Abstract
In the southern Andes mountains (27–\(39{^{\circ }}\hbox {S}\)) Azorella madreporica and Laretia acaulis, two Apiaceae cushion plant species commonly known as yaretas, conform a well-established altitudinal vegetation belt along the lower Andean zone. These species have been considered as fundamental components of several ecological dynamics within their communities; however, high-mountain ecosystems are increasingly threatened worldwide by natural and anthropogenic pressures and the southern Andes are not the exception. Recognizing that genetic information is crucial for the success of any conservation or restoration initiative in wild populations, we developed and cross-amplified 28 specifically designed microsatellite markers (14 in A. madreporica and 14 in L. acaulis), and also tested the cross amplification of 25 markers from the related species Azorella selago. In a region which is particularly vulnerable to global change trends, this new polymorphic microsatellite loci will be useful in the study of the genetic diversity of these high-mountain cushion plants, which are pivotal in the structuring of their native ecosystems.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Almost all ecological restoration programmes imply the introduction of individuals into disturbed sites. However, individual candidates for repopulation often come from diverse sources, and the success of such programmes, therefore, depends strongly on the knowledge of key natural processes, which can be inferred from genetic individual-based information (Houde et al. 2015). Regarding this, some fundamental aspects of natural populations linked to restoration success can be assessed using a population genetics approach. These include, but are not limited to, endogamy levels, individual relatedness or adaptability among potential sources of translocated individuals (Houde et al. 2015). In this context, the use of specific genetic markers is a powerful tool, since it can help to identify important ecological and evolutionary processes operating in local wild populations (Hughes et al. 2008). For this reason, we developed the first set of microsatellite markers for Azorella madreporica and its close relative Laretia acaulis, two foundational Apiaceae cushion plants native to the southern Andes (Cavieres et al. 2000). This was made to improve current knowledge of important ecological features of this key species of high-Andean ecosystems, and assist in restoration and conservation programmes in a region particularly vulnerable to global change trends.
Among high-mountain flora, cushion plants are recognized as pivotal ecological elements in almost all mountainous formations (Kikvidze et al. 2015). They typically occur in harsh environments, and in recent decades they have been recognized as one of the main structural elements of several functional dynamics in their respective ecosystems (Reid and Lortie 2012). However, high-mountain ecosystems are increasingly threatened worldwide by natural (climate change) pressures (Elsen and Tingley 2015), and by anthropogenic (mining) activities, particularly in South America (Oyarzún and Oyarzún 2011).
In the southern Andean steppe ecoregion of South America (\(27{-}39{^{\circ }}\hbox {S}\)), cushion-plant species like A. madreporica and L. acaulis are among the main structural elements of the high-Andes flora (Cavieres et al. 2000). Commonly known as yaretas, they form a well-established altitudinal vegetation belt along the lower Andean zone (Cavieres et al. 2000); therefore, they represent key foundation species for restoration programmes in this ecoregion. However, till today no specific genetic tools are available to enhance the success of conservation-related strategies for both species in these fragile ecosystems, for which this crucial aspect is still not taken into account.
Individual-based genetic data have previously been used to elucidate a wide range of biological and ecological processes, such as ancient and contemporary migration routes, reproductive isolation and even selection in wild-species (meta) populations (Orsini et al. 2013; Mandák et al. 2016), all of which are key processes affecting the success of restoration programmes. Hence, to provide the genetic tools that are needed to gather this kind of ecological information, we developed and cross-amplified 28 specifically designed microsatellite markers (14 in A. madreporica and 14 in L. acaulis), and also tested the cross amplification of 25 markers from the related species Azorella selago. A set of new polymorphic microsatellite loci will be used in this study of the altitudinal and spatial distributions of genetic diversity in both species.
Methods
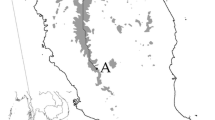
Fresh leaves from 24 individuals of each species were collected along a gradient of environmental conditions (3500–4200 m) in the high-Andean Río Estrecho basin in the Chilean Atacama region (\(70{^{\circ }}06^{\prime }\hbox {W}{-}29{^{\circ }}27^{\prime }\hbox {S}\)) (figure 1), and stored with silica-gel until dry. 100 mg of green leaves from each sample was ground to a fine powder using stainless-steel beads with a Mini BeadBeater-16 (BioSpec, USA). About 15 mg of the ground material was used for DNA extraction, applying the cetyl–trimethyl–ammonium bromide (CTAB) method (Doyle and Doyle 1987), following the protocol described in Cota-Sánchez et al. (2006). Genomic DNA was quantified with a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, USA), and its integrity was checked by electrophoresis in 1% agarose gel. Genomic libraries enriched with AC and AG motifs were constructed by Genetic Marker Services (www.geneticmarkerservices.com). Thirty-seven positive clones were isolated (A. madreporica = 21, L. acaulis = 16), and 14 primer pairs in each species were designed with Primer3 (http://biotools.umassmed.edu/bioapps/primer3_www.cgi) and optimized (Rozen and Skaletsky 2000). These specific primer pairs were first cross-tested in eight individuals from each species. In addition, we tested the cross-amplification of 25 primer pairs from the sub-Antarctic cushion A. selago, the closest taxonomic species to either species in this study, using a wide gradient of annealing temperatures (\(48{-}62{^{\circ }}\hbox {C}\)), following the protocol described by Cerfonteyn et al. (2011). Eight of these markers are published (Molecular Ecology Resources Primer Development Consortium 2010), while the remaining 17 (unpublished) were kindly provided by Dr. C. Born.
