Abstract
Circular RNAs (circRNAs) are a class of noncoding RNA molecules formed by the back splicing process. Compared to linear mRNA molecules they are more stable. CircRNA acts as miRNA sponges, regulates translation, epigenetic alterations, etc. However, the most significant aspect of circRNAs has been its role in regulating the hallmark of cancer and diabetes mellitus. Several circRNAs are extensively expressed in individuals with cancer and diabetics. Dysregulated expression of various circRNAs plays a crucial part in the development of type 2 diabetes mellitus. In the present review, we present the current understanding of cricRNAs biogenesis, regulatory mechanisms, reviews of recent findings and circRNA as potential biomarker.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Circular RNAs (circRNAs) are a family of RNA molecules where a covalent bond connects 3′ and 5′ ends and acts as mammalian gene regulator. They are noncoding, single-stranded and highly stable molecules, first recognized as viroids in plant-based viruses in 1976 and later (1979), it was also found in eukaryotic cells (Kolakofsky 1976; Sanger et al. 1976). CircRNAs are mostly formed by the precursor mRNA back-splicing process (Hsu and Coca-Prados 1979). In nature, they are less abundant than the normal linear RNA molecule and, are therefore, considered as rare events (Shan et al. 2019; Shang et al. 2019; Zhao et al. 2019). With the advancement of RNA sequencing techniques and bioinformatics, thousands of individual cricRNAs were discovered in mammalian cells. Following this, extensive research has discovered numerous functions of cricRNAs in recent years. For example, acting as scaffolds in the assembly of protein complexes, sequestrating proteins from their native subcellular localization, modulating parental gene expression, regulating alternative splicing and RNA–protein interactions and functioning as microRNA sponges (Wang et al. 2017). Most notably, certain circRNA plays a significant role in tumour initiation and cancer progression. Genomic analysis demonstrates the strong presence of circRNA in different cell types. In low-proliferating cells such as the brain, they have higher levels of expression compared to highly-proliferating liver cells (Haddad and Lorenzen 2019). Notably, enucleated cells such as red blood cells and platelets tend to exhibit higher levels of circRNAs than nucleated (hematopoietic) cells. It has been reported that platelets, in particular, express the highest number of circRNAs, almost twice as many as erythrocytes and five times more than granulocytes (Haddad and Lorenzen 2019; Nicolet et al. 2018). In this mini review, we concentrated on the circRNA’s biogenesis, functional mechanism and crucial role in the development of two human diseases, i.e. cancer and diabetes.
Characteristics and biogenesis of circRNA
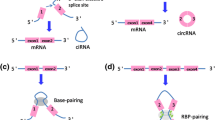
Recent studies have shown that the size of circRNA ranges from a few hundred to thousand nucleotides produced from one to five exons. Since they lack free 5′ and 3′ polarity they are not prone to exonuclease degradation and are much more stable than the linear RNA molecule. They primarily reside in the cytoplasm of the cell and some are present in the nucleus as well. Both circRNA and linear RNA originates from the precursor mRNA by back splicing of exons/introns and simple splicing, respectively. CircRNAs are of three different types depending on their origin: exon–intron circRNAs (EIciRNAs), exonic circRNAs (ecircRNAs), and circular intronic RNAs (ciRNAs) (Jeck and Sharpless 2014; Su et al. 2019b).
Three possible models of ecircRNA biogenesis have been discovered, namely circularization driven by lariat, intron pairing, and resplicing. The lariat-driven method of circularization is a form of exon skipping (cassette-on) process where the intronic lariat contains the skipped exon(s). If further splicing occurs before the lariat is unravelled by debranching enzymes within it, a stable RNA circle can be formed with the exons skipped. During the process, a linear transcript is also made, excluding the skipped exon(s) (Chen, et al. 2015). Circularization driven by intron pairing is independent of exon skipping. It differs from the lariat-driven model of circularization by selecting splice site pairs and lacks a detailed understanding of the corresponding linear product(s). Intronic motifs were proposed to edge the circularized exons(s) and thus join the circularized exons(s) (Chen et al. 2015). Resplicing-driven circularization is a two-stage cycle, initial splicing eliminates canonical splicing sites and thus resplicing makes use of cryptic splicing sites on the spliced mRNAs for exon-skipping circularization (Chen et al. 2015). Unlike ecircRNAs, EIciRNAs retain those introns that are not completely spliced out (figure 1). Pre-mRNAs contain flanking Alu complementary pairs or pairs other than Alu might facilitate EIciRNA production (Sheng, et al. 2018). CiRNAs are being derived from intron lariats that resist normal intron degradation and debranching. Lariat intron excised of reverse complementary sequences from pre-mRNA can pair to generate a close loop structure called ciRNA. The development of ciRNAs is dependent on the presence of GU-rich 7mer sequence near the 5′ splicing site and 11mer C-rich motifs near the 3′ branch site (Su, et al. 2019a).
