Abstract
A DNA region in the mitochondrial genome of the fungus Phycomyces blakesleeanus (Mucorales, Mucoromycota) was characterized in a population of wild-type strains. The region encodes a predicted protein similar to the reverse transcriptases encoded by mitochondrial retroplasmids of Neurospora species and other Sordariomycetes (Ascomycota), but is uncommon in other fungi. DNA sequences of this element, named mystique, are highly variable between the strains, having greater than 2.5% divergence, yet most of the nucleotide differences fall in codon positions that do not change the amino acid sequence. The high proportion of polymorphisms coupled to the rarity of nonsynonymous changes suggests that mystique is subject to counteracting forces of hypermutation and purifying selection. However, while evidence for negative selection may infer that the element provides a fitness benefit, some strains of P. blakesleeanus do not have the element and grow equivalently well as those strains with it. A mechanism to explain the variability between the mystique alleles is proposed, of error-prone replication through an RNA intermediate, reverse transcription and reintegration of the element into the mitochondrial genome.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mitochondrial genomes are subject to different forces of selection compared to nuclear genomes. For example, during the sexual cycle, the mitochondrial genes are commonly inherited from one parent, rather than a mix of recombination from both. Hence, the accumulation of deleterious mutations in mitochondria, through a Muller’s ratchet scenario, is balanced by the counteracting selective pressure of the requirement of mitochondria for the survival of the majority of eukaryotes. Mitochondria retain a core set of essential genes, such as transfer RNAs, ribosomal RNAs and encoding proteins for the adenosine triphosphate (ATP) synthase complex, cytochrome oxidase and NADH dehydrogenase. Other genes are more variable in distribution among species, and their presence in mitochondrial genomes raise interesting questions about their origins and what maintains them in mitochondria.
Phycomyces blakesleeanus is a filamentous fungus, in the order Mucorales of the Mucoromycota phylum, which is best known for the production of large asexual sporangiophores that sense different environmental stimuli (Cerdá-Olmedo 2001). Stable genetic modification by transformation had not been possible (Obraztsova et al. 2004), thus classical Mendelian analysis of segregating phenotypes and genotypes in progeny from crosses is one of the more powerful techniques available for gene discovery in this species. Strains with mutant phenotypes were isolated after chemical mutagenesis (e.g. Bergman et al. 1973), and subsequently characterized by genetic segregation analysis to identify the genes responsible (Alvarez et al. 1980; Orejas et al. 1987; Idnurm et al. 2006; Sanz et al. 2009; Larson and Idnurm 2010; Tagua et al. 2012; Chaudhary et al. 2013; Corrochano et al. 2016; Polaino et al. 2017).
In experiments investigating the genetic inheritance patterns of P. blakesleeanus, for the purpose of mapping genes, a region of its mitochondrial genome that had polymorphisms between two strains was identified, and the region was developed into a polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) molecular marker to track the segregation of the mitochondrial genotype during the sexual cycle. In crosses, the nuclear markers segregate in a Mendelian manner, while the mitochondrial marker is inherited only from the (\(+\)) mating type parent (Chaudhary et al. 2013; Shakya and Idnurm 2014). The mitochondrial marker was particularly useful because it exhibited polymorphisms between different wild-type strains and could be employed to study uniparental inheritance in crosses between multiple different parents (Shakya and Idnurm 2014). The finding from this research was that the uniparental inheritance of the mitochondrial gene is controlled by the genes that also control the sex of the strain; this has been shown for the first time in any eukaryote by being able to exclude the influence of cell size or gene composition in sex chromosomes or mating type loci.
As reported here, analysis of this region of the P. blakesleeanus mitochondrial genome reveals that it has unusual properties. It is most closely related to circular ‘retroplasmids’ found in Neurospora species. It is highly variable in DNA sequences between the strains, but most changes are synonymous and therefore do not change the amino acid sequence of the protein encoded.
Materials and methods
Strains and culturing
Details and origins of the wild-type P. blakesleeanus strains used are provided in table 1, and have been characterized previously for nuclear polymorphisms (Camino et al. 2015). Fifteen progenies from a cross of UBC21 (\(+\)) \(\times \) NRRL1555 (−) were previously reported (Chaudhary et al. 2013). The strains were routinely cultured on potato dextrose agar. Mycelia from strains were generated as cultures in the yeast extract (10 g/L), peptone (20 g/L) and dextrose (20 g/L) medium. For the passaging experiment, colonies were initiated from a single spore and were then sequentially subcultured by excising a growing edge of mycelium and plated it onto a minimal medium supplemented with the yeast extract as described previously (Heisenberg and Cerdá-Olmedo 1968).
