Abstract
Cacao is an economically important commodity in Jamaica. Knowledge of the genetic diversity of Jamaican cacao germplasm is essential for their conservation and management. In spite of cacao’s economic importance in Jamaica, the crop is under studied, therefore limiting sound decisions toward improving productivity. Assessment of germplasm and on-farm genetic diversity is required to assist selecting superior genotypes to propagate and distribute across the island, as well as to use them as parental clones in breeding programs. Using 94 single nucleotide polymorphism (SNP) markers, 140 Jamaican cacao samples from two germplasm collections and a farmer’s estate along with 150 reference samples were analyzed. The principal coordinate analysis demonstrated that the majority of the Jamaican cacao selections were hybrids derived from five original germplasm groups, including Criollo, Amelonado and three Upper Amazon Forastero groups. Among the Upper Amazon groups, the Bayesian clustering analysis revealed that the Parinari (PA) ancestral lineage contributed the most (29.9%) to the Jamaican cacao germplasm. The germplasm collections showed greater diversity in terms of ancestral contributions compared to the farmer’s estate. However, the genetic differentiation between the three collecting sites was small (Fst = 0.036), indicating that samples collected from the three sites were derived from a common pool of germplasm. The current study supports the historical records and clarified the ancestry of Jamaican cacao. Although the majority of the cacao genetic groups were observed in the Jamaican cacao collections, several diversity gaps were found in both germplasm collections and in the farmer’s estate, especially germplasm with disease resistance to cacao frosty pod rot that was recently found in Jamaica.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The cacao tree (Theobroma cacao L.) is a tropical perennial species which belongs to the Malvaceae family and has a chromosomal number of 2n = 20 (Alverson et al. 1999; Mossu 1992). This evergreen crop is native to the South American rainforest (Bartley 2005; Dias 2001; Motamayor et al. 2002) with the center of origin in the Upper Amazon regions where the greatest diversity is observed (Bartley 2005; Motamayor et al. 2008; Zhang et al. 2012). Theobroma cacao grows productively in many regions situated between latitudes 20°N and 20°S of the equator (Coe and Coe 1996; Beckett 2000), including West Africa, South and Central America, Asia and the Caribbean where it is a major economic commodity (Wood and Lass 1985; Becker 1999). The crop is primarily cultivated for the seeds (referred to as beans) which are used for manufacturing an assortment of products including cocoa butter, solid chocolate, beverages, confectionaries, pharmaceuticals and cosmetics (Liendoa et al. 1997; Beckett 2000; Aikpokpodion 2012). Although cacao cultivation is linked to multibillion-dollar industries, the crop is mainly cultivated by small scale farmers that provide approximately 80–90% of the global production (World Cocoa Foundation 2014).
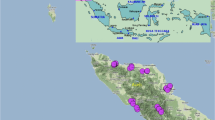
Cacao cultivation in Jamaica dates back to the early seventeenth century (Fagan 1984; Higman 2008) with the original planting materials being predominantly of the Criollo cacao type (Topper 1979). These were subsequently replaced with Amelonado cultivars, which were studier and coupled with other improved traits (Urquhart 1957). In the mid-1940s to late 1950s, several cacao selections, namely Imperial College Selections (ICS), Parinari (PA), Iquitos Mixed Calabacillo (IMC), and Scavina (SCA), were introduced to Jamaica mainly from Trinidad and St. Vincent (Fagan and Topper 1988). Most of these planting materials were established at the Orange River Agricultural Research Station located in the parish of St. Mary (Fagan and Topper 1988; see Fig. 1 for Orange River Research Station) which now maintains the majority of the cacao germplasm (Pryce et al. 2008). Various field trials were conducted and susceptibility of some cultivars to black pod disease was identified, which limited the use of these clones to breeding materials, rather than being used directly for commercial cultivation. From these planting materials, improved local hybrids were developed and distributed to farmers, including high yielding hybrid progenies derived from a cross of PA 150 and ICS 60 (Fagan and Topper 1988). Additional germplasms were again introduced to Jamaica from Trinidad, namely the high yielding black pod resistant Trinidad Select Hybrids (TSH). These selected TSH clones were established in a new research station, Montpelier Agricultural Research Station, in the parish of St. James (Fagan 1984). Breeding programs continued and hybrid seedlings with good performance as well as disease resistance were propagated at both research stations and distributed to farmers across Jamaica. Subsequently, the majority of the cacao trees found across Jamaica are a result of hybrid crosses (Fagan and Topper 1988; Topper 1979).
Cacao is an economically important commodity in Jamaica and can now be found growing in almost every parish; however, the main producing parishes are St. Mary, St. Thomas, St. Andrew and Clarendon (Jamaica Information Service [JIS] 2013). The beans, when processed, possess unique flavor attributes sought after by the international market. Consequently, the International Cocoa Council (ICCO) ranked Jamaica a fine flavored cacao exporting country, producing 100% fine and flavored cacao from 2007 to 2014. However, the percentage of fine or flavored cacao dropped slightly to 95% in the 2015 due to panel recommendations (ICCO 2008, 2016). Cacao production volumes have fluctuated drastically over the past decades in Jamaica; between 1905 to 1925, production numbers were between 2500 and 3500 metric tons (Topper 1979), while in 2010/2011 the production volume declined to 216.5 metric tons (JIS 2013), but then doubled (440 metric tons) during the 2013/2014 cropping year (JIS 2015). Several factors have contributed to the decline, including low productivity, high labor costs and inconsistency in quality. In an effort to increase cacao production, the Jamaican Government launched a project to rehabilitate farms and introduce new varieties. The new seedling selections were deposited in the Ministry’s two research stations: Orange River and Montpelier Agricultural Research Station, and seedling materials were made available to farmers (Ministry of Industry, Commerce, Agriculture and Fisheries [MICAF] 2012).
Molecular makers have had a considerable impact on cacao characterization and germplasm management. Over the past 15 years, numerous studies using various molecular markers have reported on the identification of mislabelled accessions, assessment of genetic redundancy in germplasm collections, analysis of genetic diversity and population structure in cacao, as reviewed by Zhang et al. (2012). Recently, SNPs have been used to assess the genetic diversity of cacao. The unambiguous nature of these markers allows the use of different genotyping platforms while maintaining consistency in results, thereby facilitating comparison and sharing of information across genebanks (Livingstone et al. 2010; Takrama et al. 2014). Additionally, the biallelic nature of the marker offers a very low error rate during allele calling (Rafalski 2002; Vignal et al. 2002). The use of SNP markers is not limited to genebanks, but has been extended to research conducted in farmers’ fields including an island-wide survey carried out in Puerto Rico (Cosme et al. 2016) as well as in farming communities in Honduras and Nicaragua (Ji et al. 2012).
