Abstract
A pair of stripe rust and leaf rust resistance genes was introgressed from Aegilops caudata, a nonprogenitor diploid species with the CC genome, to cultivated wheat. Inheritance and genetic mapping of stripe rust resistance gene in backcross-recombinant inbred line (BC-RIL) population derived from the cross of a wheat–Ae. caudata introgression line (IL) T291-2(pau16060) with wheat cv. PBW343 is reported here. Segregation of BC-RILs for stripe rust resistance depicted a single major gene conditioning adult plant resistance (APR) with stripe rust reaction varying from TR-20MS in resistant RILs signifying the presence of some minor genes as well. Genetic association with leaf rust resistance revealed that two genes are located at a recombination distance of 13%. IL T291-2 had earlier been reported to carry introgressions on wheat chromosomes 2D, 3D, 4D, 5D, 6D and 7D. Genetic mapping indicated the introgression of stripe rust resistance gene on wheat chromosome 5DS in the region carrying leaf rust resistance gene LrAc, but as an independent introgression. Simple sequence repeat (SSR) and sequence-tagged site (STS) markers designed from the survey sequence data of 5DS enriched the target region harbouring stripe and leaf rust resistance genes. Stripe rust resistance locus, temporarily designated as YrAc, mapped at the distal most end of 5DS linked with a group of four colocated SSRs and two resistance gene analogue (RGA)-STS markers at a distance of 5.3 cM. LrAc mapped at a distance of 9.0 cM from the YrAc and at 2.8 cM from RGA-STS marker Ta5DS_2737450, YrAc and LrAc appear to be the candidate genes for marker-assisted enrichment of the wheat gene pool for rust resistance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Stripe rust (yellow rust) caused by Puccinia striiformis Westend f. sp. tritici (Pst) is a serious disease of wheat in cooler climate (2–15 ∘C). Stripe rust reduces yield and quality of grain as seeds produced from the crop damaged by the stripe rust have low vigour and exhibit poor emergence after germination. The frequent use of limited parental genotypes in the modern wheat breeding practices and monoculture of few improved wheat varieties have resulted in narrow genetic base in the cultivated genepool. Limited variability in the host and emergence of new pathotypes render the varietal spectrum inadequate to combat the infection in almost all the wheat growing regions of the world. It also necessitates the availability of a battery of new genes in adapted backgrounds for breeding cultivars with new effective R genes.
Based on specificity, rust resistance can be classified as race-specific or race-nonspecific; and based on plant growth stage, it can be classified as seedling (also known as all-stage) resistance or adult plant resistance (APR) (Chen 2005, 2013). Seedling resistance is usually race-specific, but often provides complete control against avirulent races. Adult plant (AP) or postseedling resistance is often nonrace-specific. It is known to be ineffective during seedling stage, but effectiveness increases with plant age. Among the designated stripe rust resistance genes Yr11, Yr12, Yr13, Yr14, Yr16, Yr18, Yr29, Yr30, Yr34, Yr36, Yr39, Yr46, Yr48 and Yr52 confer APR, whereas the others confer all-stage resistance (Xu et al. 2013). Some slow rusting genes for stripe rust have also been catalogued and over 140 quantitative trait loci (QTL) for resistance to stripe rust in wheat have been published (Rosewarne et al. 2013), but it is likely that many of these QTLs are identical.
The wild progenitor and nonprogenitor species of cultivated wheat are a valuable source of additional resistance genes (Jiang et al. 1994; Chhuneja et al. 2016). Species belonging to the genus Aegilops are important sources of genetic diversity for improving the variability of cultivated bread wheat. Aegilops caudata L. ( = Ae. markgrafii = T. dichasians) is a diploid wild relative of wheat (2 n= 2 x= 14, genome CC). It carries resistance genes to powdery mildew, leaf and stripe rust, and also has high crude protein and lysine content (Valkoun et al. 1985). It is reported to be one of the most resistant representatives in the group of less closely-related species with homeologous genomes from which no rust resistance genes have been transferred to wheat (McIntosh et al. 2013). Riar et al. (2012) mapped a leaf rust resistance gene LrAc in the F 2:3 mapping population derived from one of the wheat–Ae. caudata accession pau3556 introgression lines on wheat chromosome 5DS. The present study was conducted to characterize and map a stripe rust resistance gene introgressed from same Ae. caudata acc. pau3556 into bread wheat and to study its genetic association with the leaf rust resistance gene LrAc.
