Abstract
Background and aims
HCV GT-3 has a more pronounced effect on hepatic steatosis and host lipids than other HCV genotypes and is proving less responsive to all oral interferon-free treatment with direct acting antiviral agents. As both HCV GT3 infection and NASH can result in steatosis and cirrhosis, we asked whether hepatic transcriptional profiles reflective of the host response to inflammation differed based on the etiology of injury.
Methods
Hepatic gene expression was determined for 48 pre-selected genes known to be associated with hepatic interferon signaling and lipid metabolic pathways in treatment-naïve HCV GT-3 (n = 9) and NASH (n = 14) patients.
Results
Genes with significantly higher expression in HCV included chemokines CXCL10, CXCL11 interferon IFNA2, interferon receptors IFNAR1, IL10RB negative regulators of interferon signaling SOCS3, USP18, JAK/STAT and IRF family members STAT1, STAT2, and IRF, and TGFB family members TGFB1, TGFBR1, and TGFBR2 and other ISGs like OAS2, IF127, IF144 and ISG15. HCV infection was also associated with higher expression of genes associated with lipid metabolism APOE, APOL3, SREBF1 and HMBS. Furthermore, our results suggest that, in HCV GT3-infected patients, IL28B (CC) genotype is associated with lower baseline ISG expression such as IRF9, ISG15, MX1, STAT1, CXCL10, CXCL11, and IFI27 compared to CT/TT genotype.
Conclusions
HCV GT-3 and NASH both induce hepatic steatosis and inflammation, while HCV GT-3 infection is uniquely associated with elevated transcription of hepatic ISGs and genes associated with lipid metabolism. These changes likely reflect the unique host response to HCV replication distinct from the inflammatory response induced by NASH.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hepatitis C virus (HCV) infects up to 185 million people worldwide, including 3–4 million people in the USA, and can induce fibrosis, cirrhosis, liver failure and hepatocellular carcinoma (HCC) [1]. Non-alcoholic steatohepatitis (NASH) is the most severe form of nonalcoholic fatty liver disease (NAFLD) and is characterized by inflammatory infiltration and hepatocellular damage, with or without fibrosis, which can eventually lead to cirrhosis [2, 3].
HCV genotype 3 (HCV GT3) infection and NASH share common histopathological and clinical features including hepatic steatosis, insulin resistance, and alterations in lipid metabolism, all of which are thought to contribute to advanced liver disease. HCV GT3 is associated with the highest rates of hepatic steatosis amongst HCV genotypes, which is thought to be directly related to intrahepatic HCV viral levels, suggesting a direct steatogenic effect of GT3 virus, while in other HCV genotypes and NASH, steatosis is mainly associated with metabolic syndrome in affected individuals [4–6]. As treatments for HCV have evolved, GT3 presents the greatest challenge in the treatment and management of patients with hepatitis C virus [7]. HCV GT3 has special characteristics such as more rapid progression of liver disease, higher rates of steatosis, insulin resistance and alterations in lipid metabolism and a higher risk for hepatocellular carcinoma [8–10].
Previous studies have demonstrated two important intrahepatic pathways that are associated with chronic viral hepatitis. They include the interferon signaling pathway and the lipid metabolism pathway [11, 12]. Baseline expression of ISG is inversely related to high rates of SVR with IFN-based therapy, while those with preactivated ISGs show no further gene induction with therapy, leading to interferon non-response and treatment failure [13–15]. Moreover, chronic HCV infection is also associated with metabolic complications, including insulin resistance (IR), hepatic steatosis, hypobetalipoproteinemia, and hypocholesterolemia [16, 17]. It is known that HCV utilizes host lipid metabolic pathways during replication [18, 19]. It is unclear whether these changes are reflective of a host response to ongoing inflammation or viral replication. Therefore, in the present study, we assessed the differences in hepatic ISG expression in patients infected with HCV GT-3 versus NASH in order to explore the effect of an infectious versus inflammatory processes on the hepatic transcriptional profile given similar magnitudes of inflammation and a common downstream end-result of hepatic steatosis. We have specifically evaluated the relative expression of hepatic interferon stimulated genes (ISGs) and genes associated with lipid metabolic pathways in HCV GT 3-infected patients compared to patients with NASH.