Recapturing cacao as a lucrative industry in Jamaica requires insight into the genetic diversity, which would complement phenotypic characterization of available germplasm, in order to select cacao trees with superior agronomic traits and quality attributes. Furthermore, the revealed information on genetic identity, parentage and population membership of the characterized cacao germplasm will facilitate effective use of these germplasm for the development of new hybrids to meet the market needs. The main objectives of this study were (a) to genetically characterize cacao from the two research stations and from a farmer estate consisting of three fields in Jamaica using high throughput SNP genotyping; (b) to assess the level of genetic diversity present at the three collection sites; and (c) to analyze the ancestral contributions of the genetic groups to these cacao germplasm. The resultant information will give insight into the pool of germplasm present at the two research stations and improve the efficiency for managing this cacao collection. It will also facilitate effective use of these germplasm by Jamaican cacao farmers and breeders. To our knowledge, this is the first genetic study conducted on Jamaican cacao and therefore forms the baseline on which to build, conserve and utilize the Jamaican cacao germplasm collection.
Materials and Methods
Sample Collection and DNA Extraction
Cacao trees were assessed from the two germplasm stations (Orange River Agricultural Research Station in the parish of St. Mary and Montpelier Agricultural Research Station in St. James) and a farmer’s estate in Bachelor’s Hall district located in the parish of St. Thomas (Fig. 1). At Orange River Research Station, cocoa crops are located on seven different fields of which five were assessed. Also, assessment at the farmer’s estate was carried out on three different fields. A total of 160 trees were tagged for assessment; 70 at Orange River Agricultural Research Station, 49 at Montpelier Agricultural Research Station and 41 at Bachelor’s Hall Estate. Trees were selected based on the level of productivity, varying pod morphology and the presence of newly expanded young leaves (various shades of green but soft). Three healthy young leaves were collected from each individual tree and placed in labelled zip lock bags prior to being transported on ice to The University of the West Indies Mona Campus, Biotechnology Centre. Additionally, each tree was tagged with the corresponding label for potential revisiting.
Total genomic DNA was extracted from leaf tissues using the Cetyl Trimethyl Ammonium Bromide (CTAB) procedure described by Shahzadi et al. (2010) with modifications to the extraction buffer (2.5% CTAB, 0.1 M Tris-Cl [pH 9.5], 0.025 M EDTA, 1.5 M NaCl, 4% PVP-10, 0.5% 2-β-mercaptoethanol). Furthermore, to facilitate maximum precipitation of DNA, microcentrifuge tubes containing DNA pellets were incubated overnight at 4 °C. DNA presence and quantity was assessed on 0.8% agarose gel stained with ethidium bromide then visualized with a UV trans-illuminator model M-20 (Upland, CA, USA). The DNA samples were then sent to Sustainable Perennial Crops Laboratory (SPCL) USDA Beltsville Agricultural Research Center, Maryland, USA for genotyping.
At the Sustainable Perennial Crops Laboratory, DNA concentrations were determined by measuring absorbance at 260 nm, using a NanoDrop spectrophotometer (Thermo Scientific™, Wilmington, DE, USA). DNA purity was estimated by the 260∶280 ratio and the 260∶230 ratio of absorbance maximums.
SNP Markers and Genotyping
SNP loci were identified from expressed sequence tags (ESTs) developed from cDNA sequences obtained from a wide range of cacao plant parts that displayed a good representation of the cacao transcriptome (Argout et al. 2008, 2011). The 96 SNP loci used for this panel were previously reported on by Ji et al. (2012), Fang et al. (2014) and Lukman et al. (2014). The 96 SNPs, the flanking sequences and their distribution across the ten cacao chromosomes are listed in the Supplemental Table 1. The protocol for SNP genotyping of cacao used the Fluidigm 96.96 Dynamic Array™ (Fluidigm, San Francisco, CA). Each 96.96 Dynamic Array can run 96 samples against 96 SNP assays generating a total of 9216 data points in a single experiment. One key feature of this protocol is the inclusion of a specific targeted amplification (STA) reaction (Wang et al. 2009), which allows the enrichment of template molecules for each individual Integrated Fluidic Circuit® (IFC) reaction that facilitates the multiplexing during genotyping. An advantage to STA is that it allows the use of limited or low-quality DNA samples and reduces bias that may occur when samples are loaded to the 96 sample wells of the IFC. Since cacao leaf tissues contain high levels of polysaccharides and polyphenolic compounds that can potentially inhibit PCR amplification, the STA step was strongly recommended. The STA reaction was performed as described in the Fluidigm SNP Genotyping User Guide, PN 68000098 Rev. I1 (Fluidigm 2013). The STA master mix consisted of 2.5 μL of TaqMan® Taq polymerase (Life Technologies, Carlsbad, CA), PreAmp Master Mix (2X), 1.25 μL of Pooled assay mix (0.2X), and 1.25 μL of genomic DNA for a total reaction volume of 5.0 μL.
PCR was performed with an initial denaturation step of 95 °C for 10 min, followed by 14 cycles of a 2-step amplification profile consisting of 15 s at 95 °C and 4 min at 60 °C. The resulting amplified DNA was then diluted 1:5 in TE buffer in order to reduce the concentration of any remaining PCR by-products. Samples were then genotyped using the nanofluidic 96.96 Dynamic Array™ IFC (Integrated Fluidic Circuit; Fluidigm Corp.). The 96.96 Dynamic Array IFC for SNP genotyping was described by Wang et al. (2009). End-point fluorescent images of the 96.96 IFC were acquired on an EP1™ imager (Fluidigm Corp.). The data was recorded with Fluidigm Genotyping Analysis Software (Fluidigm, San Francisco, CA).
Data Analysis
Raw data was organized in Microsoft Excel 2007 for each SNP locus and sample call. The threshold of no call was set at 10%. Samples with more than 10% no call were excluded from subsequent data analysis. For duplicate identification, pairwise multi-locus matching was applied among individual Jamaican samples using the computer program GenAlEx 6.5 (Peakall and Smouse 2006, 2012). DNA profiles of the Jamaican samples that were fully matched at all genotyped SNP loci were declared duplicates or synonymous accessions (same genotype/clone). One accession from each duplicate group was retained for subsequent analyses. Additionally, DNA fingerprint profiles of 150 known cacao samples were included as reference samples (Supplemental Table 2). These references were previously generated by USDA SPCL and have been reported in previous publications (Zhang et al. 2009; Ji et al. 2012; Cosme et al. 2016). The samples included individual genotypes representative of previously defined cacao genetic populations or germplasm. Pairwise multi-locus matching was also applied between the Jamaican samples and reference clones to decipher identification using the same program.