DNA extractions, PCR and sequencing
Genomic DNA was isolated from pulverized mycelium using a standard extraction buffer with precipitation in an equal volume of 100% isopropanol (Pitkin et al. 1996).
One location of orf511/mystique within the mitochondrial genome is defined in strain NRRL1555, the most common laboratory strain which was selected for the genome sequencing project (Corrochano et al. 2016). Inverse PCR was used to define the site of the element within the mitochondrial genome in another strain, UBC21. Genomic DNA of UBC21 was cut with an XbaI restriction enzyme, and recircularized with a T4 DNA ligase (New England Biolabs, Ipswich, USA). The primers used for the inverse PCR were ALID2185 (5\(^\prime \)-CATTCGAGGGTCATCTCC-3\(^\prime )\) and ALID2186 (5\(^\prime \)-ACGAACAGAAGCGAGAGC-3\(^\prime )\), and the amplicon was sequenced.
One region of the mitochondrial genome of P. blakesleeanus is highly variable between the strains. (a) Map of the 62,082 bp P. blakesleeanus mitochondrial genome of strain NRRL1555, redrawn from the analysis by B. F. Lang as presented in Corrochano et al. (2016). The colour scheme: genes encoding mitochondrial ribosomal RNAs in green, tRNAs in light blue, standard mitochondrial genes in dark blue and nonstandard genes in orange. The SNPs between this genome and that of strain UBC21 are indicated by red lines; two lines converge at positions 40,322 and 40,323 (Corrochano et al. 2016). (b) An enlarged view of the highly polymorphic region of the mitochondrial genome centred around the orf511/mystique gene, in which 42 nucleotide differences (red lines) are present between the strains NRRL1555 and UBC21 in less than a 1.6 kb region.
Mystique was amplified from the strains with either primers ALID2182 (5\(^\prime \)-GGTCGACAGCCAACTCCTAG-3\(^\prime )\) and ALID2183 (5\(^\prime \)-TTAGGCCTGAGGAATCATCC-3\(^\prime )\), or ALID2198 (5\(^\prime \)-CAGTTTAACTCTGTATGAGC-3\(^\prime )\) and ALID2211 (5\(^\prime \)-ATGGTGTGGTGGTCAACATG-3\(^\prime )\), depending on its location within the mitochondrial genome (table 1). For sequencing, additional primers (not listed) were designed internal to the amplicons to ensure the coverage of both DNA strands.
The PCR of a polymorphism in the rnl gene, which has a presence or absence of orf246 within it, used primers ALID2194 (5\(^\prime \)-GGCTTAACTGTAAGACTGAC-3\(^\prime )\) and ALID2195 (5\(^\prime \)-ACCAGAGGTTCATACCAC-3\(^\prime )\). The region was amplified from strains NRRL1555 and UBC21, and 15 progenies from a cross between the two strains that were previously characterized to have independent segregation using 134 nuclear markers (Chaudhary et al. 2013).
Southern and northern blotting
For Southern blotting, genomic DNA of eight strains was digested with XbaI and resolved on a 0.8% agarose 1\(\times \) TAE gel. For northern blotting, the mycelia of strain NRRL1555 were cultured for 2 days on a yeast extract peptone dextrose agar in darkness, and one set was exposed to white fluorescent light for 1 h. Total RNA was isolated using the Trizol reagent (Sigma-Aldrich, St Louis, USA) following the manufacturer’s suggestions. The RNA was resolved on a denaturing 1.4% agarose gel.
Nucleic acids were blotted from the agarose gels onto Zeta-Probe membranes (Bio-Rad, Hercules, USA). A mystique probe for hybridization was amplified with primers ALID2184 (5\(^\prime \)-AGCTCTCGCTTCTGTTCG-3\(^\prime )\) and ALID2187 (5\(^\prime \)-GGAGATGACCCTCGAATG-3\(^\prime )\). As a control probe, the homologue of the atp6 gene, which is located in the mitochondrial genome, was amplified with primers ALID2281 (5\(^\prime \)-ATGTCAACTTTAGCGACAAC-3\(^\prime )\) and ALID2282 (5\(^\prime \)-CTAGTGAAGATCAATAGCATC-3\(^\prime )\). The DNA fragments were labelled with [\(\upalpha \)-\(^{32}\)P]-dCTP using a RediPrime kit (Amersham, Pittsburgh, USA), and the mystique probe was hybridized first to the membranes. After exposure to X-ray films, the membranes were stripped, and hybridized a second time with the fragment of the atp6 homologue.
