Abstract
Sox7, Sox17 and Sox18 are the members of the Sry-related high-mobility group box family (SoxF) of transcription factors. SoxF factors regulate endothelial cell fate as well as development and differentiation of blood cells and lymphatic vessels. There is very less information about the functions of these genes in fish. We obtained the full-length cDNA sequence of SoxF genes including Sox7, Sox17 and Sox18 in Cyprinus carpio, where Sox7 and Sox18 had two copies. The construction of a phylogenetic tree showed that these genes were homologous to the genes in other species. Chromosome synteny analysis indicated that the gene order of Sox7 and Sox18 was highly conserved in fish. However, immense change in genomic sequences around Sox17 had taken place. Numerous putative transcription factor binding sites were identified in the \(5^{\prime }\) flanking regions of SoxF genes which may be involved in the regulation of the nervous system, vascular epidermal differentiation and embryonic development. The expression levels of SoxF genes were highest in gastrula, and was abundantly expressed in the adult brain. We investigated the expression levels of SoxF genes in five specific parts of the brain. The expression levels of Sox7 and Sox18 were highest in the mesencephalon, while the expression level of Sox17 was highest in the epencephalon. In carp, the expression patterns of SoxF genes indicated a potential function of these genes in neurogenesis and in vascular development. These results provide new information for further studies on the potential functions of SoxF genes in carp.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Sox genes are characterized by a DNA-binding Sry-related high mobility group (HMG) domain and they were first identified in mammals (Cui et al. 2011; Chang et al. 2017). Since the discovery of the Sox gene, a large number of Sox transcription factors have been identified in vertebrates and invertebrates (Sarkar and Hochedinger 2013; Watanabe et al. 2016). With the use of whole-genome sequencing and genomewide characterization, more than 40 members of the Sox family have been identified in mammal, birds, reptiles, amphibians and fish (She and Yang 2015; Wei et al. 2016). Over 20 Sox genes have been found in mice and humans (Schepers et al. 2002), 19 in medaka, and 27 in tilapia (Cnaani et al. 2007; Han et al. 2010).
A considerable number of evidence indicates that Sox genes participate in the regulation of a variety of developmental processes in animals. Sox transcription factors play a crucial role in neurogenesis, cardiogenesis, angiogenesis, chondrogenesis, in endoderm development, and in sex determination and differentiation (Kashimada and Koopman 2010; Jiang et al. 2012).
The family of Sox genes is subdivided into 11 groups (named from A to K) based on the sequences of both DNA and proteins (Kamachi and Kondoh 2013; Wei et al. 2016; Fu and Shi 2017). The SoxF subgroup transcription factors include three members (Sox7, Sox17 and Sox18) (Zhou et al. 2015). These three genes share some essential functions, including regulation of embryonic development, stem cell induction and early mesoderm induction (Abdelalim et al. 2014; Kinoshita et al. 2015; Banerjee and Ray 2017).
In recent years, transcription factors of the SoxF subgroup have been widely studied and their functions are characterized. These functions include the regulation of angiogenesis and regulation of endothelial cell fate (Morini and Dejana 2014; Kim et al. 2016). In mice, Sox7 is indispensable for primitive endoderm differentiation (Kinoshita et al. 2015). Sox7-enforced expression promotes the expansion of blood progenitors, impairs B lymphopoiesis and regulates cardiovascular development (Behrens et al. 2014; Cuvertino et al. 2016; Lilly et al. 2017). In addition, Sox7 promotes neuronal apoptosis by regulating \(\upbeta \)-catenin activity in mice (Wang et al. 2015).
Sox17 haploinsufficiency leads to mice female subfertility; it is a critical marker of the fate of human primordial germ cells (Hirate et al. 2016; Irie et al. 2016). Sox17-related pathways are activated in brain arteriovenous malformation (Hermanto et al. 2016).
Sox18 plays a major role in lymphangiogenesis, angiogenesis and cardiovascular development in humans and in mice (Duong et al. 2014; Bastaki et al. 2016). Sox18 hypomethylation and its interaction with other environmental and genetic factors causes neural tube defects (Rochtus et al. 2016).
It is thus clear that SoxF genes are closely linked to the functions of the cardiovascular system and nervous system. However, research on genes of the SoxF subgroup is scarce in teleost fish. Genes of the SoxF subgroup have not been characterized in the Yellow River carp.
The common carp, Cyprinus carpio, is one of the most important cyprinid species, accounting for 10% of the global freshwater aquaculture production (Peng et al. 2014). The Yellow River carp (C. carpio var.) is a popular aquaculture fish in China. The Yellow River carp has great economic value because of its nutrient content, rapid growth, and easy cultivability. After gonad differentiation, female carps significantly grow faster than males (Wohlfarth et al. 1975; Wang 2009). The nervous system is involved in the regulation of food intake, movement and reproduction (Dunn et al. 2016; Hwang et al. 2016; Zhang et al. 2016). Moreover, the central nervous system participates in the regulation of sex differentiation in fish (Lin et al. 2016). Thus, structural and functional analyses of carp SoxF genes are of great scientific value in aquaculture. In this study, we describe the identification and molecular characterization of genes of the carp SoxF subgroup and investigated the expression patterns of carp SoxF genes in adult fish and early embryo development. Our results help to understand the functions of the SoxF genes in the regulation of central nervous system and sex differentiation in carp.