Descriptive statistics measuring informativeness and quality of the 94 successfully amplified SNP loci were calculated using the GenAlEx 6.5 program (Peakall and Smouse 2006, 2012). The key descriptive statistics included Shannon’s information index (I), observed heterozygosity (HObs), expected heterozygosity (HExp), minor allele frequency (MAF), fixation index (F) and inbreeding coefficient within individuals relative to the population (FIS). For this study, the expected heterozygosity was used to estimate the genetic diversity. The parameters were computed separately for the Jamaican and reference populations, as well as for the combined data utilized.
Distance-based multivariate analysis was used to assess the relationship among the individual Jamaican cacao samples as well as their relationship with reference samples from international cacao genebanks. Pairwise genetic distances were computed using the DISTANCE procedure implemented in GenAlEx 6.5(Peakall and Smouse 2006). The same program was then used to perform Principal Coordinates Analysis (PCoA) based on the pairwise distance matrix, both distance and covariance were standardized.
A cluster analysis using the neighbour-joining method was used to further examine the genetic relationship among the Jamaican (140) and reference (150) cacao populations. First, the distances between individuals were calculated using the shared proportion of alleles distance measurement in the program Microsatellite Analyser (MSA; Dieringer and Schlötterer 2003) with 100 bootstrapping. The resulting distance matrix was used to generate a dendrogram using the Neighbour-joining algorithm (Saitou and Nei 1987) available in the program PHYLIP (Felsenstein 1989). Thereafter, the dendrogram was visualized using the FigTree program version 1.3.1 (Rambaut 2009).
The population structure of the Jamaican samples was determined using model-based Bayesian cluster analysis software STRUCTURE v2.3.4 (Pritchard et al. 2000). The analysis included seven distinctive cacao germplasm groups serving as references. The seven reference groups were selected based on the known history of cacao germplasm utilization in Central America and the Caribbean (Bartley 2005; Motamayor et al. 2008; Zhang and Motilal 2016). The full list of cacao accessions representing these seven groups is presented in Supplemental Table 2. To ensure that the assignment tests were not affected by the sample size of the tested accessions, the sample size of each reference group was brought up to 200 using the SIMULATION procedure implemented in the computer program ONCOR (Kalinowski et al. 2007). The simulated populations were then analyzed together with the 140 Jamaican cacao samples. An admixed model was selected and the number of clusters (K value) was set to 7 in correspondence to the possible genetic groups contributing to the Caribbean cacao collection. Ten independent runs were assessed for each K. All runs were carried out using 200,000 iterations after a burn-in period of 100,000. From the 10 independent runs, the highest Ln Pr (X|K) value was chosen and presented as bar plots for each sampling site.
Analysis of molecular variance (AMOVA) implemented in GenAlEx 6.5 (Peakall and Smouse 2012) was used to assess the genetic variation of the different collection sites. The codominant allelic distance was calculated, after which the statistical significance of the variances was tested using 9999 random permutations. The molecular variance was partitioned into populations within and among individuals.
Results
SNP Genotyping and Duplicate Identification
From the 96-SNP panel chosen for this study, two markers (TcSNP534 and TcSNP1484) were excluded, as they produced low repeatability for the internal controls utilized to check comparability across the plates. The remaining 94 SNP markers successfully fingerprinted the 160 Jamaican accessions.
Pairwise multilocus matching of the SNP profiles revealed duplication in the Jamaican cacao population assessed. Based on the 94 SNP markers, eight synonymous groups were identified including 28 accessions from the total Jamaican samples. Accessions assembled into the same synonymous group share identical SNP profiles at all 94 loci. (Table 1). From each synonymous group, one accession was selected as having a unique SNP profile for that group. Therefore, duplicated accessions amounted to 20 (duplicates accounting for 12.5% of the total Jamaican cacao samples). Percentage duplication was assessed for each collection site. Orange River germplasm collection recorded the highest percentage duplication (24.3%), while Montpelier germplasm collection recorded 4.1% and the farmer’s collection recorded 2.4%. The remaining 140 accessions, which accounted for 87.5% of the total samples, possessed unique SNP profiles and will hereafter be referred to as unique genotypes. Furthermore, none of the Jamaican samples matched with the reference clones used in this study.
Descriptive Statistics
The 140 Jamaican samples and 150 reference clones were used to compute the summary statistics for the 94 SNP markers and the results are presented in Table 2. The mean value for Shannon’s information index was 0.360, ranging from 0.106 for TcSNP437 locus to 0.634 for TcSNP277 locus. The mean observed heterozygosity (HObs) was 0.280, ranging from 0.555 for TcSNP437 locus to 0.775 for TcSNP277 locus. The mean genetic diversity (expected heterozygosity) was slightly lower than HObs. The analysis revealed a mean genetic diversity of 0.240, ranging from 0.054 for TcSNP437 to 0.447 for TcSNP277. Of the 94 markers, 73 (78%) revealed negative fixation index values, while 21 showed low positive values. Consequently, the mean fixation index (F) was negative (−0.114), ranging from −0.775 for TcSNP1096 locus to 0.170 for TcSNP653 locus due to the excess of heterozygotes observed. The mean minor allele frequency was 0.330, ranging from 0.078 for TcSNP1392 locus to 0.499 for TcSNP42 locus. The Jamaican population had higher observed heterozygosity (0.477) and genetic diversity (0.385) and a much lower inbreeding coefficient (−0.214) compared to the reference accessions (HObs = 0.206, HExp = 0.185, FIS = −0.080; Table 3) suggesting that the Jamaican cacao collection consists of mainly hybrids (mixing of populations). Comparison between the levels of diversity among the three collection sites revealed that Orange River germplasm collection (HExp = 0.406) was the most genetically diverse, while the farmer’s collection was the least diverse (0.373). The Montpelier germplasm collection had a similar level of genetic diversity (0.378) to that of the farmer’s collection (Supplemental Table 3).
Genetic Relationship among Jamaican Accessions and Reference Clones
The genetic relationship among the Jamaican cacao samples and the reference accessions were presented in the principal coordinates analyses (PCoA) plots (Fig. 2a-c). The three main PCoA axes, which accounted for 48.4% of the total variation, grouped the 150 reference accessions into seven clusters representing six genetic groups and one hybrid group. The first cluster consisted exclusively of reference samples from the pure Criollo group. The second cluster is the SCA group, which is comprised of wild Peruvian trees mainly from the Ucayali population and includes SCA 6 and SCA 24. The third cluster comprised of the Nacional accessions, which are not widely distributed and produce a unique “Arriba” flavour distinct to Ecuador cacao beans (Loor et al. 2009). The fourth cluster is a combination of genetic groups NA and PA which are a part of the Pound collection of natural populations originally collected from the Marañon river basin in Peru (Bartley 2005; Zhang et al. 2009). The fifth cluster also consists of Peruvian wild trees from the Pound collection but, belonged to the genetic group IMC. The sixth group comprised the Amelonado accessions and the final group consisted of the Trinitario hybrids. Most of the Jamaican samples were dispersed among the reference groups indicating admixed ancestral backgrounds for these samples. Most of the Jamaican samples overlapped with PA, NA, IMC and Trinitario clusters. They were also distributed near the Amelonado and SCA clusters. No Jamaican samples were found near the Criollo and Nacional groups, which included mostly homozygous genotypes (Fig. 2a-c).
