, but not CtBP1, was upregulated in OS tissue samples and the elevated expression level of CtBP2 was notably associated with distant metastasis. CtBP2 was demonstrated to modulate cell migration and invasion via JAK1/Stat3 signaling pathway in fetal osteoblast cells. In addition, genetic silencing of CtBP2 expression in OS cells notably reduced cell migration abilities and the phosphorylation of the JAK1/Stat3 pathway. In summary, the present studies revealed that the loss of CtBP2 constrained distant metastasis through the JAK1/Stat3 pathway in OS, suggesting that targeting CtBP2 may be a practical anti-tumor approach to prevent OS tumor progression.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The primary cause for the poor prognosis of osteosarcoma (OS) is recurrence and metastasis, and the overall 5 year survival rate is <20% (Anderson 2016; Wang et al. 2017). To date, restraining the recurrence and metastasis of OS has proven to be a limiting factor in the treatment of this disease; once tumors progress to the metastatic stage, there are currently no feasible and efficient therapeutic strategies (Salunke et al. 2016). The C-terminal of E1A binding protein (CtBP) was originally identified based on its ability to bind the carboxyl terminus of the adenovirus E1A oncoprotein (Schaeper et al. 1998). As a corepressor, CtBP binds to transcription factors (and E1A) through a conserved PXDLS peptide motif to perform its functions (Zhao et al. 2007). CtBPs are genetically coded from two DNA fragments; the mRNA products of CtBPs may also be spliced at their 5’ ends to generate two protein isoforms, CTBP1 and CTBP2 (Chinnadurai 2009). CtBPs have also been revealed to be transcriptional corepressors that interact with DNA-binding transcription factors, serving various functions in both developmental and oncogenic processes (Turner and Crossley 2001).
CtBPs are expressed at high levels during development and participate in axial patterning and cellular proliferation and differentiation in various organs, including the eyes, heart, brain and placental vasculature (Blevins et al. 2017). Genetically engineered mutations in CtBPs have adverse consequences on the development of organs/tissues, confirming the role of CtBPs as critical regulators of organogenesis and tissue morphogenesis (Dcona et al. 2017; Paredes et al. 2018). For example, CtBP2-null mice are embryonic lethal and often exhibit axial truncations, heart defects and incomplete neural development (Chinnadurai 2003; Zou et al. 2013). Knockout and gain-of-function studies have further confirmed that CtBPs are regulators of sequence-specific DNA-binding transcription factors that control segmentation, epithelial-mesenchymal transition (EMT) and apoptosis (Zhang et al. 2013; Zhao et al. 2018; Zheng et al. 2015). The importance of the CtBP corepressor complex in multiple developmental programs suggests that the overexpression of CtBPs in adult tissues may play a role in both tumorigenesis and tumor progression (Dcona et al. 2017). The target proteins regulated by CtBP have been revealed through assays that assess its binding to other gene promoters (Li et al. 2017; Shen et al. 2017). In this regard, it has been observed that the expression level of E-cadherin, a cell adhesion protein, is suppressed by CtBP through the binding of CtBP with zinc-finger E-box-binding homeobox (ZEB) to its promoter, revealing a potential molecular mechanism underlying CtBP-mediated metastasis (Ichikawa et al. 2015).
Tumorigenic cells frequently exhibit a more embryonic phenotype than normal cells, having been reprogrammed to have enhanced survival and proliferation, while also exhibiting higher activation of cancer-associated hallmark pathways, suggesting that the inhibition of developmental transcriptional pathways in cancerous tissues may constitute an effective therapeutic approach (Blevins et al. 2017). In addition to being overexpressed, CtBP can also be hyperactivated in cancer cells via other pathways (Blevins et al. 2018). Previous data have shown that CtBP has amino acid sequence homology with NADH-dependent dehydrogenases, and it is well known that tumor cells frequently have elevated NADH levels due to NADH production under hypoxic and pseudohypoxic conditions (Balasubramanian et al. 2003; Chen et al. 2009; Kumar et al. 2002). Recent reports have revealed that NADH can bind to CtBP with high affinity, leading to a structural alteration in CtBP that enables its interaction with transcriptional repressors (Bellesis et al. 2018; Verger et al. 2006; Zhang et al. 2006). A recent study also demonstrated that high NADH levels under hypoxic conditions are associated with a reduction in E-cadherin transcription in tumors, leading to the oncogenic action of CtBP in vivo (Deng et al. 2010). These properties provide a unique opportunity for cancer-specific treatment that potentially spares normal tissues. However, the impact of CtBPs on tumorigenesis in specific human malignancies, such as OS, requires further discussion and study. Therefore, the aim of this study was to evaluate the expression patterns of CtBPs and the molecular mechanisms of the impact of CtBPs in the malignancy of OS.