Phylogenetic and evolutionary analyses
The sequences of mystique from the strains were aligned with ClustalW (gap open penalty 15, gap extension penalty 6.66 and IUB weight matrix), and the alignment was imported into MEGA7 (Kumar et al. 2016) for subsequent analysis. A conserved core of 1487 bp present in all homologues was used to generate a maximum-likelihood phylogeny using the Tamura-Nei model, after selection of the most suitable evolutionary rates (gamma distributed with invariant sites; \(+G+I\) parameters), with assessment for branch supports using 100 bootstraps. Comparisons of synonymous and nonsynonymous substitutions were made using SNAP software (Korber 2000). The Z-test was used to test for evidence of selection against a null hypothesis of neutral selection in MEGA7 (Nei and Gojobori 1986; Kumar et al. 2016).
Results
A highly polymorphic region, named ‘mystique’, is in the mitochondrial genome of P. blakesleeanus
A comparison of the sequence of the mitochondrial genome of the standard laboratory wild-type strain, NRRL1555, with that of a second wild-type strain, UBC21, which was sequenced using Illumina technology during the genome sequencing project (Corrochano et al. 2016), revealed a region of this organelle’s genome as a ‘hot spot’ for polymorphisms. Aligning the sequencing reads from strain UBC21 onto the strain NRRL1555 mitochondrial genome sequence identified 46 single-nucleotide polymorphisms (SNPs) (figure 1a). Of these, four were intergenic and 42 were found within a region less than 1.6 kb (figure 1b). This region contains a putative open reading frame (ORF) of 511 amino acids (hence annotated as orf511; here named mystique after the Marvel Comics character), and BLASTx analysis against the GenBank nr database reveals that it has similarities to proteins that are reverse transcriptases encoded in mitochondrial plasmids in filamentous ascomycetes in the Sordariomycetes. The initial hypothesis was that orf511/mystique is a pseudogene in P. blakesleeanus, accumulating mutations. However, the predicted protein-coding sequences from the mystique allele in strains NRRL1555 and UBC21 both produced ORFs uninterrupted by the stop codons commonly found in pseudogenes, and, strikingly, there were few amino acid substitution differences between the two strains, in that 42 nucleotide polymorphisms corresponded to only five differences in amino acid residues.
To confirm the genome sequencing data of the two wild-type strains, the mystique alleles were amplified and resequenced. However, primer combinations used successfully to amplify mystique from strain NRRL1555 did not amplify the full region from strain UBC21, and it was therefore apparent that the position of the element in UBC21 was different from NRRL1555. Inverse PCR was therefore used to amplify the ends of the element and its surrounding regions from strain UBC21. This revealed a second position for the element in the mitochondrial genome of P. blakesleeanus, between the cob and orf311 genes. With this information, new primers were designed to be able to amplify the gene when they are in this position of the genome.
The full-length gene was amplified by PCR from 20 wild strains (table 1). The PCR amplification step indicated that the strains had the element in either of the two positions or, as in two strains, appeared to be absent.
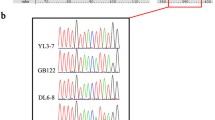
Three examples of DNA sequences that illustrate the high level of variation in mystique alleles. (a) Alignment of part of the DNA sequences of mystique from nine strains, with the predicted amino acid sequence below. *Conserved nucleotides. Of the 16 polymorphisms, only one changes an amino acid (aag to atg lysine-methionine in MI12-1a). (b) Sequencing chromatograms showing an example of a position in mystique in which all four possible nucleotides are found (*), all being synonymous to code for arginine. (c) Heterozygosity in strain NRRL2566; either an a or g at * is found, which does not alter the predicted lysine residue. The chromatograms include all the nucleotides, or omit either the a or g peaks.