Principal coordinate analysis plots for the 140 Jamaican cacao samples and 150 reference accessions. The first axis represents 22.78% of the total information, the second 18.63% and the third 6.99%. a depicts samples belonging to Orange River Agricultural Research Station, b depicts samples from Montpelier Agricultural Research Station and (c) depicts samples from the farmer’s estate in Bachelor’s Hall District
The dendrogram (Fig. 3) separated the Jamaican cacao and reference samples into two main clusters. The first cluster exclusively consisted of the Nacional and SCA reference groups. The second cluster consisted of all the Jamaican samples along with the other reference populations (Amelonado, Criollo and Trinitario) and three of the Peruvian wild cacao groups (NA, IMC and PA). Like the PCoA, the dendrogram revealed the reference group Nacional had no relationship to the Jamaican samples when compared to the other reference groups, as no Jamaican samples were found near this clade.
Neighbour-joining dendrogram depicting the relationships between and among the Jamaican cacao (140) and reference accessions (150). The first cluster exclusively consisted of the Nacional and SCA reference groups. The second cluster consisted of all the Jamaican samples along with the other reference populations (Amelonado, Criollo and Trinitario) and three of the Peruvian wild cacao groups (NA, IMC and PA). Colored branches and codes correspond to the reference samples while the Jamaican samples are displayed in black
Inference of Ancestral Background and Population Differentiation of Jamaican Accessions
The Bayesian clustering analysis separated the reference clones into the 7 respective genetic groups: IMC, NA, SCA, PA, Nacional, Amelonado and Criollo. The seven clusters represented all of the possible genetic populations that most likely contributed to the early introductions of cacao to Jamaica. Within the population structure analysis, Trinitario was not considered a distinct reference group as its genetic background is an admixture of Criollo and Amelonado. Furthermore, the new classification of the cacao genetic groups by Motamayor et al. (2008) excluded Trinitario. The bar plots (Fig. 4) and the PCoA (Fig. 2) results validated that most of the Jamaican cacao accessions are hybrid genotypes. In this study, genotypes were classified as having a single ancestral origin (belonging to a single genetic group) if the Q value was ≥0.75 for a specific genetic group and no single remaining genetic group possess a Q value of 0.25. If the above occurred, the genotype would be considered as having single ancestral origin with significant contributions from the latter genetic group. Meanwhile, genotypes with a Q value below 0.75 for any genetic group were denoted as hybrids having ancestral contributions from two or more genetic groups. Among the 140 different Jamaican genotypes identified, only 11 could be classified as having a single ancestral origin. The 11 clones were distributed among the 3 sites with 8 belonging to the Parinari reference group (ORRS_16, ORRS_70, MpRS_014, MpRS_025, MpRS_028, MpRS_040, BHE_067, and BHE_080) and 3 to the IMC group (ORRS_3, ORRS_24, ORRS_44). The remaining 129 genotypes were classified as admixed individuals or inter-population hybrids with their ancestral contribution ranging from two to seven of the reference genetic groups (Figs. 2 and 4). Among the 129 hybrid plants, 13.2% can be classified as an admixture of Amelonado and Criollo (classical Trinitario type), of which 10.8% (14) was found in the Orange River germplasm collection while the Montpelier germplasm collection and the farmer’s collection constituted 1.6% (2) and 0.78% (1), respectively (Figs. 2 and 4).
Population structure for Jamaican cacao accessions and reference clones using STRUCTURE, where k = 7. a Bar plot of reference groups from which Jamaican cacao are descendants of. Each color represents a distinct ancestral genetic group. Bar plots depicting the inferred ancestry of cacao samples belonging to (b) Orange River Agricultural Research Station, c Montpelier Agricultural Research Station and (d) farmer collection from Bachelor’s Hall District. Codes on the x-axis represents individual genotypes, and the probability (Q value) of each genotype belonging to a subpopulation(s) is plotted on the y-axis. Each vertical bar represents one genotype. Each color represents a specific ancestral population to which the genotypes most likely belong. The length of the color segments in each bar indicate the proportion of the inferred genetic group to that individual genotype. Genotypes with multiple colors are denoted as admixed genotypes or inter-group hybrids
The Orange River germplasm collection and the Montpelier germplasm collection revealed a larger ancestral genetic range (predominantly five genetic groups) than that of the farmer’s collection, which showed predominantly three genetic groups contributing to the background of the hybrids (Table 4). Overall, the Jamaican accessions were predominantly of Parinari lineage with an average population membership (Q value) of 29.9% (Fig. 5). The second largest ancestral contribution came from the Amelonado group, which accounted for 25.9% followed by the Criollo (17.0%) and IMC (13.2%) groups. The other 3 genetic groups: SCA, NA and Nacional contributed the least, 9.3%, 3.7% and 1.0% respectively, of their ancestry to the Jamaican accessions (Fig. 5). It should be noted that even though the PCoA revealed more samples overlapping with the NA than the SCA genetic group, SCA contributed more (9.3%) to the ancestral background of the Jamaican samples.
A small but significantly (p = 0.001) low level of genetic differentiation (Fst = 0.036) was observed among the collecting sites. The AMOVA indicated that majority of the genetic differentiation was attributed to individuals within the population (97%) while 3% variation occurred among collection sites. No variation was present among individuals within the different collection sites (Fig. 6).
Discussion
Genetic characterization and the presence of variation are key steps towards successful breeding and conservation of cacao cultivars. This is because morphological traits by themselves are insufficient to distinguish closely related samples and, as such, the precise variation that exists among genotypes may not be realized. The current study genetically characterized a fraction of the cacao genotypes present in Jamaica. From this information, an indication of the genetic diversity of the island’s cacao was revealed. Furthermore, quantifying and understanding the genetic groups which harbor important agronomic and disease resistant characteristics in the Jamaican cacao collection will prove useful for future breeding programs, since Jamaica’s cocoa industry now faces new disease challenges.