Materials and methods
RNA extraction and reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was isolated from cells using TRIzol® reagent (Thermo Fisher Scientific, Inc.). First-strand cDNA for CtBPs was produced using a High Capacity Reverse Transcription System kit (Takara Bio, Inc.). Then, qPCRs were performed using a Universal SYBR Green PCR Kit (Takara Bio, Inc.) and ran on an ABI 7500 Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The data were quantified with the 2-ΔΔCq method, using GAPDH or U6 small RNA as reference genes for normalization of CtBP expression. The RT-qPCR primers included primers for CtBP1 (sense, 5′-CGCCAGTGACCAGTTGTAGC-3′, and antisense, 5′-CGTGATGATGCCGTCTTCA-3’), CtBP2 (sense, 5′-ATCCACGAGAAGGTTCTAAACGA-3′, and antisense, 5′-CCGCACGATCACTCTCAGG-3′) and β-actin (sense, 5′-GCACCACACCTTCTACAATGAG-3′, and antisense, 5′-ACAGCCTGGATGGCTACGT-3′).
Antibodies
The mouse anti-human β-actin (cat. no. ab8227) were obtained from Abcam. The rabbit anti-human CtBP1(cat. no. 8684), rabbit anti-human CtBP2 (cat. no. 13256), rabbit anti-human phosphorylated (p)-Stat3 (cat. no. 9145), rabbit anti-human Stat3 (cat. no. 12640), rabbit anti-human p-JAK1 (cat. no. 74129), rabbit anti-human JAK1 (cat. no. 29261) were obtained from Cell Signaling Technology, Inc.
Western blotting
An immunohistochemistry assay was performed as previously described (Zhang et al. 2018). Protein was extracted and quantified using a bicinchoninic acid protein assay kit (Pierce; Thermo Fisher Scientific, Inc.). Equal amounts of protein (30 μg) were denatured at 98 °C for 5 min and separated via 10% SDS-PAGE gel and electro transferred to a nitrocellulose membrane (EMD Millipore). Next, the membranes were blocked overnight at 4 °C with 5% non-fat milk power and probed with the primary antibodies at 37 °C for 1 h. After washing, the membranes were further probed with horseradish peroxidase-conjugated secondary antibodies (Cell Signaling Technology, Inc.,) at a 1:1000 dilution for another 2 h. Finally, the protein bands were visualized using an enhanced chemiluminescence kit (Biorobot, Shanghai, China) on a Bio-Rad ChemiDoc MP imaging system (Bio-Rad Laboratories, Inc.).
Patients and tissue specimens
The present study was performed with the approval of the Research Ethics Committee of Qilu Hospital of Shandong University. All participants provided written informed consent. Samples were collected from 28 patients who underwent surgical resection at Qilu Hospital of Shandong University between July 2005 and June 2012. The patients were selected based on the following criteria: Pathological diagnosis of OS; no prior or secondary tumors; and no history of chemotherapy and radiotherapy. The clinicopathologic characteristics of patients with OS that participated in this study, including age, gender, distant metastasis and Tumor-Node-Metastasis (TNM) stage, are summarized in Table 1.
Immunohistochemistry
An immunohistochemistry assay was performed as previously described (Zhang et al. 2015), and the following antibodies were used in the experiment: Rabbit anti-human CtBP2 and p-JAK1. The negative control was incubated with isotype antibodies at the same dilution of the CtBP2 and p-JAK1 antibodies. Expression of CtBP2 and p-JAK1 in the cell nucleus and cytoplasm was considered a positive result. Staining assessment was performed by scoring of CtBP2 and p-JAK1 protein expression levels was classified semi quantitatively as previously described (Yang et al. 2008). Afterwards, all patients who underwent surgery or adjuvant chemotherapy were followed for up to 5 years on an outpatient basis, or through telephone interviews to confirm overall survival for 5-year Kaplan-Meier survival analysis.
Cell culture and transfection
A human fetal osteoblast cell line (hFOB.1.19) and human OS cell lines (Saos2, 143B, MG63 and U2OS) were obtained from the Institute of Basic Medical Sciences (Chinese Academy of Medical Sciences, Beijing, China). All cells were cultured in RPMI 1640 medium (Gibco; Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum (FBS, Gibco; Thermo Fisher Scientific, Inc.) at 37 °C in a humidified atmosphere of 5% CO2. The full-length fragments of CtBP2 were inserted into a pNSE-IRES2-EGFP-C1 vector to generate a CtBP2 overexpression plasmid (constructed and amplified by Nanjing KeyGen Biotech Co., Ltd.), with empty pNSE-IRES2-EGFP-C1 vectors serving as a negative control (vector). hFOB.1.19 cells were plated into 6-well plates at a density of 1.5 × 105 cells/well, followed by transfection with the above plasmids using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), and G418 (Sigma-Aldrich; Merck KGaA)-resistant clones were cultured as a monoclonal population.
Cell proliferation assay
Cells were plated at a concentration of 3 × 105 cells/well into 96-well plates in triplicate, and cultured at 37 °C overnight in an incubator. A growth curve was drawn based on the viability, which was measured every 12 h over 4 days via a Cell Counting Kit-8 (CCK-8) kit (Dojindo Molecular Technologies, Inc.), according to the manufacturer’s protocol.
Colony formation assays under 2D culture conditions
Colony formation assays were performed as described previously (Zhang et al. 2015). The cells were maintained in 60 mm cell dishes at a density of 500 cells/dish, and cultivated in RPMI-1640 complete medium at 37 °C for 14 days. The cell colonies were washed twice with phosphate-buffered saline (PBS), fixed with methanol for 20 min, and stained with 0.1% crystal violet in PBS (Beyotime Institute of Biotechnology, Haimen, China) for 15 min.