The successfully optimized microsatellite markers were then amplified by polymerase chain reaction (PCR) in 24 samples using a Veriti thermal cycler (Life Technologies, USA), with a fluorescently labelled forward primer (6FAM, HEX, NED or VIC; see table 1), in \(10\,\mu \hbox {L}\) reactions composed of 20 ng of genomic DNA, 1.5 mM \(\hbox {MgCl}_{2}\), \(1\,\mu \hbox {L}\) of AmpliTaq Gold reaction buffer (\(10\times \), Life Technologies), \(200\,\mu \hbox {M}\) each dNTP, \(0.2\,\mu \hbox {M}\) of each primer and 0.25 U of AmpliTaq Gold polymerase (Life Technologies). Thermal cycling conditions were: 10 min at \(94{^{\circ }}\hbox {C}\) for DNA denaturation and polymerase activation, followed by 30 cycles of 30 s at \(94{^{\circ }}\hbox {C}\), 30 s at specific annealing temperature (table 1) and 30 s at \(72{^{\circ }}\hbox {C}\), with a final elongation step of 15 min at \(72{^{\circ }}\hbox {C}\). Successful PCR products, as visualized in 1.2% agarose gel were sent to the sequencing unit of the Ecology Department at the Pontificia Universidad Católica de Chile for amplicon separation by capillary electrophoresis using an ABI Prism 3100 Genetic Analyzer (Applied Biosystems, USA). Allele size was determined using GeneMarker software (Softgenetics, USA), based on comparison with the migration of the GeneScan-500 Size Standard (Applied Biosystems, Chile). Polymorphic information content (PIC) was obtained for each locus in both species with the PIC function from the polysat R-package (Clark and Jasieniuk 2011). Observed and expected heterozygosities were calculated using the adegenet R-package (Jombart 2008), while departure from the Hardy–Weinberg equilibrium (HWE) and linkage disequilibrium (LD) between all loci pairs were tested in each species using GenePop v.4.2 (Raymond and Rousset 1995). The presence of null alleles at each locus was also evaluated with MICRO-CHECKER software (van Oosterhout et al. 2004).
Results
Among the 53 primer pairs tested in each species, only 11 yielded clear and reproducible amplification products, all of them were derived from the newly enriched libraries, predominantly from the A. madreporica library (see tables 1 and 2). For the 11 successfully optimized polymorphic microsatellite markers, eight microsatellite loci were polymorphic in A. madreporica and six in L. acaulis (table 1). Although some of the 25 primer pairs from A. selago also yielded positive amplification in A. madreporica (9) and L. acaulis (11), the referred markers could not be optimized since all of them consistently showed a multiband pattern (data not shown).
The average number of alleles per locus was 5.1 for A. madreporica, ranging from three (loci Azm2 and Azm12) to eight (locus Azm11) and 5.8 for L. acaulis, ranging from three (locus Azm4) to 10 (locus Azm6) (table 2). The levels of observed heterozygosity were relatively high and similar between the species, ranging between 0.166 and 0.772 in A. madreporica, and between 0.565 and 0.875 in L. acaulis. Similarly, PIC values were found between 0.334 and 0.748 and 0.494 and 0.799, respectively, and in both species the lower value corresponded to those loci with less alleles (table 2). Only the microsatellite marker Azm7 showed genotype frequencies that deviate significantly from Hardy–Weinberg proportions in A. madreporica (\(P=0.007\)). This specific marker was also the only one that showed significant evidence for the presence of null alleles in this species (\(P{<}0.05\)). However, it is important to note that genotype frequencies for Azm7 did not deviate significantly from the HWE in L. acaulis. No significant LD was found between any pair of loci in either species after sequential Bonferroni correction. One monomorphic locus (Azm14) is also reported in table 1 because it might be polymorphic in other populations, especially accounting for the fact that the surveyed individuals in this study belong to the northern distribution limit for both species.