Duplication (genetic redundancy) of germplasm accessions is a serious problem found in many cacao collections worldwide. It not only hinders the effective management of germplasm collections but also limits the genotype diversity available for utilization (Zhang et al. 2006, 2009). From the current study, a considerable amount of duplication was present at the Orange River Agricultural Research Station (24.3% duplication). This suggests that there has been substantial amount of clonal multiplication (vegetative propagation) in the propagation of materials at the research station. Clonal multiplication is rare in farmer fields, but is commonly practiced in the research station. However, it is the unrecognized duplication that poses an issue because some materials were mistakenly maintained as different germplasm accessions. The result also demonstrates that previous identification techniques based on morphological descriptors are not sufficient for accurate genotype identification. The above percentage duplication is comparable with other studies, which reported 19.6% duplication in CATIE germplasm (Zhang et al. 2009), 20.1% in TARS germplasm (Irish et al. 2010) and 14.4% in the International Cocoa Genebank, Trinidad (Motilal et al. 2011) even though, their sample size was larger. On the other hand, Montpelier Agricultural Research Station recorded only 4.1% duplication in its germplasm collection. However, it should be noted that the amount of duplication recorded in this study could be greater (not smaller). This is because genotyping error (missing data at loci) at a small number of loci may indicate that duplicate samples are different if there are missing data at a locus for one sample and data is present at the same locus for the other sample (Zhang et al. 2006).
A small percentage (2.4%) of duplication was present in the farmer’s collection. Similar findings were reported on farms surveyed in Honduras and Nicaragua (8.6%) (Ji et al. 2012). Although both farm collections gave comparable percentage duplication, the farmer’s collection in Jamaica consisted of 40 accessions, while the study by Ji et al. (2012) consisted of 115 accessions. Farms are generally composed of a mixture of materials from local germplasm and unknown origins. This is because, farmers tend to plant seeds selected from their own field based on trees identified as having desirable agronomic traits. This practice results in the generation of trees with varying genotypic and phenotypic characteristics due to the outcrossing nature of cacao. Nonetheless, duplication in the farmer’s fields is not completely unexpected as the farmer may have received cuttings of the same clone from different sources and/or at different times.
Among the three sites assessed, the farmer’s estate had least gene diversity (HExp = 0.373) while Orange River germplasm collection had the highest gene diversity (HExp = 0.406). This was expected as breeders practice intense recombination through hybridization to achieve new varieties for agronomic purposes therefore, high rate of gene flow occurring. This result coincides with previous studies by Padi et al. (2015) comparing farmers’ open pollinated trees (0.245) with breeders’ accessions (0.346). Conversely Efombagn et al. (2008) detected slightly higher gene diversity on farm (0.50) than in Genebank (0.49) collection in Cameroon.
A foundation for breeding cacao in Jamaica was created by planting materials that were imported from Trinidad. Traditionally, introduction and distribution of planting materials in Jamaica were by rooted cuttings or budding. Materials introduced to Jamaica included Trinitario selections (hybrids of Criollo and Amelonado), IMC, SCA and PA selections, among others. However, in the late 1950s, research carried out in Trinidad and Ghana prompted the need to distribute hybrid seedlings obtained from crosses of Amelonado and Trinitario varieties, as well as crosses including wild accessions from the “Pound Collection”(Anonymous 1977 in Fagan and Topper 1988). These hybrid seedlings were considered sturdier and more vigorous than the traditional rooted cuttings and budwoods (Topper 1979; Anonymous 1977 in Fagan and Topper 1988). Consequently, hybrid seedlings became the largest proportion of materials distributed across Jamaica (Topper 1979; Fagan and Topper 1988). This suggests that alleles from external populations have been contributing to the gene flow, leading to the formation of hybrid materials in Jamaica.
Since the change in planting materials from rooted cuttings and budwoods to hybrid seedlings (resulting from Upper Amazon Forastero crossed with Amelonado or Trinitario), many hybrid seeds were introduced from Trinidad with admixtures of IMC, PA, ICS, and SCA genetic groups. Additionally, breeders in Jamaica began to produce local hybrid seeds through open and hand pollination. By 1961, an appreciable number of hybrid seedlings were developed in Jamaica reducing the importation of planting materials from Trinidad (Fagan 1979; Fagan and Topper 1988). The present study is in accordance with the breeding history of cacao in Jamaica, as the PCoA revealed that the Jamaican samples were distributed among most of the reference groups indicating a mixed ancestral background. The Jamaican cacao samples were scattered among three of the Upper Amazon Forastero groups (NA, PA and IMC), and among or between the Trinitario and Amelonado clusters. The Trinitario group is formed from natural hybridization of the two “varieties” Amelonado and Criollo (Motamayor et al. 2003) and possesses improved agronomic and flavour attributes of both groups. Although the PCoA explained a small percentage (48.4%) of the total variation, the results reveal similar patterns in the Bayesian clustering analysis and the dendrogram. All analyses demonstrated that the majority of the Jamaican samples were derived from multiple genetic groups, resulting in plants of high admixed ancestry. This further indicates that there is no distinct genetic structure in the cacao population assessed. Of the 140 samples assessed, 92% (129 samples) were classified as admixed individuals (using the threshold of Q < 0.75) with their ancestral contribution ranging from two to six of the reference genetic groups, while the other 8% indicated single ancestral origin from PA and IMC. The presence of significant amount of admixture in germplasm collections and on farmers’ fields was also evident in studies conducted in the Dominican Republic (Boza et al. 2013), Cameroon (Efombagn et al. 2008), Ghana (Padi et al. 2015), Peru (Zhang et al. 2011) and Indonesia (Lukman et al. 2014; Dinarti et al. 2015). From the above studies and current findings, cacao hybrids are common on farms and can be attributed to the fact that hybrid plants are more vigorous and studier. According to Topper (1979) crosses between Upper Amazon germplasm and Trinidad Selections exhibit high yield and vigour. As a result, hybrid seedlings were the largest proportion of planting material distributed across Jamaica.
Germplasm collections serve as the principal source of plant genetic resources and therefore, should consist of the greatest possible variability. Among the three localities, the two germplasm stations revealed a wider range of ancestral contributions across the genetic groups compared to the farmer’s collection. This, therefore, reflects their greater variability in germplasm than that of farmer’s collection. Five of the seven genetic groups corresponding to PA, Amelonado, Criollo, SCA and IMC were represented principally at the germplasm stations. However, on the farmer’s estate, only three of the seven genetic groups corresponding to PA, Amelonado and Criollo made ancestral contributions to the farmer’s selections. This narrow ancestral background may be due to limited access to diverse planting materials. Also, as previously mentioned, farmers expand their farms using their own seeds, therefore limiting the introduction of new genetic groups into their fields. It may also be due to the farmer’s selections, whereby trees consisting of the three genetic groups produce large pods and favorable bean size catering to economic gains.