The sequence of amplicons and alignment of this information of each mystique allele according to the codon positions revealed that many of the changes affect the third codon position, and therefore not the protein-coding sequence. Figure 2 illustrates this on three scales. In figure 2a, a section of 108 nucleotides is aligned from 20 strains (identical sequences are excluded), showing a region with 16 polymorphisms of which only one caused a nonsynonymous change. In addition to comparisons between the strains having either of two nucleotides at a position, there were cases where all four possible nucleotides were present in four strains (figure 2b). The genetic segregation system of P. blakesleeanus is such that during the sexual cycle the genotypic diversity that can accumulate within the coenocytic hypha is reduced to effectively a single haploid recombinant progeny that carries the mitochondrial genotype from the (\(+\)) parent (Burgeff 1928; Chaudhary et al. 2013; Shakya and Idnurm 2014). Sex is likely common in nature, as zygospores can be found in nature and strains of both mating types isolated from the environmental samples (Camino et al. 2015; Lee and Idnurm 2017). With this genetic system in place, heterozygosity should be rare in strains. Thus, a third potential piece of evidence for the rapid change of mystique is the presence of heterozygosity in one strain, NRRL2566, in which either an a or g is present at the same position (figure 2c).
Alignment of the N-terminal end of the predicted amino acid sequences of mystique from strain KACC46095 (P. blake) and the reverse transcriptase encoded on the Varkud plasmid from N. intermedia (GenBank accession no. NP_058435). The positions of seven conserved blocks found in reverse transcriptases are indicated (Antal et al. 2002).
The predicted Mystique protein has similarities to reverse transcriptases encoded on mitochondrial plasmids found in a small number of other fungi
The predicted amino acid sequences encoded by mystique alleles were compared with the GenBank and Joint Genome Institute MycoCosm databases to look for potential homologues. No homologues were found in any other species in the order Mucorales. The best matches were to proteins encoded by circular mitochondrial plasmids (e.g. Varkud and Mauriceville) found in Neurospora crassa and N. intermedia (Nargang et al. 1984; Nargang 1986; Akins et al. 1988; D’Souza et al. 2005), with an ‘RT_Bac_retron_II’ domain that is found in reverse transcriptases or retron elements. Alignment of the predicted protein, from strain KACC46095, with that of N. intermedia shows a high level of conservation for the seven regions identified within reverse transcriptase domains (figure 3).
Not all P. blakesleeanus strains have mystique in their genomes
Mystique could not be amplified from two strains, RSA1739 and TN13-1a. After using internal primers and those that should amplify across the regions where mystique could lie in their genomes, the reason for unsuccessful amplification seemed likely to be due to the element being absent in those two strains. To test this, the genomic DNA of these two strains and six others was resolved on agarose gels and the presence or absence of mystique assessed by Southern blot analysis. This experiment showed no hybridization for the two strains, while, as a control of DNA loading and transfer, the homologue of the atp6 gene found in the mitochondrial genome hybridized to their DNA on the blot (figure 4).
Southern blot analysis of wild-type strains of P. blakesleeanus indicates that mystique is not present in all strains. Genomic DNA of each strain was cut with XbaI restriction enzyme, resolved on an agarose gel, blotted to a membrane and the membrane hybridized with a fragment of mystique. The membrane was stripped of the probe, and reprobed with a fragment of the mitochondrial atp6 homologue. The mystique element is absent in strains RSA1739 and TN13-1a.
Mystique is transcribed
An additional line of evidence that mystique encodes a functional gene comes from northern blot experiments. RNA from the wild-type strain NRRL1555 grown under two light regimes was resolved on agarose gels, blotted and the presence of mystique was detected by hybridization with a radioactive probe. Thus, the element is transcribed under regular vegetative growth conditions (figure 5).
Mystique is transcribed. Northern blot analysis of P. blakesleeanus mycelium from strain NRRL1555 grown under constant darkness (D) or given a 1 h pulse of white light (L), from two replicates. 15 \(\mu \)g and 10 \(\mu \)g of total RNA were used from replicates 1 and 2, respectively. RNA was resolved on agarose gels, blotted to a membrane and the membrane hybridized with a fragment of mystique. The membrane was stripped of the mystique probe and reprobed with a fragment of the atp6 homologue.