Patients and methods
Population studied
Liver biopsy specimens were collected from treatment-naive, chronic HCV GT 3 patients (n = 9) and patients diagnosed with NASH (n = 14) based on clinical, laboratory, and pathologic parameters. All subjects underwent liver biopsies at the Institute of Liver and Biliary Sciences (ILBS) between December 2012 and November 2013. Parts of liver biopsy specimens (10–15 mm), obtained in routine clinical for pathological diagnosis, were kept frozen in RNA at −80 °C until analysis. Chronic HCV patients were selected based on the inclusion criteria of age older than 18 years, treatment-naïve GT3 patients with anti-HCV antibody-positive, serum HCV RNA-positive and liver biopsy with evidence of chronic hepatitis. NASH patients were selected based on evidence with imaging technique and liver biopsy. All patients included in the study were treatment-naive and had compensated liver disease. The following patients were excluded: co-infection with hepatitis B virus or human immunodeficiency virus, pregnant or lactating female, decompensated liver disease or liver disease due to any other etiology, patients on anti-neoplastic or immunomodulatory therapies, having history of major organ transplantation, and lack of consent. Clinical characteristics of the patients are shown in Table 1. The study protocol was approved by the institutional ethical committee of the Institute of Liver and Biliary Sciences (ILBS), New Delhi, and is in accordance with the Declaration of Helsinki 1975.
Laboratory tests
Serum aspartate aminotransferase (AST), alanine aminotransferase (ALT), total bilirubin (TBILI), alkaline phosphatase (ALP), total cholesterol (CHOL), triglyceride (TRIG), and fasting plasma glucose were obtained from patients’ charts and all tests were performed within a month of liver biopsy. Hepatitis C virus RNA was measured by a real-time polymerase chain reaction assay using COBAS Amplicor v.2.0 (Roche Diagnostics, Mannheim, Germany) as per the manufacturer’s protocol. This assay has a lower limit of detection of 25 IU/mL. HCV genotyping test was performed using linear array HCV Genotyping using amplification and cDNA preparation following hybridization to a specific sequence product (Roche Molecular Diagnostics, Pleasanton, CA, USA).
IL28B genotyping
Genomic DNA was extracted using sodium per chlorate method. IL28B SNP (C/T) (rs12979860) was genotyped by the Snapshot method using ABI PRISM Snapshot Kit (Applied Biosystems, Foster City, CA, USA) and using a Roche real-time thermocycler. IL28B genotype was determined using sense (50-GCCAATTCAAGCCCAGTC-30) and antisense primers (50-GCTAGTTCACGCACAGTC-30). To validate the snapshot results, 10 % of a randomly selected PCR product was sequenced and analyzed for the C–T change.
RNA isolation and reverse transcription
Liver biopsies were collected and immediately placed in RNA at −80 °C. Total RNA was isolated using a mirVana total RNA isolation kit (Ambion, Austin, TX, USA). RNA purity (260/280 ratio) and quantity were determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). Only RNA samples with 260/280 ratios between 1.8 and 2.0 were used for expression analysis. RNA was reverse transcribed into cDNA using 1 μg total RNA and the high-capacity cDNA master kit from Applied Biosystems as per the manufacturer’s instructions.
TaqMan low density array (TLDA)
Expression analysis of selected 48 genes (“Appendix”) was performed with custom-designed TaqMan Micro Fluidic Cards assembled into the format of 384-well microfluidic cards (Life Technologies). Genes were selected based on known associations with hepatic interferon signaling and lipid metabolic pathways using previous findings from our laboratory and the literature. cDNA (100 ng RNA) was used for the array. Expression reactions in 384-well microfluidic cards were run on a ViiA™ 7 Real-Time PCR System (Life Technologies). Gene expression was determined as Ct based on 40 PCR cycles. GAPDH was used as an endogenous control, with GAPDH Ct values for all samples being distributed between 25 and 30. Relative expression of target gene was normalized by GAPDH expression and delta Ct (dCt) was calculated as d (Ct) = Ct (target) − Ct (GADPH). All quantitative data are presented as mean ± SE of delta Ct values of the genes; the higher the Ct value of a particular gene in a group of patients, the lower the expression of that gene compared to another group; conversely, a lower Ct value means a higher expression of the gene.