Material and methods
Plant material and disease evaluation
A backcross-recombinant inbred line (BC-RIL) population derived from a cross of leaf rust and stripe rust resistant wheat–Ae. caudata introgression line, T291-2 (pau16060) with a susceptible wheat cv. PBW343, consisting of 354 RILs was used. The development of the introgression line and F 2 mapping population are described in Riar et al. (2012). The introgression line T291-2 was susceptible at the seedling stage to the two most prevalent Pst pathotypes 78S84 and 46S119 of Indian subcontinent, but showed a moderate resistance reaction of 20MR at the adult plant stage against a mixture of these Pst pathotypes. The RIL population was evaluated for stripe rust at the adult plant stage against a mixture of Pst isolates for two crop seasons 2013–2014 and 2014–2015. The population was planted at 1.5 m row with a row to row distance of 20 cm and recommended agronomic practices were followed for raising the crop (Anonymous 2013). Infector rows of the susceptible cultivars, WL711 and PBW343, were planted all around the experimental plot. Artificial rust epidemic was created by spraying the infector rows and experimental material with the mixture of uredinospores of Pst isolates 78S84 and 46S119. Stripe rust assessment was according to the modified Cobb’s scale (Peterson et al. 1948). The RIL population was screened at the seedling stage against leaf rust pathotype 77-5 as described by Riar et al. (2012).
Molecular marker analysis
Genomic DNA was extracted using the CTAB method of Saghai-Maroof et al. (1984). Resistant and susceptible bulks comprising equal amounts of DNA from 10 resistant (phenotypic reaction of TR-5MR) and 10 susceptible (phenotypic reaction of 60S-80S) RILs were used for bulked segregant analysis (Michelmore et al. 1991). Riar et al. (2012) had developed an introgression profile of the donor introgression line T291-2 using whole genome SSR scan and reported Ae. caudata specific introgression on chromosomes 2D, 3D, 4D, 6D, 5D and 7D. All the introgressed markers including those linked to LrAc on 5DS were analysed in bulked segregant analysis. In addition, markers reported to be mapped on short arm of homeologous group 5A and 5B chromosomes were also genotyped on the bulks to find marker(s) cosegregating/linked with the target gene.
Polymerase chain reactions (PCR) were performed in Eppendorf and Applied Biosystems master cyclers. For SSR analysis, a 20 μL of PCR reaction mixture contained 40– 50 ng genomic DNA, 4 μL of 5 × PCR buffer, 1.2 μL of 25 mM MgCl 2, 3 μL of 1 mM dNTP mix, 1.5 μL each of 5 μM forward and reverse primers, 0.2 μL of 1U Taq polymerase. The PCR products were resolved on 6% polyacrylamide gels. For STS markers, a 20 μL of reaction mixture contained 80–100 ng genomic DNA, 4 μL of 5 × PCR buffer, 1.5 μL of 25 mM MgCl 2, 6 μL of 1 mM dNTP mix, 1.5 μL each of 5 μM forward and reverse primers and 0.2 μL of 1U Taq polymerase. PCR conditions for SSR and RGA-STS primers were same as reported by Chhuneja et al. (2015).
Development of SSR and STS markers from wheat survey sequence data
Survey sequence data of wheat chromosome 5DS was obtained from International Wheat Genome Sequence Consortium (IWGSC 2014). Transcript and protein data files of Brachypodium distachyon was also retrieved. Using a tool BLASTN, wheat 5DS survey sequence was BLAST against transcript data of Brachypodium to obtain gene containing contigs of wheat. The new SSR primers were designed from the genic contigs of short arm of 5DS using primer designing tool PerlPrimer from the sequences containing at least 10 mononucleotide or dinucleotide repeats and six tri, tetra, penta and hexa nucleotide repeats were selected.