Cell migration assay
Cell migration ability was evaluated using Transwell chambers (BD Biosciences) in 24-well plates. Briefly, 143B and hFOB.1.19 cells supplemented with serum-free medium were added into the upper chambers. The bottom chambers were filled with complete medium containing 10% FBS. Approximately 16 h after incubation, the cells on the basal side of the membrane were fixed and stained with hematoxylin (Sigma-Aldrich; Merck KGaA). The migrating and invading cells were counted in three random visual fields using a microscope (Olympus Corporation, Tokyo, Japan).
Scratch wound healing assay
Cells were cultured as a monolayer in RPMI 1640 medium with 10% FBS to 90% confluence in 6-well dishes (Costar, Corning, Cambridge, MA), and pretreated with 10 μg/ml mitomycin C for 2 h to block proliferation before scratching. Then, the monolayer was scratched using a 20 μl pipette tip and washed 3 times with PBS to create a cell-free area, and the medium was changed to FBS-free RPMI 1640 medium. The scratches were imaged in the same field of view at 0, 12 and 24 h following the scratch with a light microscope (E100; Nikon Corporation; magnification, ×200).
Short hairpin RNA (shRNA) method
The frozen bacterial stocks used for plasmid amplification and cloning were also used to generate lentiviruses. A pGCSIL-CTBP2-shRNA construct, a pGCSIL-scramble construct, a pGCSIL-EGFP, a packaging construct (pHelper 1.0) and a VSV-G-expressing construct (pHelper 2.0) were purchased from Shanghai GenePharma Co., Ltd. Briefly, 293 T cells (70–80% confluence) maintained in 6-well dishes were transfected with four plasmids: 5 μg pGCSIL CTBP2-shRNA construct or pGCSIL-scramble construct, 5 μg pGCSIL- EGFP, 5 μg packaging construct (pHelper 1.0) and 5 μg VSV-G-expressing construct (pHelper 2.0). All transfections were performed using Lipofectamine (cat. no. 11668027; Thermo Fisher Scientific, Inc.). The vector supernatant was concentrated by ultracentrifugation, and the pellet was resuspended in Hanks’ balanced salt solution. The OS cells were infected with viral stocks at a multiplicity of infection (MOI) of 200. Then, the EGFP expression in these cells was detected via FACS analysis, with transduction efficiencies estimated to be ~93.24 ± 1.07 (mean ± standard deviation). The transduced OS cells were cultured and then used in subsequent experiments.
Statistical methods
All experiments were performed independently and repeated at least three times. The data are presented as the mean ± standard deviation. The statistical significance of differences between groups was assessed by Student’s t test or analysis of variance using SPSS 20.0 software (IBM Corp.). P < 0.05 was considered to indicate a statistically significant difference.
Results
CtBP2 is markedly upregulated in OS cells compared with human fetal osteoblast cells
The expression patterns of CtBPs in OS cell lines and a human fetal osteoblast cell line were explored via RT-qPCR and western blotting assays. The data revealed that mRNA and protein levels of CtBP2 were higher in the OS cell lines Saos2, 143B, MG63 and U2OS, but were expressed at low levels in the fetal osteoblast cell line hFOB.1.19 (Fig. 1a-c). In addition, there was no difference in CtBP1 expression between the OS cell lines (Saos2, 143B, MG6, and U2OS) and a human fetal osteoblast cell line (hFOB.1.19) at the mRNA and protein levels.
mRNA and protein expression levels of CtBPs in human fetal-osteoblast line and OS cell lines. a Relative mRNA expression of CtBPs in the fetal-osteoblast and the OS cell lines. b Relative protein expression of CtBPs in the fetal-osteoblast and the OS cell lines. c Corresponding quantification and statistical analysis of CtBP protein expression. **P < 0.01 vs. fetal-osteoblast line. CtBP, C-terminal of E1A binding protein; OS, osteosarcoma
CtBP2 is upregulated in tissues of patients with OS and is associated with clinicopathologic prognosis
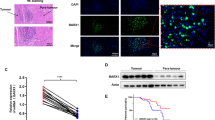
The expression pattern of CtBP2 among the 28 OS tissue samples and the 22 noncancerous tissue samples were also explored via western blotting assay and immunohistochemistry. As revealed in Fig. 2a, CtBP2 expression in OS tissues and noncancerous tissues was primarily located in the cell cytoplasm and nuclei. As depicted in Table 1, CtBP2 was expressed in 67.9% (19/28) of OS tissues and 27.8% (5/22) of noncancerous tissues (Table 1). In addition, CtBP2 was notably overexpressed in OS tissues compared with the noncancerous tissues. Moreover, the expression of CtBP2 appeared to not be associated with clinical indicators, including patient age (P = 0.742), gender (P = 0.316) and response to chemotherapy (P = 0.316). However, it was significantly associated with TNM stage (P = 0.001), p-JAK1 expression (P = 0.0023) and pulmonary metastasis (P = 0.0005).