Statistics
In this study, patient characteristics were reported as frequencies (percentages) for categorical variables and for continuous variables as median (range). Statistical significance (p value) of continuous variables was calculated by an unpaired t test with Welch’s correction and p values of categorical variables were calculated by a Fisher’s exact t test in GraphPad Prism 6.0 software. For all these tests, two-tailed p values were used and p values <0.05 were considered statistically significant.
Results
Elevated hepatic ISG expression in patients infected with HCVGT 3 compared to NASH
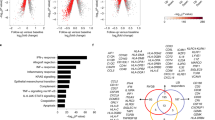
HCV GT3-infected patients had significantly higher expression of type I interferon IFNA2 (4.2 ± 1.01 vs. 10.67 ± 0.84; p = 0.01), interferon receptor IFNAR1 (4.1 ± 0.3 vs. 5.6 ± 0.4; p = 0.01) whereas no differences were observed in type II and type III interferon and interferon receptors (Fig. 1a, b). Our results showed that HCV GT-3 infection was also associated with significantly increased expression of genes involved in IFN signaling and JAK-STAT pathway (Janus kinase (JAK) and a signal transducer and activator of transcription (STAT) protein such as STAT1 (1.8 ± 0.2 vs. 4.7 ± 0.12; p = 0.0001), STAT2 (4.0 ± 0.1 vs. 5.5 ± 0.24; p = 0.003) and IRF7 (3.1 ± 0.12 vs. 5.5 ± 0.4; p = 0.004), (interferon regulatory factor-7 is the master regulator of type-I interferon-dependent immune responses); SOCS3 (6.0 ± 0.5 vs. 8.4 ± 0.3; p = 0.003), (suppressor of cytokine signaling 3 which suppresses hepatitis C virus replication); USP18 (6.1 ± 0.4 vs. 7.5 ± 0.2; p = 0.02); and ubiquitin-like specific protease 18, which is a negative regulator (Fig. 2a, b). Other important ISGs such as OAS2 (3.6 ± 0.4 vs. 6.5 ± 0.3; p = 0.01); 2′-5′-oligoadenylate synthetase 2), interferon inducible genes IFI27 (0.89 ± 0.67 vs. 4.7 ± 0.3; p = 0.0003) and IFI44 (4.2 ± 0.4 vs. 6.9 ± 0.3; p = 0.002), Mx1 (1.9 ± 0.3 vs. 4.8 ± 0.3; p = 0.0001; MX dynamin-like GTPase 1) and ISG15 (2.1 ± 0.7 vs. 7.2 ± 0.3; p = 0.0001) were also significantly increased in HCV compared to NASH (Fig. 2c). Taken together, these data suggest that intrahepatic ISG expression was profoundly higher in patients with HCV GT3 than NASH.
a Higher expression of IFNA2, IFNAR1 (type I IFN) in HCV GT3 versus NASH, b while no difference was observed in type II and type III IFNs. Values are mean ± SE of the delta Ct values of the genes; the higher the Ct value of a particular gene in a group of patients, the lower the expression of that gene compared to another group; conversely, a lower Ct value means a higher expression of the gene. p value was calculated by the unpaired t test with Welch’s correction using GraphPad Prism 6.0 software. p values less than 0.05 were considered significant
a, b IFN signaling and JAK-STAT pathway such as STAT1, STAT2 and IRF7 SOCS3, OAS2, and USP18 (negative regulator), were highly expressed in HCV GT-3 versus NASH. c Other important ISGs such as IFI27, IFI44, Mx1 and ISG15 were significantly increased in HCV GT-3 compared to NASH. Values are mean ± SE of the delta Ct values. p value was calculated by the unpaired t test with Welch’s correction using GraphPad Prism 6.0 software. p values less than 0.05 were considered significant
Elevated hepatic levels of chemokine ligands and markers for fibrosis in patients infected with HCVGT-3 compared to NASH
We observed significantly higher expression of chemokine ligand genes CXCL10 (2.06 ± 1.0 vs. 5.4 ± 0.8; p = 0.01) and CXCL11 (6.3 ± 0.5 vs. 8.1 ± 0.4; p = 0.02) (chemokines ligands to CXCR3) in HCV compared to NASH, while no differences were observed for chemokines ligands, CXCL9 and CCL5. Our results also demonstrate that HCVGT-3 is associated with higher relative gene expression of transforming growth factor beta (TGFB) family members TGFB1 (4.2 ± 0.24 vs. 5.1 ± 0.2; p = 0.007), TGFBR1 (5.1 ± 0.1 vs. 6.7 ± 0.3; p = 0.04) and TGFBR2 (2.4 ± 0.27 vs. 4.3 ± 0.3; p = 0.003), which may play a significant role in accelerating liver fibrosis and increasing risk for HCC (Fig. 3a).