For designing STS primers specific for nucleotide-binding site and leucine-rich repeats (NBS-LRR) genes on wheat chromosome 5DS, NBS-LRR protein sequences were fetched from B. distachyon protein file and BLAST searched against 5DS survey sequence. Wheat contigs containing NBS-LRR sequences were annotated to locate the positions of NBS-LRR encoding sequences using FGENESH (Solovyev et al. 2006). After getting protein and mRNA sequences of the genes, simple modular architecture research tool (SMART), an online tool for the identification and annotation of protein domains and the analysis of protein architectures (Letunic et al. 2012), was applied on protein sequence file with a default parameter of amino acid length of ≥30. The results were confirmed with the help of another online tool LRRfinder (http://www.lrrfinder.com/). RGA-STS primers were then designed from LRR regions of wheat sequence using PerlPrimer.
Genetic mapping
Mapping was conducted using MapDisto 1.7.5.1 (Lorieux 2012). A LOD score of 4.0 and recombination fraction 0.2 was used for preparing the linkage map. RILs with stripe rust reaction from 0 to 20MS were classified as resistant and those with more than 20MS as susceptible. Markers and genes were grouped and ordered using ‘find groups’ and ‘order sequence’ command, respectively. Order of markers was refined using ‘ripple order’ and ‘check inversions’ commands. The robustness of the marker order was evaluated using boostrap order with 1000 trials. The map was drawn using MAPCHART program ver. 2.1 developed by Voorrips (2002).
Results
Inheritance studies
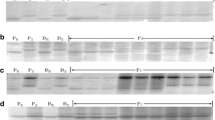
Donor introgression line was susceptible at the seedling stage, but moderately resistant at the adult plant stage implying that it carries APR gene(s) for stripe rust. The stripe rust reaction in the T291-2/PBW343 BC-RILs varied from TR to 80S in both the crop seasons (figure 1a) with overall 184 resistant (0–20MS) and 161 susceptible (>20MS) RILs with a χ 2 (1:1)= 1.53 conforming to a single gene segregation ratio. The segregating RILs were not taken into consideration for calculating the χ 2. Figure 1b exhibits stripe rust reaction of the resistant and susceptible RILs at the adult plant stage. The stripe rust resistance gene from IL T291-2 was tentatively designated as YrAc. For leaf rust resistance, 182/349 BC-RILs exhibited a hypersensitive resistant reaction with infection type ; to 1 and 159/349 were susceptible and eight BC-RILs were segregating showing goodness of fit to a single gene ratio which was consistent with the results reported by Riar et al. (2012) based on F 2:3 population.
(a) Frequency distribution of T291-2/PBW343 RIL population for stripe rust reaction in the field conditions under artificial epiphytotic conditions. (b) Stripe rust reaction of the parental lines, resistant and susceptible RILs at the adult plant stage. 1. WH868; 2. WL711; 3. IL T291-2; 4. PBW343; 5–7. resistant RILs and 8–10. susceptible RILs.
For deciphering the linkage between stripe rust resistance gene YrAc and leaf rust resistance gene LrAc (Riar et al. 2012), BC-RILs were classified into various groups based on the phenotypic reaction to leaf rust and stripe rust (table 1). A 13.3% recombination distance was observed between LrAc and YrAc taking into account only the homozygous RILs.