Materials and methods
Materials
Adult carps were obtained from Henan Provincial Research Institute of Aquaculture. Artificially fertilized eggs were incubated at \(23\,\pm \,2\,^{\circ }\hbox {C}\) in hatching tanks with an open recirculation water system and continuous aeration. Embryos of five different stages (blastula, gastrula, neurula, tail-bud and hatching) were collected. The division of embryonic developmental stages was performed according to Lin and Chapman (Lin and Weng 1986; Chapman and George 2011). Adult tissues, including heart, liver, kidney, forebrain, hindbrain, gonad, foregut, hindgut, scale, fin, muscle, eye, spleen and gill were collected. Five parts of the fish brain (diencephalon, mesencephalon, macromyelon, epencephalon and telencephalon) were carefully separated. These biological materials were stored at \(-80\,^{\circ }\hbox {C}\) until isolation of RNA.
Extraction of RNA from tissues
For adult tissues, the biological materials from three individuals were pooled together. For RNA extraction at different developmental stages, biological materials from 5 to 10 embryos were pooled. Total RNA was extracted from adult tissues and from embryos of different developmental stages using Trizol reagent (Invitrogen, Carlsbad, USA), according to the manufacturer’s instructions. Total RNA was treated with DNaseI (Promega, Shanghai, China) to eliminate contaminating DNA. The quality of tissues RNA was estimated with a 28S:18S ratio of \(\sim \)2:1, OD260/280 ratio\(\approx \)1.9–2.2. RNA concentration was determined by spectrophotometric methods. cDNA was then synthesized using Prime Script Reverse Transcriptase (TaKaRa, Shiga, Japan) from 1 \(\mu \hbox {g}\) of total RNA.
Molecular cloning of genes of the \(\varvec{SoxF subgroup}\)
Based on the sequencing data of ovarian transcriptome of carp performed in our laboratory (unpublished data), three different cDNA sequences with high homology to SoxF genes of other species were identified by BLAST software (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Homology analysis of nucleotide sequence of the genes of the SoxF subgroup was performed by BLAST software. Homologous nucleotide and protein sequences were confirmed using the BLASTn and BLASTx search algorithm in NCBI (http://www.ncbi.nlm.gov/blast).
To validate the accuracy of SoxF genes sequences from the transcriptome sequencing data, specific primers were designed on the complete Sox7 (three primer pairs), Sox17 (two primer pairs), and Sox18 (three primer pairs) genes sequences (table 1). These primers covered all the putative open reading frame (ORF) and some untranslated regions (UTRs) of the SoxF genes. Real-time PCR (RT-PCR) was performed on a C1000 Touch apparatus (Bio-Rad, Hercules, USA), in a \(25\, \mu \hbox {L}\) reaction volume that contained 2\(\times \) PCR Master Mix (TaKaRa), \(1\, \mu \hbox {L}\) of each specific forward and reverse primers, and \(0.5\, \mu \hbox {L}\) diluted cDNA. Ovary’s cDNA was used as template. Cycling conditions were as follow: \(94^{\circ }\hbox {C}\) for 3 min, followed by 34 cycles of 30 s at \(94^{\circ }\hbox {C}\), 30 s at \(55^{\circ }\hbox {C}\), and 1 min at \(72^{\circ }\hbox {C}\), plus a final extension step of 10 min at \(72^{\circ }\hbox {C}\). The PCR products were analysed by electrophoresis on 1% agarose gels stained with SYBR Green (Invitrogen). The DNA fragments were purified using an OMEGA Gel Extraction kit (OMEGA, Dalian, China), ligated into vector PMD19-T (TaKaRa), and transformed into chemically E. coli Competent Cells \(\hbox {DH5}\alpha \) (TaKaRa, Dalian, China), according to the manufacturer’s instructions. Then the recombinant plasmids were sequenced.
Sequence analysis and genomic structure analysis
CLUSTALW software (http://www.genome.jp/tools/clustalw/) was used for multiple alignments of amino acid sequences based on which a phylogenetic tree was constructed using MEGA6 (http://www.megasoftware.net). The \(5^{\prime }\)-flanking sequences of Sox7, Sox17 and Sox18 were analysed to identify potential transcription factor binding sites using the MatInspector (http://www.genomatix.de/matinspector.html), an online program. Chromosome synteny was performed on the GENE module available in NCBI (https://www.ncbi.nlm.nih.gov/gene/).
Nucleotide sequences of (a) Sox7, (b) Sox17 and (c) Sox18 in C. carpio. The deduced amino acid sequence is shown underneath the CDS. The HMG Box domain is shaded in gray and the C-TAD domain is boxed. The start and stop codons are shaded in red. The DNA-binding sites are shaded in yellow. Primers designed for qRT-PCR experiments are in a red box. Nucleotides and amino acids are numbered at the right end of the lines.