CricRNA formation. (a) Exonic cricRNA (ecricRNA) are formed by the backsplicing of 5′ splice site to a 3′ splice site. (b) The intron is removed and ecricRNA is produced by more than one exons. (c) Formation of ciRNA from intorn lariats. (d) EicricRNA are formed by the circularization of intron retained between the exons.
Functional mechanism of circRNAs
CricRNA acts as microRNA sponges (miRNA). miRNA sponges contain multiple sites complementary to a miRNA of interest. A recent study reported that circRNA is rich in miRNA response elements (MREs) and can function as miRNA sponges (Ebert and Sharp 2010; Han, et al. 2017). When cricRNA is expressed at a high level, it inhibits the activity of miRNAs sharing the common seed area. MicroRNA plays an important role in posttranscriptional regulation of gene expression by binding to the 3′ untranslated region (3′UTR) of an mRNA. CircRNA acts as sponges and binds to the miRNA complementary sites, withdrawing the miRNAs from their downstream target genes and regulates gene expression. Compared to some of the other miRNA sponges, circRNA can bind more efficiently and effectively to the target, and thus, regarded as a super sponge. The best example is CDR1as, which nurture more than 70 selectively conserved miR-7 binding sites (Schwerk and Savan 2015; Vislovukh, et al. 2014). Apart from functioning as a miRNA sponge, some circRNA also has binding sites for RNA-binding proteins and thus functions as protein sponges and regulates gene expression. For example, the PABPN1 locus originating from circRNA (circ-PABPN1) binds to human antigen R / ELAV-like protein 1 (HuR) and avertsHuR from binding to PABPN1 mRNA, thereby suppressing translation of PABPN1 (Su et al. 2019a; Zhao et al. 2019). CircRNA regulates transcription by combining with RNA polymerase II complex and translating related proteins. For example, circ-PAIP2 and circ-EIF3J were found to interrelate with the RNA polymerase II and U1 snRNPs in the promoter region of the host gene for enhanced transcription of their parental genes, such as EIF3J and PAIP2 (Su et al. 2019a; Zhao et al. 2019). Recent studies have also shown that circRNA competes with linear mRNA for selective splicing. For example, the circRNA generated from the circularization of the second exon of the muscle blind splicing factor (MBL) competes with the linear MBL mRNA (Ashwal-Fluss, et al. 2014; Wu et al. 2019).
CircRNAs in cancer
Cancer cells can produce dysregulated growth factors and corresponding receptor molecules, leading to autocrine stimulation. Epidermal growth factor receptor (EFGR) is a type of protein receptor that can trigger a cascade of other growth factors and controls cell growth. EFGR is a target of miR-7 (tumour-suppressor). CDR1as (cricRNA) acts as a sponge of miR-7 and increases the expression level of proto-oncogenes (EGFR, CCNE1 and PIK3CD). Consequently, overexpression of CDR1as results in the suppression of tumour-suppressing activity of miR-7. In addition to CDR1as, a particular kind of circRNA (has_circ_0000284) is overexpressed in CRC tissues and cell lines. The knockdown of this circRNA inhibited the proliferation and induced apoptosis of CRC cells. A recent study also showed that has_circ_0046701 promotes cell proliferation and invasion through regulating ITGB8 expression by sponging the miRNA–miR-142-3p (Du and Lovly 2018; Sever and Brugge 2015; Witsch et al. 2010).