Molecular mapping of stripe rust resistance
The SSR markers exhibiting Ae. caudata specific introgression in T291-2 were analysed on the resistant and susceptible bulks (RB and SB) pooled from stripe-rust resistant and susceptible RILs. SSR markers from the introgressed segments on chromosome 5DS only gave diagnostic polymorphism between R and S bulks. The leaf rust resistance gene LrAc introgressed from Ae. caudata was mapped on short arm of wheat chromosome 5DS. Of a total of 75 SSR markers from homeologous group 5 amplified on the bulks, only six (Xgwm205, Xcfa2104, Xgwm190, Xcfd81, Xbarc130 and Xgwm234) gave diagnostic polymorphism between R and S bulks. A pair of colocated leaf rust and stripe rust resistance genes Lr57 and Yr40 transferred from M genome of Ae. geniculata has also been mapped on 5DS (Kuraparthy et al. 2007). An STS marker Lr57-Yr40_caps16 linked to Lr57-Yr40, showed polymorphism between R and S bulks for stripe rust also. Analysis of six informative SSR markers and Lr57-Yr40_caps16 on the whole RIL population mapped YrAc at distal end of 5DS with Xgwm190 as the nearest marker and LrAc at a distance of 10.0 cM with Xgwm234 as the nearest marker.
To further enrich the 5DS region carrying YrAc and LrAc, 184 SSR markers designed from the survey sequences of 5DS were used for BSA and only 14 SSR markers were found polymorphic between the resistant and susceptible bulks. Three RGA-STS markers, two from 5DS contig Ta5DS_2737450 and one from Ta5DS_2759608, were also found to be polymorphic on bulks. The primer sequences and PCR conditions for the 14 markers mapped on 5DS are given in table 2.
A consensus data derived from the individual year’s stripe rust and leaf rust data for 2013–2014 and 2014–2015 was used for mapping stripe rust resistance gene YrAc as a major gene. Stripe rust resistance gene YrAc mapped at the distal most end of 5DS linked with five colocated SSR markers, Ta5DS_2736392, Ta5DS_2744983, Ta5DS_2744525, Ta5DS_2683050, Xgwm190 and two RGA-STS markers Ta5DS_2737450a and Ta5DS_2759608 at a distance of 5.3 cM (figure 2). Leaf rust resistance gene LrAc, mapped at the other end of the 5DS segment with Ta5DS_2737450 as the closest marker at a distance of 2.8 cM. LrAc and YrAc were thus found to be linked with a recombination distance of 9.0 cM. To determine orientation of the 5DS segment carrying YrAc and LrAc, the comparison was made with the consensus map of Somers et al. (2004) which showed that YrAc is at the distal end of 5DS and LrAc is proximal. However, no markers distal to YrAc and LrAc could be identified.
Partial linkage map of recombinant chromosome 5DS carrying an Ae. caudata introgression with stripe rust and leaf rust resistance genes YrAc and LrAc, respectively. Markers in black are the SSR markers from the map of Somers et al. (2004) and those in red are new SSR and STS markers designed from survey sequence data of 5DS. YrAc and LrAc mapped at a distance of 9.0 cM. Blue indicate the LrAc and YrAc genes and the STS markers linked to rust resistance genes Lr57 and Yr40.
Discussion
Two pairs of linked leaf rust and stripe rust all stage resistance genes (seedling resistance genes) Lr57-Yr40 and Lr76-Yr70, introgressed from Ae. geniculata (Kuraparthy et al. 2007) and Ae. umbellulata (Bansal et al. 2016), respectively, were mapped at the distal end of 5DS. YrAc characterized and mapped during present investigation is supposedly new gene as YrAc showed susceptibility to Pst isolates 78S84 and 46S119 at the seedling stage, while Yr40 and Yr70 both depicted seedling resistance for these two Pst isolates. However, it is not feasible to determine novelty of LrAc regarding Lr57 and Lr76 as all three genes show complete resistance against the leaf rust races prevalent in India.
Ae. caudata introgression, being homeologous to its wheat counterparts, was not expected to recombine but YrAc, LrAc and the markers mapped on 5DS showed recombination albeit at much lower frequency. According to consensus map by Somers et al. (2004), the SSR markers Xgwm190, Xbarc130, Xcfa2104, Xcfd81 and Xgwm205 spanned 28 cM on 5DS. However, in this study, these markers showed a recombination distance of 5.76 cM indicating suppressed recombination between Ae. caudata introgression and its wheat counterpart. Recombination between the genes and/or markers on the introgressed segment and wheat chromosomes might be due to the presence of some genes on homeologous group 5, which either partially suppress Ph1 gene (Sears 1976) or modify its action in some way. Although there is no direct evidence, but some other studies also reported spontaneous recombination leading to gene transfers from 5M chromosome of Ae. geniculata (Liu et al. 2011).
