Abstract
Genus Astyanax is well distributed in Neotropical freshwater environments and its taxonomic position is uncertain, as is the case with other Characidae genera allocated in the group incertae sedis. This study aimed to analyse the karyotype of different populations of Astyanax fasciatus (Corumbataí River basin) using Giemsa staining, C-band technique, and fluorescence in situ hybridization for the H3 histone and 5S rRNA genes, in addition we describe for the first time the chromosomal organization of H3 histone and 5S rRNA genes in A. marionae (Paraguay River basin). Chromosomes of three A. fasciatus populations were analysed (two with \(2n = 50\) and one with \(2n = 48\)) and the heterochromatin was organized in two forms (blocks with blurred boundaries and distinct blocks). H3 histone and 5S rRNA genes were observed in all the three populations of A. fasciatus on two chromosome pairs (one metacentric chromosome showing H3 histone and 5S rRNA gene clusters). In A. marionae (\(2n = 48\)), H3 histone and 5S rRNA genes were observed in one acrocentric chromosome pair (different pairs). Further, differences between karyotypes and heterochromatin, as well as the chromosomal organization of H3 histone and 5S rRNA genes in Astyanax species, focussing on chromosome evolution in the group are discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Astyanax (Characidae) is one of the genera with the largest number of species in Characidae, containing \({\sim }140\) valid species (Eschmeyer and Fong 2015). Its wide distribution from the south of USA to central Argentina (Lima et al. 2003), occupying a variety of habitats in rivers and streams, makes this group one of the most complex genera of freshwater fishes. Lima et al. (2003) allocated many characid genera into incertae sedis, e.g. Hemigrammus, Hyphessobrycon, Moenkausia and Astyanax. Recently, other authors have also showed that Astyanax does not represent a monolyphyletic group (see Javonillo et al. 2010; Mirande 2010; Oliveira et al. 2011).
Cytogenetic studies of genus Astyanax have revealed extensive variation in the diploid number, ranging from \(2n = 36\) in A. schubarti (Morelli et al. 1983) and A. correntinus (Paiz et al. 2015) to \(2n = 50\) chromosomes for most species, e.g. A. altiparanae and A. bockmanni (Fernandes and Martins-Santos 2004; Kavalco et al. 2009, respectively). Further, some species of Astyanax may have more than one diploid number e.g., the ‘fasciatus complex’ which may contain species with chromosomes \(2n = 45\) to \(2n = 50\) (Centofante et al. 2003).
Other cytogenetic papers have shown different patterns of heterochromatin C-band positive in Astyanax group (see Artoni et al. 2006; Peres et al. 2009; Tenório et al. 2013; Piscor et al. 2015; Piscor and Parise-Maltempi 2016a). For example, Artoni et al. (2006) studied three cytotypes of Astyanax aff. fasciatus (A, B and C) and verified two heterochromatin patterns. According to the authors, cytotypes A and B showed that the heterochromatin mainly distributed as very conspicuous blocks in the telomeric region of the long arms of acrocentric chromosomes, whereas few chromosomes bearing heterochromatin were observed in cytotype C.
Repetitive sequences were mapped in many Astyanax species. Piscor and Parise-Maltempi (2016b) observed the chromosomal location of 5S rRNA and H3 histone genes in eight species and identified similar chromosomes bearing H3 histone, except in A. schubarti and A. mexicanus. The chromosomal mapping of repetitives are studied here for the first time in A. marionae.
Therefore, given the karyotype complexity found in the Astyanax genus and considering A. marionae and A. fasciatus are two species with similar morphological traits, it is necessary to attest whether or not they have similar cytogenetic characteristics. Thus, this study aimed to compare the chromosomes of A. marionae with different populations of A. fasciatus, and to understand the chromosomal organization of H3 histone and 5S rRNA genes, and of heterochromatin revealed by C-banding.