Cancer cells can escape antigrowth signals by suppressing the expression of tumour suppressor genes. The antigrowth signals are necessary to block cell proliferation by arresting the cell cycle (Amin et al. 2015). The overexpression of the circularized product (Circ-ITCH) inhibits cell proliferation, migration, invasion and metastasis. Phosphatase and tensin homolog (PTEN) and p21 contributes to cell cycle suppression and recent studies have shown that circ-ITCH can sponge miR-17 and miR-224 directly, leading to increased target gene expression, PTEN and p21, respectively. Another type of circRNA known as circRNA-000425 is a target of yes-associated protein 1 (YAP1), a transcriptional coactivator factor that acts as an oncogene associated with cancer malignancy in several cancer types (Su et al. 2019a; Yang et al. 2018a). By activating CDK4 and CDK6, cyclinD1 facilitates the transition from the G1 phase of the cell cycle to S phase. Several circRNAs regulate tumour growth by regulating Cyclin D1. Xue and colleagues performed microarray research on circRNA and reported that Circ100284 is upregulated in As-HaCaT cells (arsenite-treated HaCaT cells). When icrc100284 was knocked down, it inhibited G1/S transition in As-HaCaT cells (Su et al. 2019a).
B-cell lymphoma is an anti-apoptotic molecule that protects the cell from apoptosis, whereas BCL-2 associated X protein is a proapoptotic gene. It has been documented that CircRNA Hsa circ 0007534 is overexpressed in CRC tissues (Geng et al. 2018). When it was silenced, it inhibited proliferation of CRC cells while promoting apoptosis. Microarray studies performed by Zhang et. al. (2017) showed that circUBAP2 was the most significantly overexpressed circRNA in osteosarcoma (Zhang et al. 2017a). The knockdown of circUBAP2 inhibited cell proliferation and promoted cell apoptosis. Hsa_circ_0009910 is another type of circRNA that was upregulated in osteosarcoma cells and its knockdown inhibited cell proliferation. In autophagy, circRNA also plays a very important role. Circ-Dnmt1 was found to be upregulated in the breast cancer tissue and cell lines. Overexpression of circ-Dnmt1 increases the survival and proliferation of breast cancer cells (Straten and Andersen 2010; Um 2016).
One of the major factors driving the growth and proliferation of tumour cells is angiogenesis, which creates new blood vessels to supply oxygen and nutrients to the tumour and to dispose toxic tumour waste. CircRNA-MYLK is a product of the myosin light chain kinase gene that is significantly overexpressed in breast cancer tissues and cell lines. CircRNA-MYLK acts as a sponge miRNA and binds directly to miR-29a, relieving the suppression of target vascular endothelial growth factor A (VEGFA) and activating the signalling pathway VEGFA/VEGFR2 which produces several cellular responses relevant to angiogenesis. Another kind of cirRNA, i.e. Cznf292 plays a very important role in the progression of glioma tube formation (Bielenberg and Zetter 2015; Su et al. 2019a). CircRNA is also known to regulate the stemness of cancer. Yang and colleagues reported 27 circRNA expressed in breast cancer stem cells (BCSCs) (Yan et al. 2017). They observed that circVRK1 was downregulated, inhibiting the expansion and self-renewability of BCSCs. The knockdown of circVRK1 increases the stemness of BCSCs (Yan et al. 2017). CircRNA–Hg19_circ_0005033 is upregulated in TDP cells and promotes cell proliferation and resistance to chemotherapy and radiotherapy (Ma et al. 2020; Su et al. 2019a). Liu et al. (2018a, b) found that circRNA-MTO1 is upregulated in monastrol-resistant BC cells that promoted cell cytotoxicity caused by monastrol and reversed monastrol resistance (Liu et al. 2018b). Peng and colleagues identified circRNA - circBA9.3 which contributes to increased leukaemia cell proliferation and anti-apoptotic capacity (Kun-Peng et al. 2018; Su et al. 2019b).
CricRNAs are a good candidate for biomarker studies because of their stability and tissue-specific expression. These are relatively present in high quantities in body fluids such as saliva, plasma, serum and exosomes and act as noninvasive biopsy biomarkers for cancer. CircZEB1.33 may serve as a biomarker in the prediction of HCC prognosis, as it is overexpressed in human HCC tissues. Hsa_circRNA_002059 is overexpressed in gastric cancer and thus it has been proposed as a potential biomarker. Hsa_circRNA_0067934 can also serve as a novel biomarker for disease progression or as a therapeutic target in the case of squamous cell carcinoma. Hsa_circ_0000190 was downregulated in GC tissues and plasma samples (Fang et al. 2019). In another study, hsa_circ_0000745 is lowly expressed in GC tissues and plasma samples (Huang et al. 2017). Its expression is correlated with tumour differentiation and tumour-node-metastasis stage. The stability, universality, conservativeness and specificity of circRNAs make it a better-trusted diagnostic and prognostic biomarker for cancer, and the regulatory roles of circRNAs in tumour cells make it an important candidate for cancer treatment (Su et al. 2019b). The role of different circRNAs in cancer based on recent research is shown in table 1.