Overall, the Parinari lineage made the greatest ancestral contribution to the Jamaican population, followed by Amelonado and Criollo. The Parinari group is known for its diverse morphological features, which accounts for the important agronomic and economic traits present within the population. Desirable features within this population include high yield potential, favourable level of resistance to witches’ broom and black pod diseases, large beans and favourable pod index value (Fagan 1979; Sounigo et al. 2005; Bekele et al. 2008). Consequently, this germplasm group has been widely used as a parent genotype in breeding programs worldwide. Such is the case in Jamaica, where the Parinari group is among the most important female parent material used in the production of high-yielding local hybrids (Fagan and Topper 1988). Similarly, it was the largest ancestral contributor to farms in West Africa (Aikpokpodion 2010) and Indonesia (Lukman et al. 2014). Conversely, studies conducted in neighbouring islands of the Dominican Republic and Cuba revealed Amelonado lineage as the predominant genetic group contributing to their cacao collections (Boza et al. 2013; Martínez et al. 2015). The greater contribution of Amelonado than Criollo to the Jamaican ancestral background is expected since Amelonado is more widely distributed in the Caribbean than the Criollo (Bartley 2005; Zhang and Motilal 2016).
The Nacional genetic group utilized in the reference collection consisted of pure Nacional samples which are highly homozygous. This native Nacional group found in Peru and Ecuador is known for its unique “arriba” flavour constituting various characteristics including low acidity and bitterness along with its floral and fruit taste (Deheuvels et al. 2004; Eskes et al. 2007). This group was not widely distributed in comparison with other Upper Amazon Forastero populations, and similar findings were reported in Cuba (Martínez et al. 2015), Dominican Republic (Boza et al. 2013) and Indonesia (Lukman et al. 2014). However, some Nacional type germplasm (e.g. UF 712, UF 273 and UF 12) has been used as key progenitors for breeding resistance to Frosty pod rot disease (caused by Moniliophthora roreri) in Costa Rica (Phillips-Mora et al. 2009). Given the recent occurrence of Frosty pod rot disease in Jamaica, the introduction of these germplasm accessions could be utilized to incorporate host resistance into the Jamaican cacao germplasm.
The Jamaican cocoa samples were collected from three different sites which possess different microclimates. However, there was a small but significantly low genetic differentiation among the three sites (Fst = 0.036). Nevertheless, the results indicate that most of the diversity resided among individuals rather than sites. This suggests that planting materials from the three sites were derived from a common pool of germplasm or same seed garden reservoir. This agrees with Fagan and Topper (1988), which reported that the majority of Jamaica’s planting materials were from Trinidad’s germplasm collection and distribution of the materials were in the two germplasm stations. Thereafter, local crosses were developed which showed disease resistance and good yield potential. Additionally, farmers receive planting materials from germplasm collections and thereafter use seeds from fruits of trees they consider to be superior trees to expand their farms. Consequently, many crosses occurred among different individual trees accounting for the high diversity among individuals.
In summary, the use of 94 SNP markers was useful and adequate to genetically characterize and assess the ancestral background of the existing cacao diversity in the two germplasm collections and the farmer’s collection in Jamaica. The study demonstrated that most of the Jamaican accessions sampled were hybrids consisting of PA, IMC, SCA, Amelonado and Criollo genetic groups. Additionally, the study quantified the ancestral background of the samples to each genetic group. The information obtained from the genetic diversity of each site provides insight regarding where new materials should be introduced, so as to increase diversity. Increasing diversity will enable disease resistant genes, along with economic traits, to be sustained in the Jamaican population. With the advent of Frosty pod rot disease to the island since last year (JIS 2016), the genotypes identified should now be evaluated for resistance, and breeders should introduce new varieties with disease resistance from other countries. This would broaden the genetic diversity of the Jamaican cacao collection and ensure sustainability in production for the country.
Abbreviations
- cDNA:
-
Complementary DNA
- DNA:
-
Deoxyribonucleic acid
- EST:
-
Expressed sequence tag
- PCR:
-
Polymerase chain reaction
- SNP:
-
Single nucleotide polymorphism
- TARS:
-
Tropical Agriculture Research Station
- USDA ARS:
-
United States Department of Agriculture, Agricultural Research Service
References
Aikpokpodion PO (2010) Variation in agro-morphological characteristics of cacao, Theobroma cacao L., in farmers' fields in Nigeria. J Crop Horticultural Sci 38(2):157–170. https://doi.org/10.1080/0028825X.2010.488786
Aikpokpodion PO (2012) Defining genetic diversity in the chocolate tree, Theobroma cacaol. grown in West and Central Africa. Genetic Diversity in Plants. ISBN: 978–953–51-0185-7, InTech, doi: https://doi.org/10.5772/33101
Alverson WS, Whitlock BA, Nyffeler R, Bayer C, Baum DA (1999) Phylogeny of the core Malvales: evidence from ndhF sequence data. Am J Bot 86:1474–1486
Argout X, Fouet O, Wincker P, Gramacho K et al. (2008) Towards the understanding of thecocoa transcriptome: production and analysis of an exhaustive dataset of ESTs of Theobroma cacao L. generated from various tissues and under various conditions. BMC Genomics 9:512
Argout X, Salse J, Aury JM, Guiltinan M et al (2011) The genome of Theobroma cacao. Nat Genet 43:101–109
Bartley BGD (2005) The genetic diversity of cacao and its utilization. CAB International, CABI Publishing, Wallingford
Becker H (1999) CACAO-good for agriculture and the environment. Agricultural Res Mag 47(11):2–2
Beckett ST (2000) The science of chocolate. The royal society of chemistry, London
Bekele, FL, Iwaro AD, Butler DR, Bidaisee GG (2008) Upper amazon forastero cacao (Theobroma cacao L.) 2: An overview of parinari clones from a breeder’s perspective
Boza EJ, Irish BM, Meerow AW, Tondo CL, Rodríguez OA, Ventura-López M, Gómez JA, Moore JM, Zhang D, Motamayor JC, Schnell RJ (2013) Genetic diversity, conservation, and utilization of Theobroma cacao L: genetic resources in the Dominican Republic. Genet Resour Crop Evol 60:605–619
Coe SD, Coe MD (1996) The true history of chocolate. Thames & Hudson, London
Cosme S, Cuevas HE, Zhang D, Oleksyk TK, Irish BM (2016) Genetic diversity of naturalized cacao (Theobroma cacao L.) in Puerto Rico. Tree Genet Genomes 12(5):88. https://doi.org/10.1007/s11295-016-1045-4
Deheuvels O, Decazy B, Perez R, Roch G, Amores F (2004) The first Ecuadorean ‘Nacional’ cocoa collection based on organoleptic characteristics. Trop Sci 44:23–27