Materials and methods
Sampling and classical cytogenetics
Three populations of A. fasciatus were studied: two individuals from Cabeça river tributary, five from Ribeirão Claro river tributary, and three from the Corumbataí river tributary (Corumbataí river basin, São Paulo, Brazil). The six individuals of A. marionae were captured in the Rio Claro stream (Paraguay river basin, Mato Grosso, Brazil). The metaphasic chromosomes were obtained by the methodology of Foresti et al. (1981) and stained with Giemsa (10% in phosphate buffer). The morphologies of chromosomes were determined according to the arm’s ratio, based on the more common classification system used for fish chromosomes in Brazil: the chromosomes with two arms and arm ratio (AR) of 1–1.7 were classified as metacentric (m), with AR 1.71–3 as submetacentric (sm), and with AR 3.01–7 as subtelocentric (st). Chromosomes with a single arm (\(\hbox {AR}>7\)) were considered as acrocentric (a). Heterochromatin was observed in chromosomes of all the A. fasciatus and A. marionae individuals using the C-band technique, as proposed by Sumner (1972).
DNA extraction, production of probes, and fluorescence \(\mathbf{in}\) \(\mathbf{situ}\) hybridization
Genomic DNA was extracted from fin samples of Astyanax, as described in Sambrook and Russell (2001). The 5S rDNA probe was prepared using polymerase chain reaction (PCR) with primers described by Pendás et al. (1994) and Martins and Galetti (1999) (A, \(5^\prime \)-TAC GCC CGA TCT CGT CCG ATC-\(3^\prime \), and B, \(5^\prime \)-CAG GCT GGT ATG GCC GTA AGC-\(3^\prime )\). The H3 histone probe was prepared using PCR with primers described by Cabral-de-Mello et al. (2010) (A, \(5^\prime \)-GGC NMG NAC NAA RCA RAC, and B, \(5^\prime \)-TGD ATR TCY TTN GGC ATD AT). The H3 histone genes were amplified using the genomic DNA of A. fasciatus and A. marionae. The PCR products were sequenced in Korean company (Macrogen) and the sequences were edited and aligned with BioEdit program (Hall 1999). The analysis of similarities to sequences were submitted in website GenBank (https://www.ncbi.nlm.nih.gov/genbank/) for comparison. The H3 histone gene of A. marionae was mapped for the first time, sequenced, and deposited in GenBank with the accession number KY389066.
The 5S rDNA probe was labelled using PCR with biotin-14-dATP (Invitrogen, San Diego, USA), and the H3 histone probe was labelled using PCR with digoxigenin-11-dUTP (Roche, Mannheim, Germany). Fluorescence in situ hybridization (FISH) was used in all A. fasciatus and A. marionae individuals according to Pinkel et al. (1986) with modifications described by Piscor et al. (2013). Chromosomes were counterstained with Vectashield Mounting Medium (Vector, Burlingame, USA) containing DAPI (\(4'\),6-diamidino-\(2'\)-phenylindole). Chromosomes and fluorescent signals were visualized with an Olympus BX51 microscope coupled to a digital camera (Olympus model D71), and the images were captured using the DP Controller software.
Statement of ethics
All the institutional guidelines for the care and use of laboratory animals were followed. The animals were captured with permission of Instituto Chico Mendes de Conservação da Biodiversidade – ICMBio (number 43497-1), and used for laboratory experiments approved by the Animal Experimental Ethics Committee from Universidade Estadual Paulista – UNESP (protocol number: 2335).
Results
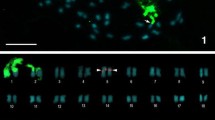
The karyotype of A. marionae showed 8 m, 24 sm, 10 st, 6 a, and the fundamental number (FN) \(=\) 90 (figure 1a). C-banded heterochromatin was observed mainly on the centromeric and proximal regions (figure 1b). Clusters of 5S rRNA and H3 histone genes were observed on pairs 22 (a) and 24 (a), respectively (figure 1c).