The reported uniparental inheritance of the mitochondrial genome is not an artefact of using mystique as a genetic marker
Uniparental inheritance of the mitochondrial genome was previously reported for P. blakesleeanus based on using polymorphisms within mystique as the marker for the mitochondrial genotype (Shakya and Idnurm 2014). Given the unusual properties of these elements, to ensure this pattern of segregation was not an artefact due to mystique, an independent polymorphic region in the mitochondrial genome was examined. In this case, a difference in the rnl gene was identified in the genome sequences of strains NRRL1555 and UBC21 where the orf246 gene is present in NRRL1555 and absent in UBC21. The region was amplified from two parents and 15 progenies of a cross between them. The progenies exhibit the independent segregation of nuclear markers, with the 15 analysed being a subset of those used to generate a genetic map of P. blakesleeanus based on PCR-RFLP markers (Chaudhary et al. 2013). In contrast to the independent assortment of the nuclear markers, all 15 progenies had the mitochondrial allele from the (\(+\)) parent (figure 6). This observation provides independent confirmation of uniparental inheritance of the mitochondrial genome in P. blakesleeanus.
Demonstration of the inheritance of the mitochondrial genome in P. blakesleeanus is uniparental, from the (\(+\)) parent, using a different marker than mystique. A region of the mitochondrial genome, the rnl gene for the large subunit of the mitochondrial ribosome which has a presence/absence polymorphism of another gene within it, was amplified from two parents NRRL1555 (−) and UBC21 (\(+\)) and 15 progenies from a cross between the two. L\(=\)1 kb ladder (Invitrogen). All progenies inherit the mitochondria from the (\(+\)) parent, while nuclear markers analysed from the same progeny segregate independently (Chaudhary et al. 2013).
Phylogenetic analyses of mystique
DNA sequences were aligned and imported into the MEGA7 program to analyse the evolution of the mystique gene. A visual inspection of the alignment, which was confirmed by a neighbour-joining analysis (data not shown) revealed three different lineages for the gene. One lineage was represented by a sole strain, KACC46095, and two other lineages, which correlate with the position of the element within the mitochondrial genome as either between trnF-nad4 L or cob-orf311. To produce a phylogeny without the bias introduced by the two different locations, the alignment was truncated to include only the interval found in all strains, and the sequence from KACC46095 was excluded. This yielded an alignment of 1487 nucleotide positions. The phylogeny shows that the gene history matches the location within the mitochondrial genome (figure 7).
Relationships between the mystique alleles in different P. blakesleeanus isolates. A total of 1487 nucleotides common to all homologues was aligned, and the homologues were compared by maximum-likelihood. Branch numbers indicate bootstrap support from 100 replicates. The position of mystique in the mitochondrial genome is provided on the right-hand side.
The proportion of nonsynonymous changes relative to all polymorphisms indicated that mystique was under unusually high levels of purifying selection (table 1 in electronic supplementary material at http://www.ias.ac.in/jgenet/). A Z-test was used to determine if the skew of synonymous versus nonsynonymous substitutions was statistically different. For this purpose, the common conserved region of the gene of 1487 bp was used. With the exception of strains with identical sequences and the comparisons between UBC24 and NRRL1555, RSA531 and RSA577, and NRRL2566 and NRRL1554, the rest were all statistically different from neutrality (table 1 in electronic supplementary material).
Presence or absence of the element within the mitochondrial genome does not correlate with strain senescence
In ascomycete fungi, mitochondrial plasmids have been implicated in the senescence of the organism, including the Neurospora plasmids most closely related to mystique (Akins et al. 1986). The number of times P. blakesleeanus can be passaged asexually before it senescences and dies is subjected to conflicting findings. Thornton did not find any signs of ageing in P. blakesleeanus, while Heisenberg and Cerdá-Olmedo could passage strains vegetatively only nine times (Heisenberg and Cerdá-Olmedo 1968; Thornton 1973). Based on the differences in strains used in these studies, one hypothesis that could explain the maintenance of mystique in the P. blakesleeanus population is if it promotes sexual reproduction, i.e. if sexual reproduction, which ensures transmission of one mitochondrial genotype, resets vegetative ageing.
The senescence property of P. blakesleeanus was investigated, including with the same strains used in those earlier studies, and the two strains identified, TN13-1a and RSA1739, that do not contain the mystique element. The expectation was that the strains without the element in the mitochondrial genome would be ‘immortal’. However, no link to senescence was found between the strains with or without the element. Considerable variation was observed between the strains, ranging from a stop in replication after six transfers for strains NRRL1555 and RSA1739 to 59 transfers for strain G5. The other strains tested had growth arrest after 18 (TN13-1a), 22 (NRRL1554) and 23 (UBC21) subcultures.