Expression pattern analysis by quantitative real-time PCR (qRT-PCR)
The expression levels of SoxF genes in embryos of different developmental stages and in adult tissues were analysed by qRT-PCR. cDNA templates were generated by the method described in the previous section. Three technical replicates were carried out for each of the three biological replicates of every sample. 40S rRNA gene was used as a reference gene in expression profiling owing to its stable expression in different tissues and in various developmental stages (Zhang et al. 2016). A standard curve was constructed using serially diluted cDNA (100, 50, 20, 10, 5, 2 and 1%) (figure 1 in electronic supplementary material at http://www.ias.ac.in/jgenet/). The correlation coefficients (\(R^{2})\) were all \(>0.99\). All samples used for qRT-PCR experiments had the same concentration. The products were further amplified with 18S primers to exclude possible DNA contamination. The primers were designed on Primer Premier5 (PREMIER, Palo Alto, USA) and synthesized by Sangon Biotech (Shanghai, China). Specificity, efficiency, and linearity ranges were established for all primer pairs using DNA electrophoresis, melting curves and standard curve analyses. The primers of Sox7 and Sox17 span \(3^{\prime }\) UTR and the ORF to ensure mRNA specificity. The primers of Sox18 were designed using sequence within the ORF but not the conserved domains to prevent nonspecific amplification (figure 1; table 2).
qRT-PCR was performed using a Light Cycler Roche 96 apparatus (Roche Diagnostics, Mannheim, Germany), in a \(20\, \mu \hbox {L}\) reaction volume containing \(2\times \) Ultra SYBR Mixture (TaKaRa), \(0.2\, \mu \hbox {M}\) of each specific forward and reverse primers, and \(1\, \mu \hbox {L}\) diluted cDNA. Cycling conditions for amplification were as follows: 10 min at \(95^{\circ }\hbox {C}\), followed by 50 cycles of 15 s at \(95^{\circ }\hbox {C}\), and 1 min at \(60^{\circ }\hbox {C}\). Cycling conditions for melting curve analysis were as follows: 10 s at \(95^{\circ }\hbox {C}\), 60 s at \(65^{\circ }\hbox {C}\), and 1 s at \(97^{\circ }\hbox {C}\).
The data collected by Light Cycler Roche 96 software was analysed using SPSS 20.0 software. All data were expressed as the mean of RQ value (\(2^{-\Delta \Delta CT})\) (\(\Delta \hbox {CT} = \hbox {CT}\) value of the target gene minus the CT value of 40S rRNA, \(\Delta \Delta \hbox {CT} = \Delta \hbox {CT}\) of any sample minus calibrator sample) (Livak and Schmittgen 2011). One-way ANOVA followed by least-significant difference (LSD) test were performed for each organ and developmental stage to identify significant differences between samples. Statistically significant differences were considered if \(P< 0.05\).
Results
Sequence analysis of C. carpio SoxF (CcSoxF)genes
The full-length cDNA sequence of carp Sox7 was 2339 bp, including a 265 bp \(5^{\prime }\) UTR and a 880 bp \(3^{\prime }\) UTR and the ORF was 1194 bp. The predicted amino acid sequence was 397 residues long and contained a 72 amino acids SOX HMG box DNA-binding domain at positions 42–113, and a SOX C-terminal transactivation domain (215 amino acids long). The amino acid sequence of the DNA-binding site was RMNFMAKRANKGWR, consisting of amino acids at positions 45, 47, 48, 50, 51, 54, 55, 58, 62, 70, 75, 78, 81 and 100 (figure 1a).
The full-length cDNA sequence of Sox17 was 1653 bp contained an ORF of 999 bp, a \(5^{\prime }\) UTR of 134 bp, and a \(3^{\prime }\) UTR of 520 bp. The predicted amino acid sequence was 332 residues long, and contained a conserved HMG box DNA-binding domain of 72 amino acids at positions 62 to 133. Within the HMG box DNA-binding domain, the amino acid component of DNA-binding sites was same as in the Sox7 (figure 1b).
Multiple alignments of SoxF subgroup proteins in different species, Sox7, Sox17, Sox18. All the sequences of SoxF homologues were retrieved from NCBI. The HMG Box characteristic of Sox proteins are in red frame. The alignment was generated by DNAMAN. GenBank accession numbers of sequences are shown in supplementary table 1.
The full-length cDNA sequence of carp Sox18 was 2458 bp contained a \(5^{\prime }\) UTR of 243 bp, a \(3^{\prime }\) UTR of 895 bp, and an ORF of 1320 bp that predicted 439 amino acids. The HMG box was composed of 72 amino acids. The sequence of Sox18 DNA-binding site was RMNFMAKRANKGWR, consisting of amino acids at positions 96, 98, 99, 101, 102, 105, 106, 109, 113, 121, 126, 129, 132 and 151. The 225 amino acids of the SOX C-terminal transactivation domain were located at positions 205–429 (figure 1c).
Based on these results, the three sequences of C. carpio were submitted to GenBank: Sox7 (KY860088), Sox17 (KY860088) and Sox18 (KY860088).
Alignment and phylogenetic analysis
Using NCBI and CLUSTALW, BLASTp analysis showed that Sox7, Sox17 and Sox18 had conserved HMG boxes (figure 2). Moreover, the deduced amino acid sequences of Sox7 and Sox18 showed high homology with zebrafish Sox7 (96.4%) and Sox18 (95.8%), but low homology with human, mouse, chicken and monkey Sox7 (46.4–56.2%) and Sox18 (50.1–38.6% homology). The amino acid sequence of Sox17 showed high homology with Sinocyclocheilus rhinocerous Sox17a (88.2%) and zebrafish Sox17 (71.5%) (table 3).
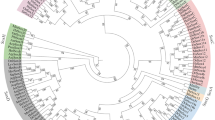
Phylogenetic tree of C. carpio Sox7, Sox17 and Sox18 in comparison with SoxF proteins in other representative vertebrates using predicted amino acid sequences. The phylogenetic tree was constructed by MEGA (ver. 6.0) using the neighbour-joining method with 1000 bootstrap replicates. The scale bar is 0.5. GenBank accession numbers of sequences are shown in supplementary table 1.