Expression of CtBP2 in OS patients. a Immunohistochemical detection of CtBP2 in primary OS tissues and non-neoplastic tissues. b Protein expression of CtBP2 in human primary OS tissues and non-neoplastic tissues. c Kaplan-Meier Survival Curves and log-tank test comparisons confirming an association between CtBP2 expression and survival time. d Corresponding quantification and statistical analysis of CtBP protein expression in human primary OS tissues and non-neoplastic tissues. **P < 0.01 vs. fetal-osteoblast line. CtBP, C-terminal of E1A binding protein; OS, osteosarcoma
Semiquantitative immunoblotting analysis was also performed to evaluate the differences in CtBP2 expression between noncancerous and OS tissues. As shown in Fig. 2b, d, the protein expression level of CtBP2 was significantly higher in the 28 cases of OS tissues compared with the 22 cases of noncancerous bone tissues (P = 0.0034). The association between survival time of patients with OS and CtBP2 expression was explored via Kaplan-Meier survival curves and compared with a log-rank test. As suggested in Fig. 2c, patients with OS whose tumors were positive for CtBP2 protein expression had significantly shorter survival times (median survival, 36.74 months) compared with patients with tumors that were negative for the CTBP2 protein (median survival, 45.27 months; χ2 = 7.854, P = 0.0024).
Overexpression of CtBP2 markedly promotes the malignant phenotype of human fetal osteoblast cells
Given that a low CtBP2 expression level was observed in the human fetal osteoblast cells, the impacts of CtBP2 overexpression on the development of malignant phenotypes were evaluated in this fetal osteoblast cell line. hFOB.1.19 cells were transfected with the pNSE-IRES2-EGFP-C1/CTBP2 plasmid (CTBP2 group) or with the pNSE-IRES2-EGFP-C1 vector as a negative control. The expression of CtBP2 and the changes in the activation state of the JAK1/Stat3 pathway in these cells were further explored via western blotting. As shown in Fig. 3a, the proportions of p-JAK1 (P = 0.0021) and p-Stat3 (P = 0.0026) were significantly increased in the hFOB.1.19 cells that overexpressed CtBP2 (P = 0.0001). To determine the impact of CtTBP2 on the malignant phenotype of fetal osteoblast cell lines, the CCK-8 method was used to draw a growth curve. As depicted in Fig. 3b, the proliferation rate of hFOB.1.19-CTBP2 cells was markedly higher than that of the vector-transfected cells (P = 0.0062). In addition, the ability of CtBP2-overexpressing cells to form colonies in 2D monolayer conditions was also explored (Fig. 3c). The results showed that the number of colonies formed by CtBP2-overexpressing cells was significantly higher than the number formed by the scramble-transfected cells (P = 0.0012). The Transwell (Fig. 3d) and wound-healing assays (Fig. 3e), further confirmed that cell invasion and migration, respectively, were enhanced following the overexpression of CtBP2 in hFOB.1.19 cells. Taken together, these observations suggest that CtBP2 expression enhances the migration and invasion abilities of fetal osteoblast cells.
Overexpression of CtBP2 promoted the malignant phenotype of the fetal-osteoblast cell line. a Western blotting was used to examine the expression of CtBP2 and the activity level of the JAK1/Stat3 signaling pathway. b Growth curve of hFOB.1.19 cells according to the Cell Counting Kit-8 assay. c The colony forming abilities of hFOB.1.19 cells in 2D culture conditions was determined via a colony formation assay. d The Transwell method was used to explore the impact of CtBP2 overexpression on the invasive ability of hFOB.1.19 cells in vitro. e The wound-healing assay was used to explore the migration ability of hFOB.1.19 cells in vitro. **P < 0.01 vs. Vector group. CtBP, C-terminal of E1A binding protein
Knockdown of CTBP2 decreases the migration and invasion ability of OS cells
Given that CtBP2 overexpression was observed in OS cells, the effects of CtBP2 silencing on OS progression were also evaluated. As presented in Fig. 4a, the activation state of the Stat3 pathway in the OS cell line 143B was explored via western blotting. As shown, the ratios of p-JAK1 (P = 0.0011 and P = 0.0026, respectively) and p-Stat3 (P = 0.0016 and P = 0.0012, respectively) were markedly decreased in the 143B cells with silenced CtBP2 (P = 0.0004 and P = 0.0003, respectively).
Loss of CtBP2 decreased the migration and invasion ability of osteosarcoma cells. a Western blotting was used to examine the effects of CtBP2 silencing and the activation of the JAK1/Stat3 signaling pathway in the 143B cell line. b Growth curve of 143B according to the Cell Counting Kit-8 assay. c The colony-forming abilities of 143B cells in 2D culture conditions were determined via a colony formation assay. d Transwell chambers were used to explore the impact of CtBP2 sliencing on the invasive ability of 143B cells in vitro. e The wound-healing assay was used to explore the migration ability of 143B cells in vitro. **P < 0.01 vs. the scramble group. CtBP, C-terminal of E1A binding protein
To determine the impact of CtBP2 knockdown on the malignancy of OS cell lines, the CCK-8 method was used to assess the growth of the 143B cell line. As depicted in Fig. 4b, the proliferation rate of CTBP2-shRNA1 or CTBP2-shRNA2 transfected cells were markedly lower than that of the scramble-transfected cells (P = 0.0013 and P = 0.0042, respectively). As data in Fig. 4c revealed, the number of colonies formed by CTBP2-shRNA1 or CTBP2-shRNA2 cells were also significantly lower than the number formed by the scramble-transfected cells (P = 0.0027 and P = 0.0013, respectively). The wound-healing and Transwell assays (Fig. 4d, e) further indicated that cell migration and invasion abilities were reduced following CtBP2 silencing in 143B cells. Taken together, these results suggested that CtBP2 silencing reduced cell migration and invasion in the human OS cell line 143B.