a HCV GT-3 was associated with higher expression of CXCL10 and CXCL11, while no differences were observed for CXCL9 and CCL5. We also observed higher relative gene expression of TGFB family members TGFB1, TGFBR1 and TGFBR2. b Lipid genes such as APOE, APOL3, SREBF1, HMBS and LDLR were highly expressed in HCV GT-3 compared to NASH, while others like APOB, APOC3 and LEPR showed no difference. Values are mean ± SE of the delta Ct values. p value was calculated by the unpaired t test with Welch’s correction using GraphPad Prism 6.0 software. p values less than 0.05 were considered significant
HCV GT-3 infection significantly alters expression of genes involved in host lipid metabolic pathways in comparison to NASH
Hepatitis C virus (HCV) is known to be critically dependent on host lipid metabolism. We therefore investigated the baseline differential gene expression signatures of genes involved in lipid metabolic pathways in HCV GT-3-infected patients versus NASH. We found that HCV infection was associated with significantly higher gene expression of some of the lipid genes such as Apo lipoprotein APOE (−3.1 ± 0.3 vs. −0.29 ± 0.4; p = 0.0001), APOL3 (2.9 ± 0.2 vs. 4.3 ± 0.4; p = 0.02), SREBF1 (3.5 ± 0.3 vs. 5.0 ± 0.37; p = 0.006): sterol regulatory element-binding protein (SREBP)-1 transcription factor, hydroxymethylbilane synthase, HMBS (6.0 ± 0.2 vs. 6.9 ± 0.1; p = 0.006); low density lipoprotein receptor, LDLR (6.0 ± 0.3 vs. 6.8 ± 0.2; p = 0.04) while others (APOB, APOC3 and LEPR) had no difference (Fig. 3b).
IL28B (CC) genotype is associated with lower baseline ISG expression compared to CT/TT genotype in HCV GT3 infected patients
During HCV infection, IL28B genotype has been strongly associated with intrahepatic ISG expression, with the favorable CC genotype patients with lower baseline ISG levels and correspondingly higher rates of SVR compared to unfavorable (CT and TT) genotypes expressing higher baseline ISG levels and poor response to therapy. Our results showed that in HCV genotype 3-infected patients, the IL28B (CC) genotype is associated with lower baseline ISG expression such as IF127 (2.5 ± 0.3 vs. −0.9 ± 0.15; p = 0.02), IRF9 (4.1 ± 0.06 vs. 3.4 ± 0.01; p = 0.02), ISG15 (3.5 ± 0.4 vs. 1.15 ± 0.08; p = 0.02), MX1 (2.8 ± 0.3 vs. 0.8 ± 0.17; p = 0.03), STAT1 (2.2 ± 0.1 vs. 0.8 ± 0.17; p = 0.02), CXCL10 (4.6 ± 1.4 vs. −2.4 ± 0.84; p = 0.02), and CXCL11 (7.4 ± 0.6 vs. 5.02 ± 0.3; p = 0.02) compared to the CT/TT genotype, while in NASH patients no significant association was observed (Fig. 4a, b).
a In HCV GT-3, the IL28B (CC) genotype was associated with lower baseline ISG expression such as IF127, IRF9, ISG15, MX1, STAT1, CXCL10, and CXCL11 compared to the CT/TT genotype, b while in NASH no significant association was observed. Values are mean ± SE of the delta Ct values. p value was calculated by the unpaired t test with Welch’s correction using GraphPad Prism 6.0 software. p values less than 0.05 were considered significant
ISG expressions and association with hepatic fibrosis in HCV GT 3 patients
In the HCV GT-3 patients, overall we did not find any significant association between advanced fibrosis stage and IL28B genotype distribution, although we observed a trend towards advanced fibrosis stage >2, with unfavorable IL28 CT/TT genotype which was further associated with higher baseline ISG expression in three HCV GT-3 patients with IL28 B CT/TT genotype (CC vs. CT/TT: 55.6 vs. 44.4 %, p > 0.05).