Genomic organization and chromosomal synteny. (a) Sox7, (b) Sox17 and (c) Sox18. Schematic presentation of genetic structure, exons (dark blue), and introns (light blue). C. carpio Sox7, Sox17 and Sox18, and their protein products. The \(5^{\prime }\) and \(3^{\prime }\) UTR (light green), and ORF (dark green) encoding the amino acid sequences are shown relative to their lengths in the cDNA sequences. Protein domains are shown relative to their lengths and positions in the amino acid sequences. N-ter, N-terminal domain (yellow); HMG box, high-mobility group box domain (pink); C-ter, C-terminal domain (purple). The following figures show the length and position of the sequence, chromosome syntenic relationships of C. carpio Sox7, Sox17 and Sox18 genes with teleostean orthologues. Conserved syntenies are shown for chromosomal segments containing Sox7, Sox17 and Sox18. Rectangles represent genes in chromosome scaffolds and arrows represent gene-coding directions. Chr, chromosome; Sca, scaffold.
A schematic diagram of putative regulatory motifs in the promoter of (a) Sox7, (b) Sox17 and (c) Sox18 in C. carpio. The scale is above and the full name of the potential transcription factor binding sites are provided at the bottom. The plus and minus signs indicate the transcription factors binding strand. Transcriptional start site (ATG) is designated as +1. Transcription factors names are shown in supplementary table 2.
Relative expression levels of C. carpio Sox7, Sox17 and Sox18 during embryonic development. Data were normalized using the reference gene 40S. Different letters indicate significant differences of the expression levels of SoxF genes at each stage which was analysed by one-way ANOVA followed by LSD test at a 0.05 probability level using SPSS software. The relative expression values are shown in table 3 in electronic supplementary material.
Relative expression levels of female C. carpio Sox7, Sox17 and Sox18 genes in different adult tissues. Data were normalized using the reference gene 40S. Different letters indicate significant differences of expression levels of SoxF genes in each organ which was analysed by one-way ANOVA followed by LSD test at a 0.05 probability level using SPSS software. The relative expression values are shown in table 3 in electronic supplementary material.
To evaluate the evolutionary relationships between carp SoxF genes and other species, a phylogenetic tree was constructed using the MEGA6 software. Sox7, Sox17 and Sox18 were split into three different branches. The sequences of amino acids between carp Sox7 and zebrafish Sox7 were highly identical; the two genes were grouped into one clade. Carp Sox18 was homologous to zebrafish Sox18, and carp Sox17 was homologous to S. rhinocerous Sox17a (figure 3) (table 1 in electronic supplementary material at http://www.ias.ac.in/jgent/).
Chromosome synteny and genomic analysis
The genomic DNA sequence of carp Sox7 (https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=7962) was interrupted by one intron between positions 497 and 1696 bp of the complete ORF, which was 1199 bp long (figure 4a). Two copies of Sox7 were found located at scaffolds 1136 and 77. The genomic DNA sequences and cDNA sequences of two Sox7 were identical. The genomic sequence of one Sox7 was flanked by the genes tdh3, pinx1, rp1l1 and blk. Another was flanked by the genes IGFBP5, ZBED4 and sirt7. Analysis of the data of whole-genome sequences and a cross-species comparison of chromosome locations showed that the genes tdh3, pinx1, sox7, rp1l1 and blk were always closely linked in fish. Except blk, these genes were also linked in mice and human.
The Sox17 genomic sequence had four introns, whose lengths were 773, 43, 134 and 72 bp (figure 4b). The Sox17 gene was detected on LG17. In medaka and in tilapia, Sox17 and Sox17a were adjoined. In Xenopus, the Sox17 and Sox17b were adjoined. There was only one Sox17 gene in carp and zebrafish, which was similar to the Sox17 gene in mammals. Among the flanking genes, only lypla1 was conserved among the analysed species.
The Sox18 genomic sequence contained one intron between 628 and 1837 bp, whose length was 1209 bp. Two copies of Sox18 were found located on scaffold 192. The two copies of Sox18 were adjacent, and flanked by tcea2 and xkr7. The cDNA sequences of the two Sox18 copies were identical (figure 4c). The arrangement of flanking genes was highly conserved in fish.
Analysis of binding sites for transcription factors
The 2000 bp \(5^{\prime }\)-flanking sequences upstream of Sox7, Sox17 and Sox18 were analysed with genomatix software (Genomatix, Ann Arbor, USA). Transcription factor binding sites with a matrix score higher than 0.9 were generally satisfied from numerous potential binding sites for transcription factors that were predicted within the \(5^{\prime }\) regulatory region. These transcription factor binding sites are drawn on the schematic diagram (see figure 5; table 2 in electronic supplementary material). Among the transcription factors binding sites, BSX and NEUROG are closely related to neurogenesis, Oct4, Nanog and FOXL1, regulate pluripotency and stem cell properties, MEF2/3 and GATA regulate the cardiovascular system, and finally MTBF and HNF6 are muscle-specific and liver-enriched. AP1, CEBPB and Sp1 are also identified.
The homology of C. carpio to D. rerio, at 2000-bp upstream 5\(^\prime \)-flanking sequences of Sox7, Sox17 and Sox18 were 43.81, 38.37 and 55.02%, respectively.
The homology of C. carpio to O. latipes at 2000 bp upstream 5\(^\prime \)-flanking sequences of Sox7, Sox17 and Sox18 were 32.96, 28.18 and 27.37%, respectively.