CTBP2 modulates cell malignancy through the JAK1/Stat3 signaling pathway in fetal osteoblast cells
To further explore the impact of the JAK1/Stat3 signaling pathway on cell invasion and migration abilities of fetal osteoblasts, these cells were treated with the JAK1 tyrosinase inhibitor AG490. As shown in Fig. 5a, after treatment with 20 nM AG490, the ratios of p-JAK1 (P = 0.0029) and p-Stat3 (P = 0.0017) were significantly decreased for 24 h in hFOB.1.19 fetal osteoblast cells that overexpressed CtBP2.
Tyrosine phosphorylation inhibitor AG490 was used to inhibit JAK1 activity in hFOB.1.19 cells. a Western blotting was used to examine the activity level of the JAK1/Stat3 signaling pathway. b Growth curve of hFOB.1.19 cells according to the Cell Counting Kit-8 assay. c The colony forming abilities of hFOB.1.19 cells in 2D culture conditions were evaluated via a colony formation assay. d The Transwell method was used to assess the invasiveness of hFOB.1.19 cells in vitro. e The wound-healing assay was used to evaluate the migration ability of hFOB.1.19 cells in vitro. **P < 0.01 vs. DMSO group
The data obtained from the CCK-8 (Fig. 5b) and colony formation (Fig. 5c) assays revealed that the cell proliferation (P = 0.0014) and colony formation (P = 0.0003) abilities in AG490-treated cells were significantly reduced compared with the DMSO-treated group. Moreover, as illustrated in Fig. 5d, e, the number of invasive cells (P = 0.0016), and the migration distance after 12 and 24 h (P = 0.0019 and P = 0.0023, respectively) of AG490-treated cells were significantly reduced compared with the DMSO-treated group.
Discussion
With a poor prognosis, the 5-year survival rate of patients with OS is <20% in the USA, China and Europe (Anderson 2016). With the increasing detection of early OS, treatment options have improved in terms of efficacy, and have also become minimally invasive, allowing a good quality of life post-treatment (Salunke et al. 2016; Wang et al. 2017). Recent work has shown that the dysregulated control of CtBP expression may be a fundamental step in the genesis and development of various human tumors (de Barrios et al. 2017; Eshelman et al. 2017; Zhao et al. 2016). CtBPs have been shown to inhibit multiple proapoptotic and epithelial genes, and overexpression of CtBP in human cancer cells may also promote EMT and cancer cell survival (Zhang et al. 2016, 2013; Zheng et al. 2015). Knockout of CtBP was also demonstrated to increase apoptosis and inhibit tumorigenesis in vivo (Corda et al. 2006; Wang et al. 2006). However, reduced CtBP2 expression has also been associated with melanoma progression, owing to a reduction of CtBP-dependent suppression of melanoma-inhibiting gene expression in melanoma cells (Deng et al. 2013; Winklmeier et al. 2009). A previous tissue chip research has suggested that overexpression of CtBP2 is highly associated with tumor progression in OS. These findings provide a unique opportunity for the development of OS-specific treatments that potentially spare normal tissues. Therefore, inhibition of CtBP function may constitute an effective anticancer therapeutic strategy, but the role of CtBP proteins in OS has not been clinically evaluated thus far. Overall, more studies on the expression patterns and roles of CtBPs in OS should be performed.
Despite earlier studies indicating that CtBP serves as a transcriptional corepressor, several studies have also revealed context-specific roles of CtBP as transcriptional activators (Blevins et al. 2018). For instance, CtBP2 has been shown to directly activate the expression of T cell lymphoma invasion and metastasis 1 in a NADH-dependent manner, and to activate the transcription factor 4 signaling pathway (Cuilliere-Dartigues et al. 2006; Valenta et al. 2003). In human multidrug-resistant (MDR) cancer cell lines, CtBP2 has been shown to directly activate the expression of the MDR1 gene, thereby increasing levels of the P-glycoprotein and drug resistance (Jin et al. 2007). Interestingly, the NADH-unbound form of CtBP has also been implicated in distinct transcriptional activities unique to apo-CtBP, including the interaction with the transcriptional regulators p300 and Hdm2, and the transcriptional activation of Wnt pathway targets (Kim et al. 2005). CtBP corepressors thus play multiple context-dependent roles in oncogenic processes (Chinnadurai 2003; Senyuk et al. 2005). Consequently, further research to expand our present understanding of these transcriptional corepressors in OS oncogenesis may positively impact the future development of new therapies. A previous gene chip study suggested that the upregulation of CtBP2 was associated with JAK1/Stat3 signaling (data not published). In the present study, the association between CtBP2 and JAK1/Stat3 signaling was explored in OS cells, revealing that CtBP2 silencing contributed to an inhibition of the Stat3 signaling pathway and suppressed OS cell migration. Besides, we also put emphasis on regulation of CtBP2 on OS cell proliferation but it seemed that the growth rate was initially affected but controls do catch up after a longer period in culture suggesting that other factors play an important role in ultimate growth and density attained. Thus, these present observations suggested that CtBP2 was most relevant to migration and invasion, which may have been achieved through the activation of Stat3 signaling via JAK1. In view of the current inefficient therapeutic options for OS, the role of CtBP2 as a novel target protein may be of great research significance.