CircRNAs in diabetes
Diabetes mellitus, commonly called diabetes, is a metabolic condition that causes high sugar in the blood, one of the most important health issue worldwide. Type 2 diabetes mellitus (T2DM) in China shows a rapid increase due to the prevalence of poor lifestyle, the ageing population and changes in dietary structure. The incidence of T2DM in adults is as high as 11.6%, which is expected to increase (Wu et al. 2014). Perusal of literature suggested that dysregulation of circRNA is one of the indicative feature of metabolic disorders (table 2). CircRNA seems to play a significant role in the pathogenesis of diabetes and related metabolic disorders thus acts as a potential biomarkers. For example, the level of hsa_circ_0054633 in peripheral blood was reported to be associated with diabetes and might serve as a diagnostic biomarker of prediabetes (Zhao et al. 2017). Another circRNA, Cdr1as, can improve the development and secretion of insulin by targeting Pax6 and Myrip in mouse β cells, respectively via miR-7 as a mediator, suggesting that this circRNA may be a new therapeutic target for diabetes. Diabetic vascular complications are the major causes of disability and high mortality among patients with diabetes (Zaiou 2020). Mitogen-activated protein kinase (MAPK) pathway plays a crucial role in gene expression and cytoplasmic activities. CircRNA may affect the status of diabetes by chipping in the MAPK pathway (Kono et al. 2006). CircRNAs are usually observed to be upregulated or downregulated in various specimens from diseased patients, such as tissue, plasma, peripheral blood, and saliva, which are easy and suitable to obtain for quantification via painless and minimally invasive methods. Some circRNA have shown efficacies as a biomarker for other types of diseases (Zhang et al. 2018b). Altered expression profile of circRNA in the blood of patients with T2DM was confirmed of which hsa_circ_0054633 was proposed to be a potential diagnostic biomarker for prediabetes and type-2 diabetes (Zhao et al. 2017). CircRNA can regulate insulin signalling and can be used as a biomarker as well for cell metabolism and T2DM. Peripheral blood has-circ000094 could be used as a diagnostic marker for T2DM. Dammann et al. (2018) found the elevated expression of has-circ000094 in patients with T2DM inhibited the expression of P21 by repressing hsa-miR-370-5p, in turn leading to the deactivation of kinase PAK1 (Dammann, et al. 2018). One of the rno_circRNA_008565 target genes, MAPK8 (JNK) had the highest score with autophagy. miRNA binding prediction found that rno-miR-504 had most binding sites (four sites) with this target and had the most stable binding structure, which suggests that it may regulate MAPK8 (JNK) and the autophagy of rat islet β-cells by inhibiting rno-miR-504 (Bai et al. 2019). CircRNA biomarkers allow early precise diagnosis, appropriate therapy selection and monitoring that can be detected effectively in body fluid, extracellular vesicles or circulating cells in various types of diseases (Zhang et al. 2018b).
Conclusion
At first, circRNA was considered to be an error during the splicing process. However, with the advancement of RNA sequencing techniques and bioinformatics, various important roles of circRNA have been highlighted, in particular their function in diseases ranging from cancerous to noncancerous pathologies and their growth and progression, which is still less understood. CircRNA is now known to be an abundant, stable and highly expressed class of mRNAs serving as miRNA sponges, interacting with RNA binding proteins, controlling splicing and translations etc. CircRNA’s most important functions are that they act as a potential biomarker in cancer and diabetes due to their stability and tissue-specific expression, making them an appropriate candidate for biomarker studies. Now it is commonly accepted that eukaryotic spliceosomes exhibit a certain degree of flexibility in the choice of splice sites, resulting in widespread alternatively spliced isoforms in transcriptomes, including circRNAs. In the coming days, enhanced bioinformatics techniques related to circRNA along with faster sequencing techniques are believed to discover more circRNA functions that will eventually enable us to better understand the origin of circRNAs. Different types of circRNA are overexpressed in different cancer and diabetic cell lines and, if guided, can regulate the growth and proliferation of these diseased cells. Further studies on this ancient RNA will provide a significant perspective in cancer and diabetes research.