The 2000-bp upstream 5\(^\prime \)-flanking sequences of Sox7, Sox17 and Sox18 in D. rerio and O. latipes were also analysed with genomatix software. Between C. carpio andD. rerio, 19, 10 and 21 same transcription factor binding sites were found in Sox7, Sox17, Sox18 respectively. Compared with C. carpio, there were 23, 10 and 20 same transcription factor binding sites respectively in the Sox7, Sox17, Sox18 of O. latipes. Among the transcription factor binding sites of Sox7, BSX, GATA4, NF-Y, Oct6, Sox4 and Sox9 were only found in C. carpio. FOXL1 and Oct6 were unique in C. carpio Sox17. For Sox18, GATA2, Oct4, Sox2 and Sox7 were unique in C. carpio (table 2 in electronic supplementary material).
Expression pattern of the CcSoxF genes during embryonic development
We studied five different developmental stages of carp embryos including blastocyst, gastrula, nerve embryonic stage, tail-bud stage and hatching stage. Sox7 had the highest expression in gastrula followed by the tail-bud stage and hatching stage. Sox7 expression was extremely low in blastocysts and in the nerve embryonic stage. The expression of Sox17 was highest in gastrula, followed by the nerve embryonic stage. The expression of Sox17 was extremely low in blastocysts, in the tail-bud stage, and in the hatching stage. The expression of Sox18 was extremely low in all developmental stages (figure 6).
Expression pattern of the CcSoxF genes in adult tissues
Sox7 had the highest expression in brain, followed by spleen, heart, eye, muscle, fin, scales, gill, kidney, gut and liver. Sox7 expressed extremely at low levels in the gonads. The expression levels of Sox17 and Sox18 were relatively low compared to Sox7. The expression level of Sox17 was highest in the eye, followed by spleen, heart, brain, gill, fin, scale and muscle. The expression level of Sox17 was extremely low in gut, kidney, liver and gonad. The expression level of Sox18 was highest in the heart, followed by brain, spleen, eye, gut, kidney, muscle and gonad. The expression level of Sox18 was extremely low in fin, gill, liver and scale (figure 7).
Expression pattern of the CcSoxF genes in adult brain
Because of the high levels of expression found in the brain, we investigated the expression levels of Sox7, Sox17 and Sox18 in five parts of the brain. The expression level of Sox7 was highest in the mesencephalon, followed by the epencephalon, telencephalon, and diencephalon. The expression level of Sox7 was lowest in the macromyelon. The expression levels of Sox17 and Sox18 were relatively low compared to Sox7. Sox17 had the highest expression levels in the epencephalon, followed by diencephalon. Sox17 expression was extremely low in the macromyelon, mesencephalon and telencephalon. The expression level of Sox18 was highest in the mesencephalon followed by the macromyelon and extremely low in the diencephalon, epencephalon and telencephalon (figure 8).
Discussion
According to the previous reports, the interplay between Sox7 and RUNX1 regulates hemogenic endothelial fate (Lilly et al. 2016). Sox18 regulates the development of blood vessels and regulates lymphangiogenesis (Wang et al. 2015). Sox17 promotes endothelial cell differentiation and hematopoiesis (Goveia et al. 2014; Clarke et al. 2015). SoxF promotes neuronal apoptosis and affects the development of the neural tube. Sox17 has been involved in brain arteriovenous vessels (Duong et al. 2014; Bastaki et al. 2016). There has been few research concerning neurogenesis. In this study, we investigated the structure, chromosome synteny, transcription factor binding sites in the \(5^{\prime }\) flanking regions and expression pattern of SoxF in carp.
Relative expression levels of C. carpio Sox7, Sox17 and Sox18 genes in different parts of the adult brain. Data were normalized using the reference gene 40S. Different letters indicate significant differences of the expression levels of SoxF gene in each organ, which was analysed by one-way ANOVA followed by LSD test at a 0.05 probability level using SPSS software. The relative expression values are shown in table 3 in electronic supplementary material.
There were two copies of Sox7 in the carp genome of which one was located in scaffold 1136 and flanked by pinx1 and rp1l1. In fish and mammals, pinx1, Sox7 and rpl11 were neighbouring genes, but there was differences in gene arrangement between fish and mammals. Another copy was located in scaffold 77 flanked by zbed4 and sirt7. However, the gene arrangement on both sides of Sox7 was not conserved. We speculate that the duplication of this gene might be due to chromosome rearrangements or gene insertions.
We found that Sox17 was located in LG17. In medaka and tilapia, Sox17 and Sox17a were adjoined. In Xenopus, Sox17 is located next to Sox17b. In carp, zebrafish and mammals, only one copy of Sox17 was found. Sox17 and lypla1 were clustered together in carp, while in other species they were separated by mrpl15. The arrangement and direction of genes around Sox17 were different. Therefore, in fish, the rearrangement of genes around Sox17 had often taken place.
We detected two copies of Sox18 in carp scaffold 192 at different positions. Sox18 was flanked by tcea2 and xkr7. These three genes are clustered together in fish. In mammals, Sox18 is flanked by tcea2 and prpf6. There are apparent differences in gene order and direction in different species.
Chromosome rearrangement (translocations and inversions) frequently occurred at the time of genome replication in fish for SoxF genes. However, conservative gene arrangement was generally consistent.