Chromosomes stained with Giemsa and C-banded chromosomes. (a–b) Karyotype and C-banded metaphase of A. fasciatus from the Cabeça river tributary. (c–d) Karyotype and C-banded metaphase of A. fasciatus from the Ribeirão Claro river tributary. (e–f) Karyotype and C-banded metaphase of A. fasciatus from the Corumbataí river tributary. The arrows indicate the blocks of C-band heterochromatin.
Populations from tributaries of the Cabeça and Corumbataí rivers showed a diploid number of \(2n=50\) chromosomes and the population from Ribeirão Claro river showed \(2n = 48\) chromosomes. Karyotypic formulae for these populations were: 16 m, 12 sm, 6 st, 16 a and FN \(=\) 84 for the Cabeça river tributary (figure 2a), 10 m, 20 sm, 8 st, 10 a and FN \(=\) 86 for the Ribeirão Claro river tributary (figure 2c), and 8 m, 26 sm, 6 st, 10 a and FN \(=\) 90 for the Corumbataí river tributary (figure 2e). Heterochromatic regions were observed in two organization forms. The first form was noted in the Cabeça and Corumbataí populations, as blocks with blurred boundaries (figure 2, b and f), and the second form was observed in the Ribeirão Claro population, as distinct blocks (figure 2d).
The H3 histone genes (sequenced from DNA of A. fasciatus and A. marionae) exhibited 95 to 100% similarity with H3 histone sequences of others Astyanax species. Gene clusters of the H3 histone were observed on two chromosome pairs in all three populations of A. fasciatus (see figure 3). The A. fasciatus population from the Cabeça river tributary showed interstitial fluorescent signals on pair 3 (m) and pericentromeric signals on pair 15 (st) (figure 3). The A. fasciatus population from the Corumbataí river tributary showed interstitial fluorescent signals on pair 3 (m) and pericentromeric signals on pair 18 (st) (figure 3). The A. fasciatus population from the Ribeirão Claro river tributary showed interstitial fluorescent signals on pair 2 (m) and pericentromeric signals on pair 16 (st) (figure 3).
Clusters of 5S rDNA were located on two chromosome pairs (see figure 3). A. fasciatus from the Cabeça river tributary showed pericentromeric fluorescent signals on pair 3 (m, same chromosome pair bearing H3 histone genes) and on pair 18 (a) (figure 3). A. fasciatus from the Corumbataí river tributary showed pericentromeric fluorescent signals on pair 3 (m, same chromosome pair that bears H3 histone genes) and on pair 21 (a) (figure 3). A. fasciatus from the Ribeirão Claro river tributary showed pericentromeric fluorescent signals on pair 2 (m, same chromosome pair that bears H3 histone genes) and on pair 20 (a) (figure 3).
A map of the hydrographic basins and distribution of A. marionae andA. fasciatus, and representative chromosome pairs bearing H3 histone and 5S rDNA clusters are shown in figure 3.
Discussion
Astyanax fasciatus populations usually differ in diploid number. According to Ferreira-Neto et al. (2012) this species can present diploid numbers \(2n = 46\), 48 and 50 chromosomes (the most common number is \(2n = 48\)). However, previous results have shown karyomorphs with \(2n = 45\), 47, and 49 chromosomes, possibly resulting from hybridizations (Artoni et al. 2006; Pazza et al. 2006). Further, chromosome variations are evident among different populations of the same species, primarily relating to the number and size of heterochromatin blocks (see, for example, Fernandes and Martins-Santos 2003, 2004).
Distinct blocks of heterochromatin have been found in other A. fasciatus populations. Pazza et al. (2008) analysed populations of A. fasciatus from three different sites along the Mogi-Guaçu river (Ouro Fino, Minas Gerais; Cachoeira de Emas, Pirassununga, São Paulo State; Barrinha, Brazil) and observed a similar pattern of constitutive heterochromatin organization on the forms with \(2n = 46\) and \(2n = 48\) chromosomes (in the telomeric region on long arms of submetacentric, subtelocentric and acrocentric chromosomes, and in the telomeric region on short arms of a submetacentric pair). Similar results were observed in the population from the Ribeirão Claro river (\(2n = 48\)) studied here.