Dias LAS (2001) Origin and distribution of Theobroma cacao L.: a new scenario. In: Dias LAS (ed) Genetic improvement of cacao. FAO. http://ecoport.org/ep?SearchType=earticleView&earticleId=197&page=−2. Accessed 25 March 2013
Dieringer D, Schlötterer C (2003) Microsatellite analyser (MSA): a platform independent analysis tool for large microsatellite data sets. Mol Ecol Notes 3:167–169
Dinarti D, Susilo AW, Meinhardt LW, Ji K, Motilal LA, Mischke S, Zhang D (2015) Genetic diversity and parentage in farmer selections of cacao from southern Sulawesi, Indonesia revealed by microsatellite markers. Breed Sci 65(5):438–446. https://doi.org/10.1270/jsbbs.65.438
Efombagn MIB, Motamayor JC, Sounigo O, Eskes AB, Nyassé S, Cilas C et al (2008) Genetic diversity and structure of farm and GenBank accessions of cacao (Theobroma cacao L.) in Cameroon revealed by microsatellite markers. Tree Genet Genomes 4:821–831
Eskes EB, Guarda D, Garci L, Garcia P (2007) Is genetic variation for sensory traits of cocoa pulp related to fine flavor cocoa traits? INGENIC Nwsl 11:22–28
Fagan HJ (1979) Growing cocoa in Jamaica. Cocoa Industry Board, Kingston
Fagan HJ (1984) An assessment of pathological research on cocoa in Jamaica from 1950 to 1980 and current research priorities. Trop Pest Manag 30:430–439
Fagan HJ, Topper BF (1988) Agronomic research on cocoa in Jamaica 1950-1980 and current research trends. Trop Agric (Trinidad) 65:290–294
Fang WP, Meinhardt LW, Mischke S, Bellato CM, Motilal L, Zhang D (2014) Accurate determination of genetic identity for a single cacao bean, using molecular markers with a nanofluidic system, ensures cocoa authentication. J Agric Food Chem 62(2):481–487
Felsenstein J (1989) PHYLIP-phylogeny inference package (version 3.2). Cladistics 5:164–166
Fluidigm (2013) Fluidigm® SNP genotyping analysis software user guide. Rev I1, PN 68000098, South San Francisco, CA, Fluidigm Corporation https://wwwmsciencecomau/upload/pages/fluidigmtech/fluidigm-snp-genotyping-user-guide-151112pdf. Accessed 10 July 2013
Higman BW (2008) Jamaica Food. University of the West Indies Press, Kingston
International Cocoa Organization (ICCO) (2008) ICCO document: Annual report 2007/2008. International Cocoa Organization, London U.K
ICCO (2016) Fine or flavour cocoa. http://www.icco.org/about-cocoa/fine-or-flavour-cocoa.html. Accessed 7 Sept 2016
Irish BM, Goenaga R, Zhang D, Schnell R, Brown JS, Motamayor JC (2010) Microsatellite fingerprinting of the USDA-ARS tropical agriculture Research Station cacao (L.) germplasm collection. Crop Sci 50:656–667
Jamaica Information Service (JIS) (2013) CIB reports 100% increase in cocoa production. Jamaica Information Service, Kingston http://jis.gov.jm/cib-reports-100-increase-in-cocoa-production/. Accessed 11 March 2015
Ji K, Zhang D, Motilal LA, Boccara M, Lachenaud P, Meinhardt LW (2012) Genetic diversity and parentage in farmer varieties of cacao (Theobroma cacao L.) from Honduras and Nicaragua as revealed by single nucleotide polymorphism (SNP) markers. Genet Resour Crop Ev 60:441–453
Jamaica Information Service (JIS) (2015) Cocoa industry returning to path of growth. Kingston, Jamaica http://jis.gov.jm/cocoa-industry-returning-path-growth/. Accessed 7 Sept 2016
Jamaica Information Service (JIS) (2016) Agricultural minister to spend $150 million to eradicate cocoa disease http://jis.gov.jm/agriculture-ministry-spend-150-million-eradicate-cocoa-disease/. Accessed 7 Sept 2016
Kalinowski ST, Manlove KR, Taper ML (2007) ONCOR A computer program for genetic stock identification. Department of ecology, Montana State University, Bozeman MT 59717. Available: http://www.montana.edu/kalinowski
Liendoa R, Padilla FC, Quintana A (1997) Characterization of cocoa butter extracted from Criollo cultivars of Theobroma cacao L. Food ResInt 30:727–731
Livingstone DS, Motamayor JC, Schnell RJ, Cariaga K, Freeman B, Meerow AW, Brown JS, Kuhn DN (2010) Development of single nucleotide polymorphism markers in Theobroma cacao and comparison to simple sequence repeat markers for genotyping of Cameroon clones. Mol Breed 27:93–106
Loor RG, Risterucci AM, Courtois B, Fouet O, Jeanneau M, Rosenquist E, Amores F, Vasco A, Medina M, Lanaud C (2009) Tracing the native ancestors of the modern Theobroma cacao L. population in Ecuador. Tree Genet Genomes 5:421–4733
Lukman ZD, Susilo AW, Dinarti D, Bailey B, Mischke S, Meinhardt LW (2014) Genetic identity, ancestry and parentage in farmer selections of cacao from aceh, Indonesia revealed by single nucleotide polymorphism (SNP) markers. Trop Plant Biol 7:133–143
Martínez IB, Nelson MR, Flamand MC, Bertin P (2015) Genetic diversity and population structure of anciently introduced Cuban cacao Theobroma cacao plants. Genet Resour Crop Evol 62(1):67–84
Ministry of Industry, Commerce, Agriculture and Fishers (MICAF) (2012) Agriculture minister says cocoa industry must create value-added activities. Kingston Jamaica http://www.moa.gov.jm/News/2012/2012-03-20_Agriculture_Minister_says_cocoa_industry_must_create_value-added_activities.php Accessed 11 March 2015
Mossu G (1992) Cocoa. The tropical Agriculturalist. CTA. The Macmillan Press Ltd, London
Motamayor JC, Risterucci AM, Lopez PA, Ortiz CF, Moreno A, Lanaud C (2002) Cacao domestication I: the origin of the cacao cultivated by the Mayas. Heredity 89:380–386
Motamayor JC, Risterucci AM, Heath M, Lanaud C (2003) Cacao domestication II: progenitor germplasm of the Trinitario cacao cultivar. Heredity 91:322–330
Motamayor JC, Lachenaud P, da Silva e Mota JW, Loor R, Kuhn DN, Brown JS, Schnell RJ (2008) Geographic and genetic population differentiation of the Amazonian chocolate tree (Theobroma cacao L). PLoS One 3(10):e3311. https://doi.org/10.1371/journal.pone.0003311
Motilal LA, Zhang D, Umaharan P, Mischke S, Pinney S, Meinhardt LW (2011) Microsatellite fingerprinting in the International cocoa Genebank Trinidad: accession and plot homogeneity information for germplasm management. Plant Genet Resour 9:430–438. https://doi.org/10.1017/S147926211100058X
Padi FK, Ofori A, Takrama J, Djan E, Opoku SY, Dadzie AM, Bhattacharjee R, Motamayor JC, Zhang D (2015) The impact of SNP fingerprinting and parentage analysis on the effectiveness of variety recommendations in cacao. Tree Genet Genomes 11(3):1–14
Peakall R, Smouse PE (2006) GENALEX 6.5: genetic analysis in excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Phillips-Mora W, Castillo J, Arciniegas A, Mata A, Sánchez A, Leandro M, Astorga C, Motamayor J, Guyton B, Seguine E, Schnell R (2009) Overcoming the main limiting factors of cacao production in central America through the use of improved clones developed at CATIE. Proceedings of the 16th International cocoa research conference, COPAL; 16-21th Nov. 2009, Bali, pp 93–99
Pritchard JK, Stephens M, Donnelly P (2000) Inference population structure using multilocus genotype data. Genetics 155:945–959
Pryce M, Mitchell S, Burke A, McKenzie C, Stirling S, Ryan J, Simpson W, McGlashan D (2008) Jamaica: country report to the FAO International technical conference on plant genetic resources for food and agriculture. Kingston. URL: www.moa.gov.jm/jam/jamaica2.pdf.