Two copies of Sox7 were detected in C. carpio genome. One was located at scaffold 77, another at scaffold 1136. The mature mRNA sequences, transcription factor binding sites, UTR sequences, and expression patterns of the two copies were identical, but the sequence of their introns was different. The flanking genes of Sox7 in scaffold 1136 and in scaffold 77 were also different. It is noteworthy that the flanking genes of Sox7 in scaffold 1136 were conserved among different species but the flanking genes of Sox7 in scaffold 77 were not. Therefore, we hypothesized that the copy of Sox7 in scaffold 77 was caused by the insert. A similar example also appeared in Sox18. Based on the karyotype analysis of carp, Yu et al. (1987) suggested that carp were likely to be tetraploid. The tetraploid underwent a long process and gradually evolved into diploid. In this process, some segments of chromosomes and genes were inserted, or deleted, and their positions changed frequently. We suggest that it was possible that variation in gene arrangement around SoxF along chromosomes were caused in this process.
Analysis of the patterns of gene expression is the basis of the study of gene function. In the previous report, the SoxF transcription factors play a complex role in regulating cardiovascular and vascular development in mice and zebrafish, and in regulating Xenopus embryonic development (Lilly et al. 2017). SoxF promotes the proliferation and differentiation of lymphatic vessels (Francois et al. 2011). Sox7, Sox17 and Sox18 regulate vascular development in mouse retina (Zhou et al. 2015). SoxF genes are dispensable for primitive endoderm differentiation. In this study, SoxF genes were expressed in each developmental stage of carp. The expression level was highest in gastrula. SoxF genes were expressed in all adult tissues. The expression level was highest in eye, spleen and heart. These results indicated that the SoxF genes seemed to possess functions similar to those previously reported in other animals.
However, the expression level of SoxF genes was high in the brain. Therefore, a meticulous analysis of the expression of SoxF was performed in five regions of the carp brain. Sox7 and Sox18 exhibited the highest expression in the mesencephalon. Sox17 was highly expressed in the epencephalon. We hypothesized that SoxF genes might be associated with neurological development.
Sequences at \(5^{\prime }\) UTRs play an important role in the regulation of gene expression. We analysed the 2000-bp upstream \(5^{\prime }\) flanking sequences of each member of the SoxF subgroup using bioinformatics software. We identified several transcription factor binding sites related to neural development.
BRN4 induces differentiation of neural stem cells into neurons and promotes maturation of new neurons and maintains cells survival (Tan et al. 2010). BRN3 regulates the development of the central nervous system; it is an important factor that regulates the normal development and differentiation of retinal ganglion cells (Huang et al. 2011). HNF induces pluripotent stem cells to differentiate into hepatic cells (Yahoo et al. 2016). BSX plays an important role in the early stages of vertebrate neuronal determination and neurogenesis (Takahashi and Holland 2004). In addition, RUSH, FOXL1, MYT1 and GSH2 might interact with SoxF genes to regulate their functions in the nervous system. Oct4, TAL1 and Nanog might be attributed to neural stem/progenitor (Gabut et al. 2011).
Other transcription factor binding sites were found. The GATA family of transcription factors plays an indispensable role in ectoderm differentiation, in the hematopoietic system, and in the development of the heart, thymus and intestine (Jin and Liu 2009; Tarradas et al. 2016). MEF2 regulates cardiac development and the cardiovascular system (Desjardins and Naya 2016; Sacilotto et al. 2016). These results correspond to the previous study about the function of SoxF on vascular development (Morini and Dejana 2014; Kim et al. 2016). AP1, CEBPB and Sp1 are widely expressed in eukaryotes and play an important role in various cells processes.
The discovery of these transcription factor binding sites and the expression pattern analysis of SoxF genes were consensus. These results further verify that SoxF genes might participate in neurological development and are important for maintaining neurological functions.
In summary, we obtained the full-length cDNA sequence of SoxF genes including Sox7, Sox17 and Sox18 in carp. Both Sox7 and Sox18 have two copies. The construction of a phylogenetic tree showed that these genes were homologous to genes in other species. Chromosome synteny analysis indicated that the gene order of Sox7 and Sox18 was highly conserved in the fish. However, genomic sequences around Sox17 in fish was rearranged during evolution. Numerous putative transcription factor binding sites were identified in the \(5^{\prime }\) upstream flanking regions of SoxF genes, which may be involved in the regulation of the nervous system, vascular epidermal differentiation and embryonic development. The expression patterns of SoxF genes indicated a potential function of SoxF genes in neurogenesis and vascular development in carp. These results provide new information for further studies on the potential functions of SoxF genes in carp.
References
Abdelalim E. M., Emara M. M. and Kolatkar P. R. 2014 The SOX transcription factors as key players in pluripotent stem cells. Stem. Cells Dev. 23, 2687–2699.
Bastaki F., Mohamed M., Nair P., Saif F., Tawfiq N., Al-Ali M. T. et al. 2016 A novel SOX18 mutation uncovered in Jordanian patient with hypotrichosis-lymphedema-telangiectasia syndrome by Whole Exome Sequencing. Mol. Cell. Probes. 30, 18–21.
Banerjee A and Ray S. 2017 Structural insight, mutation and interactions in human Beta-catenin and SOX17 protein: a molecular-level outlook for organogenesis. Gene 610, 118–126.
Behrens A. N., Zierold C., Shi X., Ren Y., Koyano-Nakagawa N., Garry D. J. et al. 2014 Sox7 is regulated by ETV2 during cardiovascular development. Stem. Cells Dev. 23, 2004–2013.
Chang Y. K., Srivastava Y., Hu C., Joyce A., Yang X., Zuo Z. et al. 2017 Quantitative profiling of selective Sox/POU pairing on hundreds of sequences in parallel by Coop-seq. Nucleic Acids Res. 45, 832–845.