Moreover, the presence of two heterochromatic organization forms in all three A. fasciatus populations analysed in this paper points to the conclusion that distinct evolutionary events may have influenced the organization of heterochromatic segments. According to Peres et al. (2009), three A. fasciatus populations from the São Francisco river basin (MG) showed \(2n = 48\) chromosomes, two of which have shown heterochromatic blocks. The authors suggest that such characteristics may be due to the endemism of populations with discrete heterochromatin blocks. Differing from that, the present work shows three populations that are part of the Corumbataí river basin and are able to maintain contact with each other.
Astyanax marionae presented heterochromatic blocks especially on the centromeric and pericentromeric regions in almost all chromosomes in this study. Krinski and Miyazawa (2014) also evidenced similar heterochromatin location in A. marionae and discussed resembling morphological characteristics between A. marionae and A. fasciatus.
On the other hand, our molecular cytogenetic data pointed to two different chromosomal organization patterns of repetitive sequences: (i) one metacentric pair bearing an H3 histone cluster in the interstitial region on the short arm and a 5S rDNA cluster in the proximal region on the long arm in A. fasciatus, as one subtelocentric pair with a proximal signal for the H3 histone and one acrocentric pair with a proximal signal for the 5S rDNA are evident for all three populations; (ii) both genes are organized on the proximal regions of two different acrocentric pairs in A. marionae.
Therefore, H3 histone sequences may be located on homeologous pairs in both A. fasciatus karyomorphs (\(2n = 48\) and \(2n = 50\)) observed in this work, and in the karyomorph with \(2n = 46\) evidenced by other authors (Hashimoto et al. 2011; Pansonato-Alves et al. 2013; Silva et al. 2015; Piscor and Parise-Maltempi 2016b). Here, for the first time, location of H3-5S clusters in A. marionae is described, showing a particular form of organization. Other peculiar forms have also been observed in A. jordani by Silva et al. (2015), A. schubarti and A. mexicanus by Piscor and Parise-Maltempi (2016b).
According to Piscor and Parise-Maltempi (2016b), A. fasciatus and other Astyanax species (A. altiparanae, A. abramis, A. asuncionensis, A. bockmanni and A. eigenmanniorum) present H3 histone clusters on two chromosome pairs (one m/sm and one sm/st) with similar morphologies (which contribute to the conservation hypothesis for the H3 histone genes), with the exception of A. schubarti, and A. mexicanus, that show different forms. In this study, A. marionae showed a different system of H3 histone cluster organizations.
In this context, despite presenting similar morphological traits and chromosomal macrostructure as discussed by Krinski and Miyazawa (2014), A. marionae and A. fasciatus show remarkable differences in the organization of repetitive sequences. In such respect, we suggest that these distinct organization forms of 5S rDNA and H3 histone clusters does not exclude the phylogenetic closeness between them.
References
Artoni R. F., Shibatta O. A., Gross M. C., Schneider C. H., Almeida M. C., Vicari M. R. and Bertollo L. A. C. 2006 Astyanax aff. fasciatus Cuvier, 1819 (Teleostei; Characidae): evidences of a species complex in the upper rio Tibagi basin (Paraná, Brazil). Neotrop. Ichthyol. 4, 197–202.
Cabral-de-Mello D. C., Moura R. C. and Martins C. 2010 Chromosomal mapping of repetitive DNAs in the beetle Dichotomius geminatus provides the first evidence for an association of 5S rRNA and histone H3 genes in insects, and repetitive DNA similarity between the B chromosome and A complement. Heredity 104, 393–400.
Centofante L., Bertollo L. A. C., Justi A. J. and Moreira-Filho O. 2003 Correlation of chromosomal and morphologic characters in two Astyanax species (Teleostei: Characidae). Ichthyol. Explor. Freswaters 14, 361–368.
Eschmeyer W. N. and Fong J. D. 2015 Catalog of fishes (http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp).