References
Amin A. R. M. R., Karpowicz P. A., Carey T. E., Arbiser J., Nahta R., Chen Z. G. et al. 2015 Evasion of anti-growth signaling: A key step in tumorigenesis and potential target for treatment and prophylaxis by natural compounds. Semin. Cancer Biol. 35 (Suppl), S55–S77.
Ashwal-Fluss R., Meyer M., Pamudurti N. R., Ivanov A., Bartok O., Hanan M. et al. 2014 circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 56, 55–66.
Bai C., Yang W., Lu Y., Wei W., Li Z. and Zhang L. 2019 Identification of circular RNAs regulating islet β-cell autophagy in type 2 diabetes mellitus. BioMed. Res. Int. 1–11. (https://doi.org/10.1155/2019/4128315).
Bi J., Liu H., Dong W., Xie W., He Q., Cai Z. et al. 2019 Circular RNA circ-ZKSCAN1 inhibits bladder cancer progression through miR-1178-3p/p21 axis and acts as a prognostic factor of recurrence. Mol. Cancer 18, 1–14.
Bielenberg D. R. and Zetter B. R. 2015 The contribution of angiogenesis to the process of metastasis. Cancer J. 21, 267–273..
Chen B., Wei W., Huang X., Xie X., Kong Y., Dai D. et al. 2018 circEPSTI1 as a prognostic marker and mediator of triple-negative breast cancer progression. Theranostics 8, 4003.
Chen I., Chen C.-Y. and Chuang T.-J. 2015 Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip. Rev. RNA 6, 563–579.
Chen J., Cui L., Yuan J., Zhang Y. and Sang H. 2017 Circular RNA WDR77 target FGF-2 to regulate vascular smooth muscle cells proliferation and migration by sponging miR-124. Biochem. Biophy. Res. Commun. 494, 126–132.
Cheng Z., Yu C., Cui S., Wang H., Jin H., Wang C. et al. 2019 CircTP63 functions as a ceRNA to promote lung squamous cell carcinoma progression by upregulating FOXM1. Nat. Commun. 10, 1–13.
Dai X., Zhang N., Cheng Y., Yang T., Chen Y., Liu Z. et al. 2018 RNA-binding protein trinucleotide repeat-containing 6A regulates the formation of circular RNA circ0006916, with important functions in lung cancer cells. Carcinogenesis 39, 981–992.
Dammann K., Khare V., Coleman C., Berdel H. and Gasche C. 2018 p-21 Activated kinase as a molecular target for chemoprevention in diabetes. Geriatrics (Basel) 3, 73.
Du Z. and Lovly C. M. 2018 Mechanisms of receptor tyrosine kinase activation in cancer. Mol. Cancer 17, 58.
Ebert M. S. and Sharp P. A. 2010 MicroRNA sponges: progress and possibilities. RNA (New York) 16, 2043–2050.
Fang X., Wen J., Sun M., Yuan Y. and Xu Q. 2019 CircRNAs and its relationship with gastric cancer. J. Cancer 10, 6105.
Geng Y., Jiang J. and Wu C. 2018 Function and clinical significance of circRNAs in solid tumors. J. Hematol. Oncol. 11, 98.
Gong Y., Mao J., Wu D., Wang X., Li L., Zhu L. et al. 2018 Circ-ZEB1. 33 promotes the proliferation of human HCC by sponging miR-200a-3p and upregulating CDK6. Cancer Cell Int. 18, 1–9.
Gu Y., Ke G., Wang L., Zhou E., Zhu K. and Wei Y. 2017 Altered expression profile of circular RNAs in the serum of patients with diabetic retinopathy revealed by microarray. Ophthalmic Res. 58, 176–184.
Haddad G. and Lorenzen J. M. 2019 Biogenesis and function of circular RNAs in health and in disease. Front. Pharmacol. 10, 428.
Han C., Seebacher N. A., Hornicek F. J., Kan Q. and Duan Z. 2017 Regulation of microRNAs function by circular RNAs in human cancer. Oncotarget 8, 64622–64637.
Hsiao K.-Y., Lin Y.-C., Gupta S. K., Chang N., Yen L., Sun H. S. et al. 2017 Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 77, 2339–2350.