The APR genes have been known to provide moderate level of resistance, which is more desirable as it puts less selection pressure on the pathogen. The donor introgression line T291-2 showed stripe rust response of 20MR but some of the RILs depicted much higher level of resistance indicating that some minor QTL from the recipient parent or the bridging parents might also be segregating in the population and as a whole population showed a quantitative type of resistance. RILs showing transgressive segregation will be used to develop a subpopulation for the genetic dissection of the other resistance loci.
Identification of markers closely linked with disease resistance has progressed in the last decade through the development of high-throughput and cost-effective genotyping facilities. Advances in wheat genome sequencing and NGS technologies have led to immense data which can be used to develop markers for wheat (Varshney et al. 2014; Jordan et al. 2015). All these technologies individually or in combination can be used to fine map the genes of interest. In the present investigation, the target region was enriched using survey sequence data of 5DS chromosome of Chinese Spring (IWGSC 2014). SSR markers designed from the contigs carrying genic sequences and mapped on 5DS, indicate the location of potential genes. Mapping of three RGA_STS markers designed from NBS-LRR genes identified the putative disease-resistance gene loci present on 5DS. APR gene for stripe rust resistance YrAc identified in the present study is a putatively new gene as per the published evidence, no other stripe rust resistance gene has been transferred from the C genome of Ae. caudata to bread wheat.
SSR and STS markers linked to LrAc and YrAc genes are suitable for marker-assisted transfer of the Ae. caudata genes to elite wheat backgrounds. Selected RILs will be used as donor for marker-assisted mobilization of LrAc and YrAc to advance breeding lines.
References
Anonymous 2013 The package of practices for crops of Punjab, rabi 2013–14. Punjab Agricultural University, Ludhiana, India.
Bansal M., Kaur S., Dhaliwal H., Chhuneja P., Bariana H. and Bansal U. 2016 Introgression of linked rust resistance genes Lr76 and Yr70 from Aegilops umbellulata to wheat chromosome 5DS. Plant Pathol. (http://dx.doi.org/10.1111/ppa.12549).
Chen X. M. 2005 Epidemiology and control of stripe rust (Puccinia striiformis f.sp. tritici) on wheat. Can. J. Plant Pathol. 27, 314–337.
Chen X. M. 2013 High-temperature adult-plant resistance, key for sustainable control of stripe rust. Am. J. Plant Sci. 4, 608–627.
Chhuneja P., Yadav B., Stirnweis D., Hurni S., Kaur S., Elkot A. F. et al. 2015 Fine mapping of powdery mildew resistance genes PmTb7A.1 and PmTb7A.2. In Triticum boeoticum (Boiss.) using the shotgun sequence assembly of chromosome 7AL. Theor. Appl. Genet. 128, 2099–2111.
Chhuneja P., Kaur S. and Dhaliwal H. S. 2016 Introgression and exploitation of biotic stress tolerance from related wild species in wheat cultivars. In Molecular breeding for sustainable crop improvement (ed. V. R. Rajpal, S. Rama Rao and S. N. Raina), pp. 269–324. Springer International Publishing, Switzerland.
IWGSC 2014 A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345, 1251788–1–1251788-11.
Jiang J., Friebe B. and Gill B. S. 1994 Recent advances in alien gene transfer in wheat. Euphytica 73, 199–212.
Jordan K. W., Wang S., Lun Y., Gardiner L. -J., MacLachlan R., Hucl P. et al. 2015 A haplotype map of allohexaploid wheat reveals distinct patterns of selection on homeologous genomes. Genome Biol. 16, 48–1-18.
Kuraparthy V., Chhuneja P., Dhaliwal H. S., Kaur S., Bowden R. L. and Gill B. S. 2007 Characterization and mapping of cryptic alien introgression from Aegilops geniculata with novel leaf rust and stripe rust resistance genes Lr57 and Yr40 in wheat. Theor. Appl. Genet. 114, 1379–1389.