Rafalski A (2002) Applications of single nucleotide polymorphisms in crop genetics. Curr Opin Plant Biol 5:94–100
Rambaut A (2009) Molecular evolution, phylogenetics and epidemiology: FigTree v1.3.1 2006–2009: http://tree.bio.ed.ac.uk/software/figtree/
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Shahzadi I, Ahmed R, Hassan A, Shah MM (2010) Optimization of DNA extraction fromseeds and fresh tissues of wild marigold (Tagetes minuta) for polymerase chain reaction analysis. Genet Mol Res 9(1):386–393
Sounigo O, Bekele F, Iwaro D, Thévenin J-M, Bidaisee G, Umaharan P, Sankar A, Sukha D, Boccara M, Butler DR, Eskes AB (2005) Description of the CFC/ICCO/IPGRI project collection. Proceedings of the 14th International cocoa research conference, October 13–17 2003, Accra, Ghana, Nigeria, Cocoa Producers’ Alliance, 21–32
Takrama J, Ji K, Meinhardt L, Mischke S, Opoku SY, Padi FK, Zhang D (2014) Verification of genetic diversity of introduced cacao germplasm in Ghana using single nucleotide polymorphism (SNP) markers. Afr J Biotechnol 13:2127–2136
Topper BF (1979) Growing cocoa in Jamaica. Cocoa Industry Board, Kingston
Urquhart DH (1957) Report on the expansion of the cocoa industry in Jamaica. Publications. Cadbury Brothers Ltd, Bournville
Vignal A, Milan D, SanCristobal M, Eggen A (2002) A review on SNP and other types of molecular markers and their use in animal genetics. Genet Sel Evol 34(3):275–306
Wang J, Lin M, Crenshaw A, Hutchinson A, Hicks B, Yeager M, Berndt S, Huang WY, Hayes RB, Chanock SJ, Jones RC, Ramakrishnan R (2009) High throughput single nucleotide polymorphism genotyping using nanofluidic dynamic arrays. BMC Genomics 10:561
Wood GAR, Lass RA (1985) Cocoa, 4th edn. Longman, London
World Cocoa Foundation (2014) http://www.worldcocoafoundation.org/wpcontent/uploads/Cocoa-Market-Update-as-of-4-1-2014.pdf Accessed 31 Aug 2016
Zhang D, Motilal L (2016) Origin, dispersal and current global distribution of cacao genetic diversity. In: Bailey BA, Meinhardt LW (eds) Cacao Diseases. Springer International Publishing, New York
Zhang D, Mischke S, Goenaga R, Hemeida AA, Saunders JA (2006) Accuracy and reliability of high-throughput microsatellite genotyping for cacao clone identification. Crop Sci 46:2084–2092
Zhang D, Mischke BS, Johnson ES, Mora A, Phillips-Mora W, Meinhardt LW (2009) Molecular characterization of an International cacao collection using microsatellite markers. Tree Genet Genomics 5:1–10
Zhang D, Gardini EA, Motilal L, Baligar V, Bailey B, Zuñiga-Cernades L et al (2011) Dissecting genetic structure in farmer selections of Theobroma cacao in the Peruvian Amazon: implications for on farm conservation and rehabilitation. Trop Plant Biol 4(2):106–116
Zhang D, Figueira A, Motilal L, Lachenaud P, Meinhardt LW (2012) Theobroma. In: Kole C (ed) Wild crop relatives: genomic and breeding resources, plantation and ornamental crops. Springer, Berlin, pp 277–296. https://doi.org/10.1007/978-3-642-21201-7_13
Acknowledgements
We would like to give special thanks to Stephen Pinney (USDA-ARS) and Lin Zhou (Nanjing Agricultural University, China) for assistance with SNP genotyping using the nanofluidic array system. We also thank Sue Mischke (USDA-ARS) for editing the manuscript. References by USDA to a company and/or product are only for the purposes of information and do not imply approval or recommendation of the product to the exclusion of others that may also be suitable. This project was supported in part by grants from The University of the West Indies, Mona Campus Research and Publications Committee and Inter-American Development Bank Multilateral Investment Fund (IDB MIF) through the Jamaica Cocoa Farmers’ Association (JCFA). Special thanks to Mr. Desmond Jadusingh owner of Bachelor’s Hall Estate who graciously accommodated the research on his farm.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no conflict of interest.
Additional information
Communicated by: Philippe Lashermes
Electronic supplementary material
ESM 1
(DOCX 46.0 kb)
Rights and permissions
About this article
Cite this article
Lindo, A.A., Robinson, D.E., Tennant, P.F. et al. Molecular Characterization of Cacao (Theobroma cacao) Germplasm from Jamaica Using Single Nucleotide Polymorphism (SNP) Markers. Tropical Plant Biol. 11, 93–106 (2018). https://doi.org/10.1007/s12042-018-9203-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12042-018-9203-5