Hsu M. T. and Coca-Prados M. 1979 Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 280, 339–340.
Hu W., Han Q., Zhao L. and Wang L. 2019 Circular RNA circRNA_15698 aggravates the extracellular matrix of diabetic nephropathy mesangial cells via miR-185/TGF-β1. J. Cell. Physiol. 234, 1469–1476.
Hu Z. Q., Zhou S. L., Li J., Zhou Z. J., Wang P. C., Xin H. Y. et al. 2020 Circular RNA sequencing identifies CircASAP1 as a key regulator in hepatocellular carcinoma metastasis. Hepatology. 72, 906–922.
Huang M., He Y.-R., Liang L.-C., Huang Q. and Zhu Z.-Q. 2017 Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer. World J. Gastroenterol. 23, 6330.
Jeck W. R. and Sharpless N. E. 2014 Detecting and characterizing circular RNAs. Nat. Biotechnol. 32, 453–461.
Jiang G., Ma Y., An T., Pan Y., Mo F., Zhao D. et al. 2017 Relationships of circular RNA with diabetes and depression. Sci. Rep. 7, 1–8.
Jiang Q., Liu C., Li C., Xu S., Yao M., Ge H. et al. 2020 Circular RNA-ZNF532 regulates diabetes-induced retinal pericyte degeneration and vascular dysfunction. J. Clin. Invest. 130, 3833–3847.
Kolakofsky D. 1976 Isolation and characterization of Sendai virus DI-RNAs. Cell 8, 547–555.
Kong Z., Wan X., Lu Y., Zhang Y., Huang Y., Xu Y. et al. 2020 Circular RNA circFOXO3 promotes prostate cancer progression through sponging miR-29a-3p. J. Cell. Mol. Med. 24, 799–813.
Kono M., Dunn I. S., Durda P. J., Butera D., Rose L. B., Haggerty T. J. et al. 2006 Role of the mitogen-activated protein kinase signaling pathway in the regulation of human melanocytic antigen expression. Mol. Cancer Res. 4, 779–792.
Kun-Peng Z., Xiao-Long M. and Chun-Lin Z. 2018 Overexpressed circPVT1, a potential new circular RNA biomarker, contributes to doxorubicin and cisplatin resistance of osteosarcoma cells by regulating ABCB1. Int. J. Biol. Sci. 14, 321–330.
Li G., Yang H., Han K., Zhu D., Lun P. and Zhao Y. 2018 A novel circular RNA, hsa_circ_0046701, promotes carcinogenesis by increasing the expression of miR-142-3p target ITGB8 in glioma. Biochem. Biophy. Res. Commun. 498, 254–261.
Li K., Wan C.-L. and Guo Y. 2020 Circular RNA circMTO1 suppresses RCC cancer cell progression via miR9/LMX1A Axis. Technol. Cancer Res. Treat. 19, 1533033820914286.
Li Y., Zheng F., Xiao X., Xie F., Tao D., Huang C. et al. 2017 Circ HIPK 3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 18, 1646–1659.
Li Z., Hu Y., Zeng Q., Wang H., Yan J., Li H. et al. 2019 Circular RNA MYLK promotes hepatocellular carcinoma progression by increasing Rab23 expression by sponging miR-362-3p. Cancer Cell Int. 19, 1–11.
Liu T., Liu S., Xu Y., Shu R., Wang F., Chen C. et al. 2018a Circular RNA-ZFR inhibited cell proliferation and promoted apoptosis in gastric cancer by sponging miR-130a/miR-107 and modulating PTEN. Cancer Res. Treat. 50, 1396.
Liu Y., Dong Y., Zhao L., Su L. and Luo J. 2018b Circular RNA-MTO1 suppresses breast cancer cell viability and reverses monastrol resistance through regulating the TRAF4/Eg5 axis. Int. J. Oncol. 53, 1752–1762.
Ma Z., Shuai Y., Gao X., Wen X. and Ji J. 2020 Circular RNAs in the tumour microenvironment. Mol. Cancer 19, 8.
Meng J., Chen S., Han J.-X., Qian B., Wang X.-R., Zhong W.-L. et al. 2018 Twist1 regulates vimentin through Cul2 circular RNA to promote EMT in hepatocellular carcinoma. Cancer Res. 78, 4150–4162.