Chapman D. C. and George A. E. 2011 Developmental rate and behavior of early life stages of bighead carp and silver carp. U. S. Geological Survey of Scientific Investigations Report 62. SIR11-5076, pp. 11. Reston, Virginia.
Clarke R. L., Robitaille A. M., Moon R. T. and Keller G. 2015 A quantitative proteomic analysis of hemogenic endothelium reveals differential regulation of hematopoiesis by SOX17. Stem. Cell Rep. 5, 291–304.
Cnaani A., Lee B. Y., Ozouf-Costaz C., Bonillo C., Baroiller J. F., Cotta H. et al. 2007 Mapping of Sox2 and Sox14 in tilapia (Oreochromis spp.). Sex. Dev. 1, 207–210.
Cui J., Shen X., Zhao H. and Nagahama Y. 2011 Genome-wide analysis of Sox genes in Medaka (Oryzias latipes) and their expression pattern in embryonic development. Cytogenetic. Genome Res. 134, 283–294.
Cuvertino S., Lacaud G. and Kouskoff V. 2016 SOX7-enforced expression promotes the expansion of adult blood progenitors and blocks B-cell development. Open Biol. 6, 160070.
Desjardins C. A. and Naya F. J. 2016 The Function of the MEF2 Family of transcription factors in cardiac development, cardiogenomics, and direct reprogramming. J. Cardiovasc. Dev. Dis. 3, 26.
Duong T., Koltowska K., Pichol-Thievend C., Le Guen L., Fontaine F., Smith K. A. et al. 2014 VEGFD regulates blood vascular development by modulating SOX18 activity. Blood 123, 1102–1112.
Francois M., Harvey N. L. and Hogan B. M. 2011 The transcriptional control of lymphatic vascular development. Physiology 26, 146–155.
Fu L. and Shi Y. B. 2017 The Sox transcriptional factors: Functions during intestinal development in vertebrates. Semin. Cell Dev. Biol. 3, 58–67.
Gabut M., Samavarchi-Tehrani P., Wang X., Slobodeniuc V., O’Hanlon D. X., Sung H. K. et al. 2011 An alterna-tive splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell 147, 132–146.
Goveia J., Zecchin A., Rodriguez F. M., Moens S., Stapor P. and Carmeliet P. 2014 Endothelial cell differentiation by SOX17: promoting the tip cell or stalking its neighbor instead. Circ. Res. 115, 205–207.
Han F., Wang Z., Wu F., Liu Z., Huang B. and Wang D. 2010 Characterization, phylogeny, alternative splicing and expression of Sox30 gene. BMC Mol. Biol. 11, 1–11.
Hermanto Y., Takagi Y., Ishii A., Yoshida K., Kikuchi T., Funaki T. et al. 2016 Immunohistochemical Analysis of Sox17 Associated Pathway in Brain Arteriovenous Malformations. World Neurosurg. 87, 573–583.
Hirate Y., Suzuki H., Kawasumi M., Takase H. M., Igarashi H., Naquet P. et al. 2016 Mouse Sox17 haploinsufficiency leads to female subfertility due to impaired implantation. Sci. Rep. 6, 24171.
Huang L., Hu F. and Zhang X. Y. 2011 Recent advanced in Brn3 family transcription facter regulation on retinal ganglion cell development. Rec. adv. Ophthalmol. 31, 488–489.
Hwang J. A., Goo I. B., Kim J. E., Kim M. H., Kim D. H., Im J. H. et al. 2016 Growth comparison of israeli carp (Cyprinus carpio) to different breeding combination. Dev. Reprod. 20, 275–281.
Irie N., Weinberger L., Tang W. W., Kobayashi T., Viukov S., Manor Y. S. et al. 2016 SOX17 is a critical specifier of human primordial germ cell fate. Cell 160, 253–268.
Jiang T., Hou C. C., She Z. Y. and Yang W. X. 2012 The SOX gene family: function and regulation in testis determination and male fertility maintenance. Mol. Biol. Rep. 40, 2187–2194.
Jin C. Q. and Liu F. 2009 The research progress of the transcription factor GATA-1. Int. J. immunology. 1, 1673–4394.
Kamachi Y. and Kondoh H. 2013 Sox proteins: regulators of cell fate specification and differentiation. Development 140, 4129–4144.
Kashimada K. and Koopman P. 2010 Sry: The master switch in mammalian sex determination. Development 137, 3921–3930.
Kim K., Kim I. K., Yang J. M., Lee E., Koh B. I., Song S. et al. 2016 Sox F transcription factors are positive feedback egulators of VEGF signaling. Circ. Res. 119, 839–852.
Kinoshita M., Shimosato D., Yamane M. and Niwa H. 2015 Sox7 is dispensable for primitive endoderm differentiation from mouse ES cells. BMC Dev. Biol. 15, 37.
Lilly A. J., Costa G., Largeot A., Fadlullah M. Z., Lie-A-Ling M., Lacaud G. et al. 2016 Interplay between SOX7 and RUNX1 regulates hemogenic endothelialfate in the yolk sac. Development 143, 4341–4351.
Lilly A. J., Lacaud G. and Kouskoff V. 2017 SOXF transcription factors in cardiovascular development. Semin. Cell. Dev. Biol. 63, 50–57.