In summary, the present study revealed that CtBP2 was upregulated in OS tissues and that its upregulation was associated with the occurrence of pulmonary metastasis. This observation was validated in a CtBP2-overexpressing fetal osteoblast cell line and in a CtBP2-knockout OS cell line. The results revealed that the loss of CtBP2 inhibited the migration of OS cells by inhibiting the JAK1/Stat3 signaling pathway.
Data availability
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Abbreviations
- CTBP:
-
C-terminus of the E1A binding protein
- IHC:
-
immunohistochemical analysis
- CCK-8:
-
cell counting kit-8
- JAK1:
-
Janus Kinase 1
- Stat3:
-
signal transducer and activator of transcription 3
- OS:
-
osteosarcoma
References
Anderson ME (2016) Update on Survival in Osteosarcoma. Orthop Clin North Am 47:283–292. https://doi.org/10.1016/j.ocl.2015.08.022
Balasubramanian P, Zhao LJ, Chinnadurai G (2003) Nicotinamide adenine dinucleotide stimulates oligomerization, interaction with adenovirus E1A and an intrinsic dehydrogenase activity of CtBP. FEBS Lett 537:157–160
Bellesis AG, Jecrois AM, Hayes JA, Schiffer CA, Royer WE Jr (2018) Assembly of human C-terminal binding protein (CtBP) into tetramers. J Biol Chem 293:9101–9112. https://doi.org/10.1074/jbc.RA118.002514
Blevins MA, Huang M, Zhao R (2017) The role of CtBP1 in oncogenic processes and its potential as a therapeutic target. Mol Cancer Ther 16:981–990. https://doi.org/10.1158/1535-7163.MCT-16-0592
Blevins MA, Zhang C, Zhang L, Li H, Li X, Norris DA, Huang M, Zhao R (2018) CPP-E1A fusion peptides inhibit CtBP-mediated transcriptional repression. Mol Oncol 12:1358–1373. https://doi.org/10.1002/1878-0261.12330
Chen S, Whetstine JR, Ghosh S, Hanover JA, Gali RR, Grosu P, Shi Y (2009) The conserved NAD(H)-dependent corepressor CTBP-1 regulates Caenorhabditis elegans life span. Proc Natl Acad Sci U S A 106:1496–1501. https://doi.org/10.1073/pnas.0802674106
Chinnadurai G (2003) CtBP family proteins: more than transcriptional corepressors. BioEssays : news and reviews in molecular, cellular and developmental biology 25:9–12. https://doi.org/10.1002/bies.10212
Chinnadurai G (2009) The transcriptional corepressor CtBP: a foe of multiple tumor suppressors. Cancer Res 69:731–734. https://doi.org/10.1158/0008-5472.CAN-08-3349
Corda D, Colanzi A, Luini A (2006) The multiple activities of CtBP/BARS proteins: the Golgi view. Trends Cell Biol 16:167–173. https://doi.org/10.1016/j.tcb.2006.01.007
Cuilliere-Dartigues P, el-Bchiri J, Krimi A, Buhard O, Fontanges P, Fléjou JF, Hamelin R, Duval A (2006) TCF-4 isoforms absent in TCF-4 mutated MSI-H colorectal cancer cells colocalize with nuclear CtBP and repress TCF-4-mediated transcription. Oncogene 25:4441–4448. https://doi.org/10.1038/sj.onc.1209471
Dcona MM, Morris BL, Ellis KC, Grossman SR (2017) CtBP- an emerging oncogene and novel small molecule drug target: advances in the understanding of its oncogenic action and identification of therapeutic inhibitors. Cancer Biol Ther 18:379–391. https://doi.org/10.1080/15384047.2017.1323586
de Barrios O, Győrffy B, Fernández-Aceñero MJ, Sánchez-Tilló E, Sánchez-Moral L, Siles L, Esteve-Arenys A, Roué G, Casal JI, Darling DS, Castells A, Postigo A (2017) ZEB1-induced tumourigenesis requires senescence inhibition via activation of DKK1/mutant p53/Mdm2/CtBP and repression of macroH2A1. Gut 66:666–682. https://doi.org/10.1136/gutjnl-2015-310838
Deng Y, Liu J, Han G, Lu SL, Wang SY, Malkoski S, Tan AC, Deng C, Wang XJ, Zhang Q (2010) Redox-dependent Brca1 transcriptional regulation by an NADH-sensor CtBP1. Oncogene 29:6603–6608. https://doi.org/10.1038/onc.2010.406
Deng H, Liu J, Deng Y, Han G, Shellman YG, Robinson SE, Tentler JJ, Robinson WA, Norris DA, Wang XJ, Zhang Q (2013) CtBP1 is expressed in melanoma and represses the transcription of p16INK4a and Brca1. J Invest Dermatol 133:1294–1301. https://doi.org/10.1038/jid.2012.487
Eshelman MA, Shah M, Raup-Konsavage WM, Rennoll SA, Yochum GS (2017) TCF7L1 recruits CtBP and HDAC1 to repress DICKKOPF4 gene expression in human colorectal cancer cells. Biochem Biophys Res Commun 487:716–722. https://doi.org/10.1016/j.bbrc.2017.04.123