Nicolet B. P., Engels S., Aglialoro F., van den Akker E., von Lindern M. and Wolkers M. C. 2018 Circular RNA expression in human hematopoietic cells is widespread and cell-type specific. Nucleic Acids Res. 46, 8168–8180.
Pan Y., Lou J., Wang H., An N., Chen H., Zhang Q. et al. 2018 CircBA9. 3 supports the survival of leukaemic cells by up-regulating c-ABL1 or BCR-ABL1 protein levels. Blood Cell. Mol. Dis. 73, 38–44.
Sanger H. L., Klotz G., Riesner D., Gross H. J. and Kleinschmidt A. K. 1976 Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA 73, 3852–3856.
Schwerk J. and Savan R. 2015 Translating the untranslated region. J. Immunol. 195, 2963–2971.
Sever R. and Brugge J. S. 2015 Signal transduction in cancer. Cold Spring Harb. Perspect. Med. 5, a006098.
Shan C., Zhang Y., Hao X., Gao J., Chen X. and Wang K. 2019 Biogenesis, functions and clinical significance of circRNAs in gastric cancer. Mol. Cancer 18, 136.
Shan K., Liu C., Liu B.-H., Chen X., Dong R., Liu X. et al. 2017 Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation 136, 1629–1642.
Shang Q., Yang Z., Jia R. and Ge S. 2019 The novel roles of circRNAs in human cancer. Mol. Cancer 18, 6.
Sheng J.-Q., Liu L., Wang M.-R. and Li P.-Y. 2018 Circular RNAs in digestive system cancer: potential biomarkers and therapeutic targets. Am. J. Cancer Res. 8, 1142.
Stoll L., Sobel J., Rodriguez-Trejo A., Guay C., Lee K., Venø M. T. et al. 2018 Circular RNAs as novel regulators of β-cell functions in normal and disease conditions. Mol. Metab. 9, 69–83.
Straten P. t. and Andersen M. H. 2010 The anti-apoptotic members of the Bcl-2 family are attractive tumor-associated antigens. Oncotarget 1, 239–245.
Su M., Xiao Y., Ma J., Tang Y., Tian B., Zhang Y. et al. 2019 Circular RNAs in Cancer: emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol. Cancer 18, 90.
Tang W., Ji M., He G., Yang L., Niu Z., Jian M. et al. 2017 Silencing CDR1as inhibits colorectal cancer progression through regulating microRNA-7. Onco. Targets Ther. 10, 2045.
Um H.-D. 2016 Bcl-2 family proteins as regulators of cancer cell invasion and metastasis: a review focusing on mitochondrial respiration and reactive oxygen species. Oncotarget 7, 5193–5203.
Vislovukh A., Vargas T. R., Polesskaya A. and Groisman I. 2014 Role of 3’-untranslated region translational control in cancer development, diagnostics and treatment. World J. Biol. Chem. 5, 40–57.
Wang J.-J., Liu C., Shan K., Liu B.-H., Li X.-M., Zhang S.-J. et al. 2018a Circular RNA-ZNF609 regulates retinal neurodegeneration by acting as miR-615 sponge. Theranostics 8, 3408.
Wang L., Tong X., Zhou Z., Wang S., Lei Z., Zhang T. et al. 2018b Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-β-induced epithelial-mesenchymal transition and metastasis by controlling TIF1γ in non-small cell lung cancer. Mol. Cancer 17, 140.
Wang M., Yu F., Wu W., Zhang Y., Chang W., Ponnusamy M. et al. 2017 Circular RNAs: a novel type of non-coding RNA and their potential implications in antiviral immunity. Int. J. Biol. Sci. 13, 1497–1506.
Witsch E., Sela M. and Yarden Y. 2010 Roles for growth factors in cancer progression. Physiology (Bethesda) 25, 85–101.
Wu J., Qi X., Liu L., Hu X., Liu J., Yang J. et al. 2019 Emerging epigenetic regulation of circular RNAs in human cancer. Mol. Therapy. Nucleic Acids 16, 589–596.
Wu Y., Ding Y., Tanaka Y. and Zhang W. 2014 Risk factors contributing to type 2 diabetes and recent advances in the treatment and prevention. J. Int. J. Med. Sci. 11, 1185–1200.