Discussion
We identified two distinct hepatic transcriptional responses to infectious and inflammatory processes in this study (Fig. 5). First, there was an enhanced hepatic transcriptional profile of ISG expression in patients with HCV GT3 when compared to NASH. Second, we observed that IL28B impacts the magnitude of hepatic ISG expression only in HCV GT3 rather than in NASH, implicating the effect of viral replication rather than the inflammation itself. Finally, we were able to demonstrate a differential expression of genes for the lipid biosynthetic pathway in the liver of patients with GT3 versus NASH. Overall, for the first time we are able to demonstrate a discriminatory hepatic transcriptional response to NASH versus HCV GT3, which is a significant step toward a better understanding of the pathologic processes of liver injury in these two major public health problems.
HCV GT 3 patients displayed increased intrahepatic expression of several interferon and ISGs. In this regard, interferon (IFNA2), interferon receptor (IFNAR1) genes, interferon signaling and JAK/STAT pathway genes STAT1, STAT2 and IRF7, which is a master regulator of IFN-a production, and negative regulators such as SOCS3, USP18 and other ISGs like OAS2, IF127, IF144 and ISG 15 were expressed higher in HCV patients compared to patients with NASH. It is clear that the innate immune response plays a critical role in the control of viral infection. The type I IFN system is the most potent arm of innate antiviral immunity and signals predominantly through the Jak/STAT kinase pathway. Conceivably, a robust type I interferon response is helpful in elimination of HCV. However, more robust preactivation of ISGs is associated with treatment failure during IFN-based regimes [13–16]. Katsounas et al. have previously established that higher baseline ISG-15 expression negatively affects HCV treatment outcome, and is a prognostic indicator and resistance factor to IFN-α in patients co-infected with HCV and HIV [20]. Also, it is documented in other studies that higher baseline expression of IFI27, IFI44 and Mx1 is associated with poor response to IFN-α treatment of HCV infection [21].
During chronic HCV infection, chemokine ligands CXCL10 (interferon-γ-inducible protein-10; IP-10) and CXCL11 (interferon-inducible T cell α chemo attractant; I-TAC) are produced in the liver, inducing migration of activated T cells from the periphery to infected liver parenchyma [22]. Our data also showed increased expression of CXCL10 and CXCL11 in HCV GT3 patients compared to NASH, which may implicate the specific role of these ISGs chemokine in pathophysiology of HCV infection. In summary, the pattern of hyper-activation of innate immunity in chronic hepatitis C GT3 patients over NASH reflects host response to an intracellular replicating viral pathogen rather than a chronic inflammatory reaction that follows HCV replication.
The lipid biosynthetic pathway also seems to be influenced by HCV rather than hepatic inflammation resulting from NASH. These are novel observations and are likely due to specific interactions between HCV viral target proteins and molecules involved in the lipid biosynthetic pathway, possibly during intracellular replication and exocytosis from infected hepatocytes. These observations are unique and could explain the abnormal clinical and laboratory parameters with HCV GT3 infection compared to NASH.
Earlier studies suggest a host genetic influence on antiviral response to HCV infection. In this regard, the IL28B CC genotype has been shown to be associated with spontaneous clearance of HCV and a favorable response to treatment with interferon-based HCV therapy [23]. During HCV genotype 1 infection, the IL28B genotype has been shown to be strongly associated with intrahepatic ISG expression, with the unfavorable genotypes (CT and TT) expressing higher baseline ISG levels and poor responses to therapy compared with the favorable CC genotype patients with lower baseline ISG levels and correspondingly higher rates of SVR [24]. To explore the relationship between these two strong predictors of disease outcome in HCV genotype-3 patients, we further analyzed the correlation between the corresponding baseline ISG expression and the IL28B genotype. Our results showed that in HCV GT 3-infected patients the favorable IL28B (CC) genotype is associated with lower baseline ISG expression compared to the unfavorable CT/TT genotype, but this was not seen in NASH. These results suggest a genetic influence on host intrahepatic response to viral pathogens such as HCV.