Letunic I., Doerks T. and Bork P. 2012 SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 40, 302–305.
Liu W., Rouse M., Friebe B., Jin Y., Gill B. S. and Pumphrey M. O. 2011 Discovery and molecular mapping of a new gene conferring resistance to stem rust, Sr53, derived from Aegilops geniculata and characterization of spontaneous translocation stocks with reduced alien chromatin. Chromosome Res. 19, 669– 682.
Lorieux M. 2012 MapDisto: fast and efficient computation of genetic linkage maps. Mol. Breed. 30, 1231–1235.
McIntosh R. A., Yamazaki Y., Dubcovsky J., Rogers J., Morris C., Somers J. et al. 2013 Catalogue of gene symbols for wheat. In KOMUGI-integrated wheat science database at http://www.shigen.nig.ac.jp/wheat/komugi/genes/download.jsp.
Michelmore R. W., Paran I. and Kesseli R. V. 1991 Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 88, 9828–9832.
Peterson R. F., Campbell A. B. and Hannah A. E. 1948 A diagnostic scale for estimating rust severity on leaves and stem of cereals. Can. J. Res. 26, 496–500.
Riar A. K., Kaur S., Dhaliwal H. S., Singh K. and Chhuneja P. 2012 Introgression of a leaf rust resistance gene from Aegilops caudata to bread wheat. J. Genet. 91, 155–161.
Rosewarne G. M., Herrera-Foessel S. A., Singh R. P., Huerta-Espino J., Lan C. X. and He Z. H. 2013 Quantitative trait loci of stripe rust resistance in wheat. Theor. Appl. Genet. 126, 2427–2449.
Saghai-Maroof M. A., Soliman K. M., Jorgensen R. A. and Allard R. W. 1984 Ribosomal DNA spacer-length polymorphism in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. USA 81, 8014–8019.
Sears E. R. 1976 Genetic control of chromosome pairing in wheat. Annu. Rev. Genet. 10, 31–51.
Solovyev V., Kosarev P., Seledsov I. and Vorobyev D. 2006 Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 7, 1–10.
Somers D. J., Isaac P. and Edwards K. 2004 A high density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 109, 1105–1114.
Valkoun J., Hammer K., Kucerova D. and Bartos P. 1985 Disease resistance in the genus Aegilops L.—stem rust, leaf rust, stripe rust and powdery mildew. Kulturpflanze 33, 133–153.
Varshney R. K., Terauchi R. and McCouch S. R. 2014 Harvesting the promising fruits of genomics: applying genome sequencing technologies to crop breeding. PLoS Biol. 12, e1001883.
Voorrips R. E. 2002 Map Chart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78.
Xu L. S., Wang M. N., Cheng P., Kang Z. S., Hulbert S. H. and Chen X. M. 2013 Molecular mapping of Yr53, a new gene for stripe rust resistance in durum wheat accession PI 480148 and its transfer to common wheat. Theor. Appl. Genet. 126, 523–533.
Acknowledgements
Financial assistance provided by the USDA-ARS under the Project IN-ARS-842 and various grants from the Department of Biotechnology, Ministry of Science and Technology, Government of India, New Delhi, are gratefully acknowledged. The continuous supply of the rust inoculum from Regional Research Station, Indian Institute of Wheat and Barley Research is also gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Additional information
Corresponding editor: Arun Joshi
[Toor P. I., Kaur S., Bansal M., Yadav B. and Chhuneja P. 2016 Mapping of stripe rust resistance gene in an Aegilops caudata introgression line in wheat and its genetic association with leaf rust resistance. J. Genet. 95, xx–xx]
Rights and permissions
About this article
Cite this article
TOOR, P.I., KAUR, S., BANSAL, M. et al. Mapping of stripe rust resistance gene in an Aegilops caudata introgression line in wheat and its genetic association with leaf rust resistance. J Genet 95, 933–938 (2016). https://doi.org/10.1007/s12041-016-0718-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12041-016-0718-y