Wu Y., Zhang Y., Niu M., Shi Y., Liu H., Yang D. et al. 2018 Whole-transcriptome analysis of CD133+ CD144+ cancer stem cells derived from human laryngeal squamous cell carcinoma cells. Cell. Physiol. Biochem. 47, 1696–1710.
Xu H., Guo S., Li W. and Yu P. 2015 The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci. Rep. 5, 1–12.
Yan N., Xu H., Zhang J., Xu L., Zhang Y., Zhang L. et al. 2017 Circular RNA profile indicates circular RNA VRK1 is negatively related with breast cancer stem cells. Oncotarget 8, 95704.
Yan Q., He X., Kuang G. and Ou C. 2020 CircRNA cPWWP2A: an emerging player in diabetes mellitus. J. Cell Commun. Signal. 1-3.
Yang C., Yuan W., Yang X., Li P., Wang J., Han J. et al. 2018a Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21 PTEN expression. Mol. Cancer 17, 19–19.
Yang Q., Du W. W., Wu N., Yang W., Awan F. M., Fang L. et al. 2017 A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 24, 1609–1620.
Yang Y., Gao X., Zhang M., Yan S., Sun C., Xiao F. et al. 2018b Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J. Natl. Cancer Ins. 110, 304–315.
Yu J., Xu Q.-G., Wang Z.-G., Yang Y., Zhang L., Ma J.-Z. et al. 2018 Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J. Hepatol. 68, 1214–1227.
Zaiou M. 2020 circRNAs signature as potential diagnostic and prognostic biomarker for diabetes mellitus and related cardiovascular complications. Cells 9, 659.
Zeng K., Chen X., Xu M., Liu X., Hu X., Xu T. et al. 2018 CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 9, 1–15.
Zhang H., Wang G., Ding C., Liu P., Wang R., Ding W. et al. 2017a Increased circular RNA UBAP2 acts as a sponge of miR-143 to promote osteosarcoma progression. Oncotarget 8, 61687.
Zhang M., Huang N., Yang X., Luo J., Yan S., Xiao F. et al. 2018a A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene 37, 1805–1814.
Zhang S.-J., Chen X., Li C.-P., Li X.-M., Liu C., Liu B.-H. et al. 2017b Identification and characterization of circular RNAs as a new class of putative biomarkers in diabetes retinopathy. Invest. Ophthalmol. Visual Sci. 58, 6500–6509.
Zhang X. and l., Xu L. l. and Wang F. 2017c Hsa_circ_0020397 regulates colorectal cancer cell viability, apoptosis and invasion by promoting the expression of the miR-138 targets TERT and PD-L1. Cell Biol. Int. 41, 1056–1064.
Zhang Z., Yang T. and Xiao J. 2018c Circular RNAs: promising biomarkers for human diseases. EBioMedicine 34, 267–274.
Zhao M., Ma W. and Ma C. 2020 Circ_0067934 promotes non-small cell lung cancer development by regulating miR-1182/KLF8 axis and activating Wnt/β-catenin pathway. Biomed. Pharmacother. 129, 110461.
Zhao X., Cai Y. and Xu J. 2019 Circular RNAs: biogenesis, mechanism, and function in human cancers. Int. J. Mol. Sci. 20, 3926.
Zhao Z., Li X., Jian D., Hao P., Rao L. and Li M. 2017 Hsa_circ_0054633 in peripheral blood can be used as a diagnostic biomarker of pre-diabetes and type 2 diabetes mellitus. Acta Diabetol. 54, 237–245.
Zhong L., Wang Y., Cheng Y., Wang W., Lu B., Zhu L. et al. 2018 Circular RNA circC3P1 suppresses hepatocellular carcinoma growth and metastasis through miR-4641/PCK1 pathway. Biochem. Biophys. Res. Commun. 499, 1044–1049.
Acknowledgments
Authors are thankful to University of Science and Technology, Meghalaya, India for providing the necessary facilities to carry out the review work. We are also thankful to Negenome Bio Solutions Pvt Ltd, CSIR-IIIM Jammu for their support.
Author information
Authors and Affiliations
Corresponding author
Additional information
Corresponding editor: Upendra Nongthomba
Rights and permissions
About this article
Cite this article
Hatibaruah, A., Rahman, M., Agarwala, S. et al. Circular RNAs in cancer and diabetes. J Genet 100, 21 (2021). https://doi.org/10.1007/s12041-021-01268-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12041-021-01268-4