Discussion
A region in the mitochondrial genome of the Mucorales species P. blakesleeanus is highly variable between the strains at the DNA sequence level, yet it is nevertheless subject to high levels of purifying selection as illustrated by the low proportion of nonsynonymous compared with synonymous changes.
BLAST analysis against the GenBank database identified the closest matches to a gene on mitochondrial plasmids, termed retroplasmids, in N. intermedia and N. crassa. These elements were characterized from the 1980s onwards with the discovery of mitochondrial plasmids in this genus (Collins et al. 1981; Galligan and Kennell 2007). Analysis of the DNA sequences of the plasmids revealed the presence of a single ORF within them that encodes a reverse transcriptase enzyme (Nargang et al. 1984; Nargang 1986; Akins et al. 1988; D’Souza et al. 2005). Subsequent research characterized the replication process of these elements (for a review, see Galligan and Kennell 2007). The role for the plasmids in fungal biology is still unknown even after three decades although the plasmids are associated with organism senescence when they integrate into the mitochondrial genome (Akins et al. 1986; Fox and Kennell 2001).
Two pieces of evidence suggest the potential for rapid change of the mystique element of P. blakesleeanus. One is the presence of two variants in strain NRRL2566 (figure 2c). The second is the presence of three different sequences for strain NRRL1555 or strains derived from it. For example, the Illumina data of phototropism mutants (Corrochano et al. 2016) reveals a difference between the sequenced strain NRRL1555, in which those mutants were isolated, and these. The NRRL1555 strain used here (obtained from the University of Salamanca) has three differences compared with the sequenced strain (from the University of Seville). A third potential example is the isolation of strains from the same location two months apart. RSA531 and RSA557 were isolated from Evey Canyon in the San Gabriel mountains of California on the 5th March 1957 and 30th April 1957, respectively (Ootaki and Miyazaki 1993), and likely originate from the same population yet their copies of mystique differ in sequence.
Counter to the hypothesis that mystique mutates at a high rate is the strains that have the identical sequence of this element. This is represented by the same sequence seen in strains represented by TX11-1a and TX11-1b, G5 and UBC21, and NRRL1554, NRRL2565 and K1. For the two strains from Texas, they were isolated from the same sample, although are of different sex. For the other combinations, these are not clones—either in nature or as a laboratory mix up—as they have differences in their nuclear genomes (Camino et al. 2015). However, G5 and UBC21 could well be both from North America, and NRRL1554 could potentially be from Germany, as are NRRL2565 and K1.
An additional unknown factor for mystique is in the annotation of the encoded protein. The use of the first methionine codon for the genome sequence strain NRRL1555 results in a predicted protein that appears to be truncated at the N-terminal end because the key amino acids within the reverse transcriptase domain are present before this atg. However, the mitochondrial genome has a number of other proteins without the atg start codon, and it is possible that other codons would initiate translation of the protein.
While the start codon of mystique is ambiguous, some strains (TX12-1b, RSA499 and VA12-1a) have a tga stop codon within mystique. The tga triplet can code for either a stop signal in most nuclear genomes or as a tryptophan in mitochondrial genomes. None of the genes predicted within the mitochondrial genome of P. blakesleeanus have this codon, either internally or as a potential stop codon (Corrochano et al. 2016). The polymorphisms in mystique suggest that in P. blakesleeanus tga codes for the tryptophan, as occurs in other organisms, and has been proposed for other Mucoromycotina mitochondrial genomes (Seif et al. 2005).
One hypothesis that can account for the unusual features of mystique is that it was a retroplasmid component that has integrated into the mitochondrial genome and it still undergoes an error-prone replication process that involves reverse transcription of its cDNA and retrohoming, that is reinsertion into the genome, at a certain frequency. To expand, the gene can be copied by the standard DNA replication of the mitochondrial genome. However, it is transcribed (figure 5) and presumably translated to encode a reverse transcriptase. That reverse transcriptase recognizes its own mRNA, produces a DNA copy, albeit the enzyme would have low fidelity to bring about the high proportion of mutations in the DNA. The DNA recombines back into that region of the genome to replace the original copy with one now of different sequence from the original. The reverse transcriptase enzyme must be prone to loss of function when amino acids are substituted.