Discussion
The polymorphic microsatellite loci reported in this study for A. madreporica and L. acaulis harboured high levels of genetic diversity, as well as a wide range of allelic richness, making them useful for individual-based genetic analyses. In addition, although it was impossible to successfully amplify the A. selago primer pairs in either of the studied species, cross-amplification between A. madreporica and L. acaulis was in fact successful. Thus, the use of these microsatellite markers in other taxa of this group remains a possibility. Nevertheless, despite we chose to work with dinucleotide microsatellite markers instead of trinucleotide markers because of their great variability within the plant individual genome (Scotti et al. 2002), greater efforts are required in the case of L. acaulis, for which the development of polymorphic primer pairs was found to be more complex.
Despite their wide variability, if compared with other studies of genetic diversity involving alpine plant species, the obtained \(H_{\mathrm{e}}\) values for all loci in A. madreporica and L. acaulis appear to be higher than most mean \(H_{\mathrm{e}}\) estimations previously reported (Stöcklin et al. 2009). Further, if the estimated \(H_{\mathrm{e}}\) values are compared specifically with those from other cushion plants (Mortimer et al. 2008; Cerfonteyn et al. 2011) they are still higher in average. This could be in part the result of the designed sampling along the complete altitudinal gradient of both species, in which strong ecological changes are expected, with potential influences over the distribution of individual genotypes (Ohsawa and Ide 2008). In a similar way, average PIC values in both A. madreporica and L. acaulis loci were found to be similar, or even higher, to those usually showed by most plant species (Varshney et al. 2005).
Climate change is driving an upward range displacement of plant communities in mountain ecosystems globally, and high-mountain plants are particularly affected (Elsen and Tingley 2015). Located above the upper altitudinal limit of trees, A. madreporica and L. acaulis are typically restricted to extreme elevations (Cavieres et al. 2000). These landscapes are both highly fragmented and of limited spatial extent, offering few opportunities for colonization (Elsen and Tingley 2015). Further, in addition to climate change, these ecosystems are also impacted in northern Chile by important anthropogenic threats linked to large-scale mining activities (Oyarzún and Oyarzún 2011). These activities usually cause intense, localized habitat disturbances, and a common compensatory measure in restoration and conservation programmes includes propagating and/or translocating individuals of the most vulnerable plant species (Batson et al. 2015).
Unfortunately for most wild species, the lack of basic biological and ecological knowledge has resulted in very low reintroduction success (Wortley et al. 2013). Nevertheless, since now the combination of ecological, genetic and spatial data permits analysis and monitoring of complex ecological processes (Hughes et al. 2008), the use of genetic data in restoration programmes could significantly improve the efficiency of these initiatives (Mijangos et al. 2015). In this context, the potential threats faced by high-mountain ecosystems under the current global trends provide a strong argument for genetic characterization and future monitoring of high-Andean plant species. Therefore, the new set of microsatellite markers developed in this study will particularly be useful to assess spatial genetic structure in both cushion-plant species, to assist restoration programmes in one of the most active mining regions of the world.
References
Batson W. G., Gordon I. J., Fletcher D. B. and Manning A. D. 2015 Review: translocation tactics: a framework to support the IUCN Guidelines for wildlife translocations and improve the quality of applied methods. J. Appl. Ecol. 52, 1598–1607.
Cavieres L. A., Peñaloza A. and Arroyo M. T. K. 2000 Altitudinal vegetation belts in the high-Andes of central Chile (\(33^{\circ } \text{ S }\)). Rev. Chil. Hist. Nat. 73, 331–344.
Cerfonteyn M. E., Le P. C. R., Van B. J. V. and Born C. 2011 Cryptic spatial aggregation of the cushion plant Azorella selago (Apiaceae) revealed by a multilocus molecular approach suggests frequent intraspecific facilitation under sub-Antarctic conditions. Am. J. Bot. 98, 909–914.
Clark L. and Jasieniuk M. 2011 Polysat: an R package for polyploid microsatellite analysis. Mol. Ecol. Res. 11, 562–566.
Cota-Sánchez J. H., Remarchuk K. and Ubayasena K. 2006 Ready-to-use DNA extracted with a CTAB method adapted for herbarium specimens and mucilaginous plant tissue. Plant. Mol. Biol. Rep. 24, 161–167.
Doyle J. and Doyle J. L. 1987 Genomic plant DNA preparation from fresh tissue-CTAB method. Phytochem. Bull. 19, 11–15.
Elsen P. R. and Tingley M. W. 2015 Global mountain topography and the fate of montane species under climate change. Nat. Clim. Change 5, 772–776.