Our study has a few limitations that are important to acknowledge. First, the sample size was small, and this might have reduced our ability to detect relevant differences in the gene expression. A second limitation includes the absence of healthy liver tissue for comparison of normal gene expression with HCV and NASH. Future larger studies are important to further validate our findings.
Collectively, these results implicate that viral replication consistently triggers ISG expression, which is further influenced by genetic (IL28B) status. Previous data suggest that increased ISG expression is a poor prognostic factor for therapeutic response to HCV therapy (SVR). Similarly, a host response primarily resulting from the IL28B response is associated with high ISG expression and often results in skewed host response to IFN-based therapy and an inability to achieve SVR. In this study, we demonstrate for the first time the differential hepatic transcriptional response to viral replication versus inflammation as observed in NASH patients. These suggest specific effects of viral replication rather than the inflammation itself.
Abbreviations
- ALP:
-
Alkaline phosphatase
- ALT:
-
Alanine aminotransferase
- AST:
-
Serum aspartate aminotransferase
- CHOL:
-
Total cholesterol
- HCC:
-
Hepatocellular carcinoma
- HCV GT-3:
-
Hepatitis C virus GT3
- IL28 B:
-
Interleukin 28 B
- ISG:
-
Interferon stimulated genes
- NAFLD:
-
Nonalcoholic fatty liver disease
- NASH:
-
Non-alcoholic steatohepatitis
- SVR:
-
Sustained virological response
- TBILI:
-
Total bilirubin
- TRIG:
-
Triglyceride
References
MohdHanafiah K, Groeger J, Flaxman AD, Wiersma ST. Global epidemiology of hepatitis C virus infection: new estimates of age specific antibody to HCV seroprevalence. Hepatology 2013;57:1333–1342
Ekstedt M, Franzen LE, Mathiesen UL, Thorelius L, Holmqvist M, Bodemar G, et al. Long-term follow-up of patients with NAFLD and elevated liver enzymes. Hepatology 2006;44:865–873
Teli MR, James OF, Burt AD, Bennett MK, Day CP. The natural history of nonalcoholic fatty liver: a follow-up study. Hepatology 1995;22:1714–1719
Lomonaco R, Chen J, Cusi K. An endocrine perspective of nonalcoholic fatty liver disease (NAFLD). Ther Adv Endocrinol Metabol 2011;2:211–225
Lonardo A, Loria P, Adinolfi LE, Carulli N, Ruggiero G. Hepatitis C and steatosis: a reappraisal. J Viral Hepat 2006;13(2):73–80
Milić S, Lulić D, Štimac D. Non-alcoholic fatty liver disease and obesity: biochemical, metabolic and clinical presentations. World J Gastroenterol 2014;20(28):9330–9337.