Fernandes C. A. and Martins-Santos I. C. 2003 Cytogenetic characterization of two populations of Astyanax scabripinnis (Pisces, Characiformes) of the Ivaí basin PR Brazil. Cytologia 68, 289–293.
Fernandes C. A. and Martins-Santos I. C. 2004 Cytogenetic studies in two populations of the Astyanax altiparanae (Pisces, Characiformes). Hereditas 141, 328–332.
Ferreira-Neto M., Artoni R. F., Vicari M. R., Moreira-Filho O., Camacho J. P. M., Bakkali M. et al. 2012 Cytogenetics three sympatric karyomorphs in the fish Astyanax fasciatus (Teleostei, Characidae) do not seem to hybridize in natural populations. Comp. Cytogenet. 6, 29–40.
Foresti F., Almeida-Toledo L. F. and Toledo-Filho S. A. 1981 Polymorphic nature of nucleous organizer regions in fishes. Cytogenet. Cell Genet. 31, 134–141.
Hashimoto D. T., Ferguson-Smith M. A., Rens W., Foresti F. and Porto-Foresti F. 2011 Chromosome mapping of H1 histone and 5S rRNA gene clusters in three species of Astyanax (Teleostei, Characiformes). Cytogenet. Genome Res. 134, 64–71.
Javonillo R., Malabarba L. R., Weitzman S. H. and Burns J. R. 2010 Relationships among major lineages of characid fishes (Teleostei: Ostariophysi: Characiformes), based on molecular sequence data. Mol. Phylogenet. Evol. 54, 498–511.
Kavalco K. F., Pazza R. and Almeida-Toledo L. F. 2009 Astyanax bockmanni Vari and Castro, 2007: an ambiguous karyotype in the Astyanax genus. Genetica 136, 135–139.
Krinski D. and Miyazawa C. S. 2014 Cytogenetic analysis of three species of Astyanax genus (Pisces, Characidae, Incertae sedis) from freshwaters of upper Paraguay basin, Mato Grosso state, Brazil. J. Life Sci. 8, 51–57.
Lima F. C. T., Malabarba L. R., Buckup P. A., Silva J. F. P., Vari R. P., Harold A. et al. 2003 Genera Incertae Sedis in Characidae. In Check list of the freshwater fishes of South and Central America (ed. R. Reis, E. S. O. Kullander and C Jr Ferraris), pp. 106–169. Edipucrs, Porto Alegre, Brazil.
Martins C. and Galetti Jr P. M. 1999 Chromosomal localization of 5S rDNA genes in Leporinus fish (Anostomidae, Characiformes). Chromosome Res. 7, 363–367.
Mirande J. M. 2010 Phylogeny of the family Characidae (Teleostei: Characiformes): from characters to taxonomy. Neotrop. Ichthyol. 8, 385–568.
Morelli S., Bertollo L. A. C., Foresti F., Moreira-Filho O. and Toledo-Filho S.A. 1983 Cytogenetic considerations on the genus Astyanax (Pisces, Characidae). I. Karyotypic variability. Caryologia 36, 235–244.
Oliveira C., Avelino G. S., Abe K. T., Mariguela T. C., Benine R. C., Ortí G. et al. 2011 Phylogenetic relationships within the speciose family Characidae (Teleostei, Ostariophysi, Characiformes) based on multilocus analysis and extensive ingroup sampling. BMC Evol. Biol. 11, 275.
Paiz L. M., Baumgärtner L., da Graça W. J. and Margarido V. P. 2015 Basic cytogenetics and physical mapping of ribosomal genes in four Astyanax species (Characiformes, Characidae) collected in Middle Paraná River, Iguassu National Park: considerations on taxonomy and systematics of the genus. Comp. Cytogenet. 9, 51–65.
Pansonato-Alves J. C., Hilsdorf A. W. S., Utsunomia R., Silva D. M. Z. A., Oliveira C. and Foresti F. 2013 Chromosomal mapping of repetitive DNA and cytochrome C oxidase I sequence analysis reveal differentiation among sympatric samples of Astyanax fasciatus (Characiformes, Characidae). Cytogenet. Genome Res. 141, 133–142.