Mystique is peculiar because (i) the gene is maintained in the genome while not being essential and (ii) its origin is a mystery given its absence from all other Mucorales species and the presence of homologues in only a small number of Sordariomycetes species. In addition to the strains described here, the element was successfully amplified from P. blakesleeanus strains NRRLA6737, NRRL1465, UBC1, UBC25, UBC31 and UBC33 (details of their originals are found in Camino et al. 2015), yet could not be amplified from DNA extracted from the strains of P. nitens, the second species within the Phycomyces genus. At present, horizontal gene transfer either into or out of P. blakesleeanus seems to be the most likely explanation. The transfer of mitochondrial plasmids by direct contact between two ascomycete species, from Ascobolus immersus into Podospora anserina, has been demonstrated (Kempken 1995). However, P. blakesleeanus is particularly difficult to transform with foreign DNA, in that it has never been accomplished under laboratory conditions to yield a strain with a stable integration of the DNA (Obraztsova et al. 2004). It is these types of genetic elements that appear to defy our understanding of evolution that make analysis of mitochondrial genes particularly interesting.
References
Akins R. A., Kelley R. L. and Lambowitz A. M. 1986 Mitochondrial plasmids of Neurospora: integration into mitochondrial DNA and evidence for reverse transcription in mitochondria. Cell 47, 505–516.
Akins R. A., Grant D. M., Stohl L. L., Bottorff D. A., Nargang F. E. and Lambowitz A. M. 1988 Nucleotide sequence of the Varkud mitochondrial plasmid of Neurospora and synthesis of a hybrid transcript with a \(5^{\prime }\) leader derived from mitochondrial RNA. J. Mol. Biol. 204, 1–25.
Alvarez M. I., Peláez M. I. and Eslava A. P. 1980 Recombination between ten markers in Phycomyces. Mol. Gen. Genet. 179, 447–452.
Antal Z., Manczinger L., Kredics L., Kevei F. and Nagy E. 2002 Complete DNA sequence and analysis of a mitochondrial plasmid in the mycoparasitic Trichoderma harzianum strain T95. Plasmid 47, 148–152.
Bergman K., Eslava A. P. and Cerdá-Olmedo E. 1973 Mutants of Phycomyces with abnormal phototropism. Mol. Gen. Genet. 123, 1–16.
Burgeff H. 1928. Variabilität, Vererbund and Mutation bei Phycomyces blakesleeanus Bgff. Z. Vererbungsl. 49, 26–94.
Camino L. P., Idnurm A. and Cerdá-Olmedo E. 2015 Diversity, ecology, and evolution in Phycomyces. Fungal Biol. 119, 1007–1021.
Cerdá-Olmedo E. 2001. Phycomyces and the biology of light and color. FEMS Microbiol. Rev. 25, 503–512.
Chaudhary S., Polaino S., Shakya V. P. S. and Idnurm A. 2013. A new genetic map for the zygomycete fungus Phycomyces blakesleeanus. PLoS One 8, e58931.
Collins R. A., Stohl L. L., Cole M. D. and Lambowitz A. M. 1981 Characterization of a novel plasmid DNA found in mitochondria of N. crassa. Cell 24, 443–452.
Corrochano L. M., Kuo A., Marcet-Houben M., Polaino S., Salamov A., Villalobos-Escobedo J. M. et al. 2016 Expansion of signal transduction pathways in fungi by extensive genome duplication. Curr. Biol. 26, 1577–1584.
D’Souza A. D., Sultana S. and Maheshwari R. 2005 Characterization and prevalence of a circular mitochondrial plasmid in senescence-prone isolates of Neurospora intermedia. Curr. Genet. 47, 182–193.
Fox A. N. and Kennell J. C. 2001. Association between variant plasmid formation and senescence in retroplasmid-containing strains of Neurospora spp. Curr. Genet. 39, 92–100.
Galligan J. T. and Kennell J. C. 2007 Retroplasmids: linear and circular plasmids that replicate via reverse transcription. In Microbiology monographs (ed F. Meinhardt and R. Klassen), pp. 163–185. Springer-Verlag, Berlin Heidelberg.
Heisenberg M. and Cerdá-Olmedo E. 1968 Segregation of heterokaryons in the asexual cycle of Phycomyces. Mol. Gen. Genet. 102, 187–195.
Idnurm A., Rodríguez-Romero J., Corrochano L. M., Sanz C., Iturriaga E. A., Eslava A. P. and Heitman J. 2006 The Phycomyces madA gene encodes a blue-light photoreceptor for phototropism and other light responses. Proc. Natl. Acad. Sci. USA 103, 4546–4551.