Ampuero J, Romero-Gómez M, Reddy KR. Review article: HCV genotype 3—the new treatment challenge. Aliment Pharmacol Ther 2014;39(7):686–698
Jhaveri R, McHutchison J, Patel K, Qiang G, Diehl AM. Specific polymorphisms in hepatitis C virus genotype 3 core protein associated with intracellular lipid accumulation. J Infect Dis 2008;197:283–291
Rubbia-Brandt L, Quadri R, Abid K, Giostra E, Malé PJ, Mentha G, et al. Hepatocyte steatosis is a cytopathic effect of hepatitis C virus genotype 3. J Hepatol 2000;33:106–115
Nkontchou G, Ziol M, Aout M, Lhabadie M, Baazia Y, Mahmoudi A et al. HCV genotype 3 is associated with a higher hepatocellular carcinoma incidence in patients with ongoing viral C cirrhosis. J Viral Hepat 2011; 18: e516–e522
Gale M Jr, Foy EM. Evasion of intracellular host defence by hepatitis C virus. Nature 2005; 436: 939–945
Negro F, Sanyal AJ. Hepatitis C virus, steatosis and lipid abnormalities: clinical and pathogenic data. Liver Int 2009;29(Suppl 2):26–37
Rosen HR. Emerging concepts in immunity to hepatitis C virus infection. J Clin Invest 2013;123(10):4121–4130
Lau DT, Negash A, Chen J, Crochet N, Sinha M, Zhang Y, et al. Innate immune tolerance and the role of kupffer cells in differential responses to interferon therapy among patients with HCV genotype 1 infection. Gastroenterology 2013; 144(2): 402–413
Feld JJ, Nanda S, Huang Y, Chen W, Cam M, Pusek SN, et al. Hepatic gene expression during treatment with peginterferon and ribavirin: Identifying molecular pathways for treatment response. Hepatology 2007;46(5):1548–1563
Felmlee DJ, Hafirassou ML, Lefevre M, Baumert TF, Schuster C. Hepatitis C virus, cholesterol and lipoproteins impact for the viral life cycle and pathogenesis of liver disease. Viruses 2013;5:1292–1324
Negro F. Hepatitis C in 2013: HCV causes systemic disorders that can be cured. Nat Rev Gastroenterol Hepatol 2014;11:77–78
Bartenschlager R, Lohmann V, Penin F. The molecular and structural basis of advanced antiviral therapy for hepatitis C virus infection. Nat Rev Microbiol 2013;11(482–496):13
Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, et al. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol 2007;9:1089–1097
Katsounas A, Hubbard JJ, Wang CH, Zhang X, Dou D, Shivakumar B, et.al. High interferon-stimulated gene ISG-15 expression affects HCV treatment outcome in patients co-infected with HIV and HCV. J Med Virol 2013; 85(6): 959–993
Onomoto K, Morimoto S, Kawaguchi T, Toyoda H, Tanaka M, Kuroda M, Uno K, Kumada T, Matsuda F, Shimotohno K, Fujita T, Murakami Y. Dysregulation of IFN system can lead to poor response to pegylated interferon and ribavirin therapy in chronic hepatitis C. PLoS ONE 2011;6(5):e19799
Larrubia JR, Benito-Martínez S, Calvino M, Sanz-de-Villalobos E, Parra-Cid T. Role of chemokines and their receptors in viral persistence and liver damage during chronic hepatitis C virus infection. World J Gastroenterol 2008;14(47):7149–7159
Stättermayer AF, Scherzer T, Beinhardt S, Rutter K, Hofer H, Ferenci P. Review article: genetic factors that modify the outcome of viral hepatitis. Aliment Pharmacol Ther 2014;39(10):1059–1070
Naggie S, Osinusi A, Katsounas A, Lempicki R, Herrmann E, Thompson AJ, et al. Dysregulation of innate immunity in hepatitis C virus genotype 1 IL28B-unfavorable genotype patients: impaired viral kinetics and therapeutic response. Hepatology 2012;56(2):444–454
Acknowledgements
We would like to thank all the patients who participated in this study.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Funding sources
National Institute of Allergy and Infectious Diseases (NIH), Bethesda, United States and Institute of Liver and biliary Sciences, New Delhi, India, supported this study.
Conflict of interest
Shikha Shrivastava, Eric G. Meissner, Emily Funk, Seerat Poonia, Virender Shokeen, Arun Thakur, Bhawna Poonia, Shiv Kumar Sarin, NirupmaTrehanpati and Shyamasundaran Kottilil declare that they have no conflicts of interest to disclose.
Ethical approval
The study protocol was approved by institutional ethical committee of Institute of Liver and Biliary Sciences (ILBS), New Delhi, and is in accordance with Declaration of Helsinki 1975.
Informed consent
All patients provided written informed consent to participate in the study.
Additional information
N. Trehanpati and S. Kottilil contributed equally to this manuscript.
Appendix
Appendix
See Table 2.
Rights and permissions
About this article
Cite this article
Shrivastava, S., Meissner, E.G., Funk, E. et al. Elevated hepatic lipid and interferon stimulated gene expression in HCV GT3 patients relative to non-alcoholic steatohepatitis. Hepatol Int 10, 937–946 (2016). https://doi.org/10.1007/s12072-016-9733-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12072-016-9733-6