Pazza R., Kavalco K. F. and Bertollo L. A. C. 2006 Chromosome polymorphism in Astyanax fasciatus (Teleostei, Characidae). 1 – Karyotypic analysis, Ag-NORs and mapping of the 18S and 5S ribosomal genes in sympatric karyotypes and their possible hybrid forms. Cytogenet. Genome Res. 112, 313–319.
Pazza R., Kavalco K. F. and Bertollo L. A. C. 2008 Chromosome polymorphism in Astyanax fasciatus (Teleostei, Characidae). 2 – Chromosomal location of a satellite DNA. Cytogenet. Genome Res. 122, 61–66.
Pendás A. M., Morán P., Freije J. P. and García-Vázquez E. 1994 Chromosomal location and nucleotide sequence of two tandem repeats of the Atlantic salmon 5S rDNA. Cytogenet. Cell Genet. 67, 31–36.
Peres W. A. M., Buckup P. A., Kantek D. L. Z., Bertollo L. A. C. and Moreira-Filho O. 2009 Chromosomal evidence of downstream dispersal of Astyanax fasciatus (Characifiomes, Characidae) associated with river shed interconnection. Genetica 137, 305–311.
Pinkel D., Straume T. and Gray J. W. 1986 Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc. Natl. Acad. Sci. USA 83, 2934–2938.
Piscor D., Ribacinko-Piscor D. B., Fernandes C. A. and Parise-Maltempi P. P. 2013 Cytogenetic analysis in three Bryconamericus species (Characiformes, Characidae): first description of the 5S rDNA-bearing chromosome pairs in the genus. Mol. Cytogenet. 6, 13.
Piscor D., Alves A. L. and Parise-Maltempi P. P. 2015 Chromosomal microstructure diversity in three Astyanax (Characiformes, Characidae) species: comparative analysis of the chromosomal locations of the 18S and 5S rDNAs. Zebrafish 12, 81–90.
Piscor D. and Parise-Maltempi P. P. 2016a Microsatellite organization in the B chromosome and A chromosome complement in Astyanax (Characiformes, Characidae) species. Cytogenet. Genome Res. 148, 44–55.
Piscor D. and Parise-Maltempi P. P. 2016b Chromosomal mapping of H3 histone and 5S rRNA genes in eight species of Astyanax (Pisces, Characiformes) with different diploid numbers: syntenic conservation of repetitive genes. Genome 59, 167–172.
Sambrook J. and Russell D. W. 2001 Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York, USA.
Silva D. M. Z. d. A., Utsunomia R., Pansonato-Alves J. C., Oliveira C. and Foresti F. 2015 Chromosomal mapping of repetitive DNA sequences in five species of Astyanax (Characiformes, Characidae) reveals independent location of U1 and U2 snRNA sites and association of U1 snRNA and 5S rDNA. Cytogenet. Genome Res. 146, 144–152.
Sumner A. T. 1972 A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 75, 304–306.
Tenório R. C. C. O., Vitorino C. A., Souza I. L., Oliveira C. and Venere P. C. 2013 Comparative cytogenetics in Astyanax (Characiformes: Characidae) with focus on the cytotaxonomy of the group. Neotrop. Ichthyol. 11, 553–564.
Acknowledgements
The authors are grateful to Coordenadoria de Aperfeiçoamento de Pessoal de Nível Superior for the financial support (CAPES).
Author information
Authors and Affiliations
Corresponding author
Additional information
Corresponding editor: Indrajit Nanda
Rights and permissions
About this article
Cite this article
Piscor, D., Centofante, L. & Parise-Maltempi, P.P. Distinct classical and molecular cytogenetics of Astyanax marionae and A. fasciatus (Characiformes: Characidae): a comparative study of the organization of heterochromatin and repetitive genes. J Genet 96, 665–671 (2017). https://doi.org/10.1007/s12041-017-0813-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12041-017-0813-8