Ichikawa K, Kubota Y, Nakamura T, Weng JS, Tomida T, Saito H, Takekawa M (2015) MCRIP1, an ERK substrate, mediates ERK-induced gene silencing during epithelial-mesenchymal transition by regulating the co-repressor CtBP. Mol Cell 58:35–46. https://doi.org/10.1016/j.molcel.2015.01.023
Jin W, Scotto KW, Hait WN, Yang JM (2007) Involvement of CtBP1 in the transcriptional activation of the MDR1 gene in human multidrug resistant cancer cells. Biochem Pharmacol 74:851–859. https://doi.org/10.1016/j.bcp.2007.06.017
Kim JH, Cho EJ, Kim ST, Youn HD (2005) CtBP represses p300-mediated transcriptional activation by direct association with its bromodomain. Nat Struct Mol Biol 12:423–428. https://doi.org/10.1038/nsmb924
Kumar V, Carlson JE, Ohgi KA, Edwards TA, Rose DW, Escalante CR, Rosenfeld MG, Aggarwal AK (2002) Transcription corepressor CtBP is an NAD(+)-regulated dehydrogenase. Mol Cell 10:857–869
Li L, Liu X, He L, Yang J, Pei F, Li W, Liu S, Chen Z, Xie G, Xu B, Ting X, Zhang Z, Jin T, Liu X, Zhang W, Yuan S, Yang Z, Wu C, Zhang Y, Yang X, Yi X, Liang J, Shang Y, Sun L (2017) ZNF516 suppresses EGFR by targeting the CtBP/LSD1/CoREST complex to chromatin. Nat Commun 8:691. https://doi.org/10.1038/s41467-017-00702-5
Paredes R, Schneider M, Stevens A, White DJ, Williamson AJK, Muter J, Pearson S, Kelly JR, Connors K, Wiseman DH, Chadwick JA, Löffler H, Teng HY, Lovell S, Unwin R, van de Vrugt HJ, Smith H, Kustikova O, Schambach A, Somervaille TCP, Pierce A, Whetton AD, Meyer S (2018) EVI1 carboxy-terminal phosphorylation is ATM-mediated and sustains transcriptional modulation and self-renewal via enhanced CtBP1 association. Nucleic Acids Res 46:7662–7674. https://doi.org/10.1093/nar/gky536
Salunke AA, Shah J, Gupta N, Pandit J (2016) Pathologic fracture in osteosarcoma: association with poorer overall survival. Eur J Surg Oncol: the journal of the European Society of Surgical Oncology and the British Association of Surgical Oncology 42:889–890. https://doi.org/10.1016/j.ejso.2016.02.255
Schaeper U, Subramanian T, Lim L, Boyd JM, Chinnadurai G (1998) Interaction between a cellular protein that binds to the C-terminal region of adenovirus E1A (CtBP) and a novel cellular protein is disrupted by E1A through a conserved PLDLS motif. J Biol Chem 273:8549–8552
Senyuk V, Sinha KK, Nucifora G (2005) Corepressor CtBP1 interacts with and specifically inhibits CBP activity. Arch Biochem Biophys 441:168–173. https://doi.org/10.1016/j.abb.2005.06.024
Shen Z, Asa SL, Ezzat S (2017) Ikaros and its interacting partner CtBP target the metalloprotease ADAMTS10 to modulate pituitary cell function. Mol Cell Endocrinol 439:126–132. https://doi.org/10.1016/j.mce.2016.10.032
Turner J, Crossley M (2001) The CtBP family: enigmatic and enzymatic transcriptional co-repressors. BioEssays : news and reviews in molecular, cellular and developmental biology 23:683–690. https://doi.org/10.1002/bies.1097
Valenta T, Lukas J, Korinek V (2003) HMG box transcription factor TCF-4's interaction with CtBP1 controls the expression of the Wnt target Axin2/Conductin in human embryonic kidney cells. Nucleic Acids Res 31:2369–2380
Verger A, Quinlan KGR, Crofts LA, Spano S, Corda D, Kable EPW, Braet F, Crossley M (2006) Mechanisms directing the nuclear localization of the CtBP family proteins. Mol Cell Biol 26:4882–4894. https://doi.org/10.1128/MCB.02402-05
Wang SY, Iordanov M, Zhang Q (2006) c-Jun NH2-terminal kinase promotes apoptosis by down-regulating the transcriptional co-repressor CtBP. J Biol Chem 281:34810–34815. https://doi.org/10.1074/jbc.M607484200
Wang W, Yang J, Wang Y, Wang D, Han G, Jia J, Xu M, Bi W (2017) Survival and prognostic factors in Chinese patients with osteosarcoma: 13-year experience in 365 patients treated at a single institution. Pathol Res Pract 213:119–125. https://doi.org/10.1016/j.prp.2016.11.009
Winklmeier A, Poser I, Hoek KS, Bosserhoff AK (2009) Loss of full length CtBP1 expression enhances the invasive potential of human melanoma. BMC Cancer 9:52. https://doi.org/10.1186/1471-2407-9-52