Kempken F. 1995 Horizontal transfer of a mitochondrial plasmid. Mol. Gen. Genet. 248, 89–94.
Korber B. 2000 HIV Signature and sequence variation analysis. In Computational analysis of HIV molecular sequences (ed A. G. Rodrigo and G. H. Learn), pp. 55–72. Kluwer Academic Publishers, Dordrecht, Netherlands.
Kumar S., Stecher G. and Tamura K 2016 MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874.
Larson E. M. and Idnurm A. 2010 Two origins for the gene encoding \(\upalpha \)-isopropylmalate synthase in fungi. PLoS One 5, e11605.
Lee S. C. and Idnurm A. 2017. Fungal sex: the Mucoromycota. Microbiol. Spectrum 5, FUNK-0041-2017.
Nargang F. E. 1986 Conservation of a long open reading frame in two Neurospora mitochondrial plasmids. Mol. Biol. Evol. 3, 19–28.
Nargang F. E., Bell J. B., Stohl L. L. and Lambowitz A. M. 1984 The DNA sequence and genetic organization of a Neurospora mitochondrial plasmid suggest a relationship to introns and mobile elements. Cell 38, 441–453.
Nei M. and Gojobori T. 1986 Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3, 418–426.
Obraztsova I. N., Prados N., Holzmann K., Avalos J. and Cerdá-Olmedo E. 2004 Genetic damage following introduction of DNA in Phycomyces. Fungal Genet. Biol. 41, 168–180.
Ootaki T. and Miyazaki A. 1993 Genetic nomenclature and strain catalogue of Phycomyces. Tohoku University, Sendai, Japan.
Orejas M., Peláez M. I., Alvarez M. I. and Eslava A. P. 1987 A genetic map of Phycomyces blakesleeanus. Mol. Gen. Genet. 210, 69–76.
Pitkin J. W., Panaccione D. G. and Walton J. D. 1996 A putative cyclic peptide efflux pump encoded by the TOXA gene of the plant-pathogenic fungus Cochliobolus carbonum. Microbiology 142, 1557–1565.
Polaino S., Villalobos-Escobedo V. M., Shakya V. P. S., Miralles-Durán A., Chaudhary S., Sanz C., Shahriari M., Luque E. M., Eslava A. P., Corrochano L. M., Herrera-Estrella A. and Idnurm A. 2017 A Ras GTPase associated protein is involved in the phototropic and circadian photobiology responses in fungi. Sci. Rep. 7, 44790.
Sanz C., Rodríguez-Romero J., Idnurm A., Christie J. M., Heitman J., Corrochano L. M. and Eslava A. P. 2009 Phycomyces MADB interacts with MADA to form the primary photoreceptor complex for fungal phototropism. Proc. Natl. Acad. Sci. USA 106, 7095–7100.
Seif E., Leigh J., Liu Y., Roewer I., Forget L. and Lang B. F. 2005 Comparative mitochondrial genomics in zygomycetes: bacteria-like RNase P RNAs, mobile elements and a close source of the group I intron invasion in angiosperms. Nucleic Acids Res. 33, 734–744.
Shakya V. P. S. and Idnurm A. 2014. Sex determination directs uniparental mitochondrial inheritance in Phycomyces. Eukaryotic Cell 13, 186–189.
Tagua V. G., Medina H. R., Martín-Domínguez R., Eslava A. P., Corrochano L. M., Cerdá-Olmedo E. and Idnurm A. 2012 A gene for carotene cleavage required for pheromone synthesis and carotene regulation in the fungus Phycomyces. Fungal Genet. Biol. 49, 398–404.
Thornton R. M. 1973 New photoresponses of Phycomyces. Plant Physiol. 51, 570–576.
Acknowledgements
This work was initiated at the University of Missouri-Kansas City, and I thank the UMKC undergraduates in the LS 399 and LS 499 courses for their technical assistance in DNA amplification and sequencing. The research was supported by the University of Melbourne and Australian Research Council.
Author information
Authors and Affiliations
Corresponding author
Additional information
Corresponding editor: Qingpo Liu
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Idnurm, A. Mystique of Phycomyces blakesleeanus is a peculiar mitochondrial genetic element that is highly variable in DNA sequence while subjected to strong negative selection. J Genet 97, 1195–1204 (2018). https://doi.org/10.1007/s12041-018-1014-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12041-018-1014-9