Lin C. J., Fan-Chiang Y. C., Dufour S and Chang C. F. 2016 Activation of brain steroidogenesis and neurogenesis during the gonadal differentiation in protandrous black porgy, Acanthopagrus schlegelii. Dev. Neurobiol. 76, 121–136.
Lin G. H. and Weng S. C. 1986 The embryo development of Cyprinus carpio var. singuonensis (in Chinese). J.Jiangxi Uni. (Nat. Sci.) 10, 1–9.
Livak K. J. and Schmittgen T. D. 2011 Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) Method. Methods 25, 402–408.
Morini M. F. and Dejana E. 2014 Transcriptional regulation of arterial differentiation via Wnt, Sox, and Notch. Curr. Opin. Hematol. 21, 229–234.
Peng X., Xiofeng Z. and Xumin W. 2014 Genome sequence and genetic diversity of the common carp, Cyprinus carpio. Nat. Genet. 46, 1212–1219.
Rochtus A., Winand R., Laenen G., Vangeel E., Izzi B., Wittevrongel C. et al. 2016 Methylome analysis for spina bifida shows SOX18 hypomethylation as a risk factor with evidence for a complex (epi) genetic interplay to affect neural tube development. Clin. Epigenet. 8, 108.
Sacilotto N., Chouliaras K. M., Nikitenko L. L, Lu Y. W., Fritzsche M., Wallace M. D. et al. 2016 MEF2 transcription factors are key regulators of sprouting angiogenesis. Genes Dev. 30, 2297–2309.
Sarkar and Hochedinger. 2013 The Sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell 12, 15–30.
Schepers G. E., Teasdale R. D. and Koopman P. 2002 Twenty pairs of Sox: extent, homology, and nomenclature of the mouse and human Sox transcription factor gene families. Dev. Cell 3, 167–170.
She Z. Y. and Yang W. X. 2015 SOX family transcription factors involved in diverse cellular events during development. Eur. J. Cell Biol. 94, 547–563.
Takahashi T. and Holland P. W. 2004 Amphioxus and ascidian Dmbx homeoboxgenes give clues to the vertebrate origins of midbrain development. Development 131, 3285–3294.
Tan X. F., Qin J. B., Jin G. H, Tian M. L, Li H. M. and Zhu H. X. 2010 The effect and mechanism of Brn4 on the neuronal differentiation of neural stem cells. Cell Biol. Int. 34, 877–882.
Tarradas A., Pinsach-Abuin M. L., Mackintosh C., Llorà-Batlle O., Pérez-Serra A., Batlle M. et al. 2016 Transcriptional regulation of the sodium channel gene (SCN5A) by GATA4 in human heart. J. Mol. Cell. Cardiol. 102, 74–82.
Wang C. H. 2009 Quantitative genetic estimates of growth-related traits in the common carp (Cyprinus carpio L.): a review. Front. Biol. China 4, 298–304.
Wang C., Qin L., Min Z., Zhao Y., Zhu L., Zhu J. et al. 2015 SOX7 interferes with b-catenin activity to promote neuronal apoptosis. Eur. J. Neurosci. 41, 1430–1437.
Watanabe M., Kawasaki K., Kawasaki M., Portaveetus T.,Oommen S., Blackburn J. et al. 2016 Spatio-temporal expression of Sox genes in murine palatogenesis. Gene. Expr. Patterns 21, 111–118.
Wei L., Yang C., Tao W. and Wang D. 2016 Genome-wide identification and transcriptome-based expression profiling of the Sox gene family in the nile tilapia (Oreochromis niloticus). Int. J. Mol. Sci. 17, 270.
Wohlfarth G., Moav R. and Hulata G. 1975 Genetic differences between the Chinese and European races of the common carp. II. Multi-character variation-a response to the diverse methods of fish cultivation in Europe and China. Heredity 34, 341–350.
Yahoo N., Pournasr B., Rostamzadeh J., Hakhamaneshi M. S., Ebadifar A., Fathi F. et al. 2016 Forced expression of Hnf1b/Foxa3 promotes hepatic fate of embryonic stem cells. Biochem. Biophys. Res. Commun. 474, 199–205.
Yu X., Zhou T., Li K., Li Y. and Zhou M. 1987 On the karyosystematics of cyprinid fishes and a summary of fish chromosome studies in China. Genetica 72, 225–235.
Zhang W. W., Jia Y. F., Ji X. L., Zhang R. H., Liang T. T. and Chang Z. J. 2016 Optimal reference genes in different tissues, gender, and gonad of Yellow River Carp (Cyprinus carpio var) at various developmental periods. Pak. J. Zool. 48, 1615–1622.
Zhou Y, Williams J., Smallwood P. M. and Nathans J. 2015 Sox7, Sox17, and Sox18 cooperatively regulate vascular development in the mouse retina. PLoS One 10, e0143650.
Acknowledgements
This work is supported by grants from the National Natural Science Foundation of China (no. U1204329), Innovative Research Team (Science and Technology) in University of Henan Province (no. 17IRTSTHN017), the Henan Scientific and Technological Research Projects (no. 172102110098).
Author information
Authors and Affiliations
Corresponding author
Additional information
Corresponding editor: Indrajit Nanda
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liang, T., Jia, Y., Zhang, R. et al. Identification, molecular characterization and analysis of the expression pattern of \({\varvec{SoxF}}\) subgroup genes the Yellow River carp, \({\varvec{Cyprinus} \varvec{carpio}}\). J Genet 97, 157–172 (2018). https://doi.org/10.1007/s12041-018-0898-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12041-018-0898-8