Yang Y, Yang H, McNutt MA, Xiong F, Nie X, Li L, Zhou R (2008) LAPTM4B overexpression is an independent prognostic marker in ovarian carcinoma. Oncol Rep 20:1077–1083
Zhang Q, Wang SY, Nottke AC, Rocheleau JV, Piston DW, Goodman RH (2006) Redox sensor CtBP mediates hypoxia-induced tumor cell migration. Proc Natl Acad Sci U S A 103:9029–9033. https://doi.org/10.1073/pnas.0603269103
Zhang XL, Huang CX, Zhang J, Inoue A, Zeng SE, Xiao SJ (2013) CtBP1 is involved in epithelial-mesenchymal transition and is a potential therapeutic target for hepatocellular carcinoma. Oncol Rep 30:809–814. https://doi.org/10.3892/or.2013.2537
Zhang X, Ruan Y, Li Y, Lin D, Quan C (2015) Tight junction protein claudin-6 inhibits growth and induces the apoptosis of cervical carcinoma cells in vitro and in vivo. Med Oncol 32:148. https://doi.org/10.1007/s12032-015-0600-4
Zhang G, Kang L, Chen J, Xue Y, Yang M, Qin B, Yang L, Zhang J, Lu H, Guan H (2016) CtBP2 regulates TGFbeta2-induced epithelial-mesenchymal transition through notch signaling pathway in Lens epithelial cells. Curr Eye Res 41:1057–1063. https://doi.org/10.3109/02713683.2015.1092554
Zhang X, Wang H, Li Q, Li T (2018) CLDN2 inhibits the metastasis of osteosarcoma cells via down-regulating the afadin/ERK signaling pathway. Cancer Cell Int 18:160. https://doi.org/10.1186/s12935-018-0662-4
Zhao LJ, Subramanian T, Vijayalingam S, Chinnadurai G (2007) PLDLS-dependent interaction of E1A with CtBP: regulation of CtBP nuclear localization and transcriptional functions. Oncogene 26:7544–7551. https://doi.org/10.1038/sj.onc.1210569
Zhao C, Shen Y, Tao X, Xu J, Lu J, Liu C, Xu Z, Tang Q, Tao T, Zhang X (2016) Silencing of CtBP1 suppresses the migration in human glioma cells. J Mol Histol 47:297–304. https://doi.org/10.1007/s10735-016-9678-z
Zhao Z, Hao D, Wang L, Li J, Meng Y, Li P, Wang Y, Zhang C, Zhou H, Gardner K, di LJ (2018) CtBP promotes metastasis of breast cancer through repressing cholesterol and activating TGF-beta signaling. Oncogene 38:2076–2091. https://doi.org/10.1038/s41388-018-0570-z
Zheng X, Song T, Dou C, Jia Y, Liu Q (2015) CtBP2 is an independent prognostic marker that promotes GLI1 induced epithelial-mesenchymal transition in hepatocellular carcinoma. Oncotarget 6:3752–3769. https://doi.org/10.18632/oncotarget.2915
Zou F, Xu J, Fu H, Cao J, Mao H, Gong M, Cui G, Zhang Y, Shi W, Chen J (2013) Different functions of HIPK2 and CtBP2 in traumatic brain injury. J Mol Neurosci: MN 49:395–408. https://doi.org/10.1007/s12031-012-9906-2
Acknowledgements
We would like to thank American Journal Experts (AJE) for help with this manuscript.
Author information
Authors and Affiliations
Contributions
PW performed the experiments and analyzed the data. SZ contributed to the conception and design of the study. BY and CW revised the manuscript critically for important intellectual content. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Disclosure of potential conflicts of interest
The authors declare that they have no competing interests.
Research involving human participants and/or animals
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 9864 Helsinki declaration and its later amendments or comparable ethical standards. Ethics approval (approval no. SD02872) was approved by the Ethics Committee of Shandong University. This article does not contain any studies with animals performed by any of the authors. The informed consent for participation was obtained from all patients and their parents who participated in this study in an appropriate method.
Informed consent
Written informed consent for publication was obtained. A copy of the consent form is available for review by the Editor of this journal.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, P., Yu, B., Wang, C. et al. C-terminal of E1A binding protein 2 promotes the malignancy of osteosarcoma cells via JAK1/Stat3 signaling. J. Cell Commun. Signal. 14, 67–76 (2020). https://doi.org/10.1007/s12079-019-00523-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12079-019-00523-9