Houde A. L. S., Garner S. R. and Neff B. D. 2015 Restoring species through reintroductions: strategies for source population selection. Restor. Ecol. 23, 746–753.
Hughes A. R., Inouye B. D., Johnson M. T., Underwood N. and Vellend M. 2008 Ecological consequences of genetic diversity. Ecol. Lett. 11, 609–623.
Jombart T. 2008 Adegenet: a R-package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403–1405.
Kikvidze Z., Brooker R. W., Butterfield B. J., Callaway R. M., Cavieres L. A., Cook B. J. et al. 2015 The effects of foundation species on community assembly: a global study on alpine cushion plant communities. Ecology 96, 2064–2069.
Mandák B., Havrdová A., Krak K., Hadincová V., Vít P., Zákravský P. and Douda J. 2016 Recent similarity in distribution ranges does not mean a similar postglacial history: a phylogeographical study of the boreal tree species Alnus incana based on microsatellite and chloroplast DNA variation. New Phytol. 210, 1395–1407.
Mijangos J. L., Pacioni C., Spencer P. and Craig M. D. 2015 Contribution of genetics to ecological restoration. Mol. Ecol. 24, 22–37.
Molecular Ecology Resources Primer Development Consortium 2010 Permanent genetic resources added to Molecular Ecology Resources Database 1 August 2009–30 September 2009. Mol. Ecol. Res. 10, 232–236.
Mortimer E., McGeoch M. A., Daniels S. R. and van Vuuren B. J. 2008 Growth form and population genetic structure of Azorella selago on sub-Antarctic Marion Island. Antarct. Sci. 20, 381–390.
Ohsawa T. and Ide Y. 2008 Global patterns of genetic variation in plant species along vertical and horizontal gradients on mountains. Glob. Ecol. Biogeogr. 17, 152–163.
Orsini L., Mergeay J., Vanoverbeke J. and Meester L. 2013 The role of selection in driving landscape genomic structure of the water flea Daphnia magna. Mol. Ecol. 22, 583–601.
Oyarzún J. and Oyarzún R. 2011 Sustainable development threats, inter-sector conflicts and environmental policy requirements in the arid, mining rich, northern Chile territory. Sustain. Dev. 19, 263–274.
Raymond M. and Rousset F. 1995 GENEPOP (version 1.2): population genetics software for exact tests and ecumenism. Heredity 86, 248–249.
Reid A. M. and Lortie C. J. 2012 Cushion plants are foundation species with positive effects extending to higher trophic levels. Ecosphere 3, art96.
Rozen S. and Skaletsky H. 2000 Primer3 on the www for general users and for biologist programmers. Methods Mol. Biol. 132, 365–386.
Scotti I., Magni F., Paglia G. and Morgante M. 2002 Trinucleotide microsatellites in Norway spruce (Picea abies): their features and the development of molecular markers. Theor. Appl. Genet. 106, 40–50.
Stöcklin J., Kuss P. and Pluess A. R. 2009 Genetic diversity, phenotypic variation and local adaptation in the alpine landscape: case studies with alpine plant species. Bot. Helv. 119, 125–133.
van Oosterhout C., Hutchinson W. F., Wills D. P. and Shipley P. 2004. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes. 4, 535–538.
Varshney R. K., Graner A. and Sorrells M. E. 2005 Genic microsatellite markers in plants: features and applications. Trends Biotechnol. 23, 48–55.
Wortley L., Hero J. M. and Howes M. 2013 Evaluating ecological restoration success: a review of the literature. Restor. Ecol. 21, 537–543.
Acknowledgements
The authors thank Rasme Hereme and Maria A. Montoya for their valuable laboratory assistance, Dr Celine Born for kindly sharing the A. selago microsatellite library with additional information for optimization and Craig Weideman for checking the language of the manuscript. This study was funded by Compañía Minera Nevada (project: NEVA0606c) and the Institute of Ecology and Biodiversity (IEB) through the projects: P05-002 (Iniciativa Científica Milenio) and PFB 23 (CONICYT, Chile). The Ph.D. scholarship to IA was also funded by Compañía Minera Nevada.
Author information
Authors and Affiliations
Corresponding author
Additional information
Corresponding editor: Manoj Prasad
Rights and permissions
About this article
Cite this article
Acuña-Rodríguez, I.S., Gouin, N., Cifuentes-Lisboa, L. et al. Isolation and cross-amplification of the first set of polymorphic microsatellite markers of two high-Andean cushion plants. J Genet 97 (Suppl 1), 95–100 (2018). https://doi.org/10.1007/s12041-018-0999-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12041-018-0999-4