Abstract
Evidence suggests that long non-coding RNAs (lncRNAs) play a significant role in autism. Herein, we explored the functional role and possible molecular mechanisms of NEAT1 in valproic acid (VPA)-induced autism spectrum disorder (ASD). A VPA-induced ASD rat model was constructed, and a series of behavioral tests were performed to examine motor coordination and learning-memory abilities. qRT-PCR and western blot assays were used to evaluate target gene expression levels. Loss-and-gain-of-function assays were conducted to explore the functional role of NEAT1 in ASD development. Furthermore, a combination of mechanistic experiments and bioinformatic tools was used to assess the relationship and regulatory role of the NEAT1-YY1-UBE3A axis in ASD cellular processes. Results showed that VPA exposure induced autism-like developmental delays and behavioral abnormalities in the VPA-induced ASD rat model. We found that NEAT1 was elevated in rat hippocampal tissues after VPA exposure. NEAT1 promoted VPA-induced autism-like behaviors and mitigated apoptosis, oxidative stress, and inflammation in VPA-induced ASD rats. Notably, NEAT1 knockdown improved autism-related behaviors and ameliorated hippocampal neuronal damage. Mechanistically, it was observed that NEAT1 recruited the transcription factor YY1 to regulate UBE3A expression. Additionally, in vitro experiments further confirmed that NEAT1 knockdown mitigated hippocampal neuronal damage, oxidative stress, and inflammation through the YY1/UBE3A axis. In conclusion, our study demonstrates that NEAT1 is highly expressed in ASD, and its inhibition prominently suppresses hippocampal neuronal injury and oxidative stress through the YY1/UBE3A axis, thereby alleviating ASD development. This provides a new direction for ASD-targeted therapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Autism spectrum disorder (ASD) is a neurodevelopmental disorder characterized by a complex etiology and severe clinical symptoms. It manifests in deficits in social interaction and communication, as well as repetitive, stereotyped behaviors [1]. Additionally, co-morbidities such as narrow interests, intellectual disability, depression, and anxiety are often present. ASD typically emerges in early childhood and has lifelong, irreversible effects [2, 3]. The prevalence of ASD is increasing each year due to the advances in screening and diagnosis, with males being 8.4 times more likely to be diagnosed than females [4]. Most individuals with ASD struggle to adapt to society and live independently, and there is a notable lack of effective therapeutic drugs [5]. Therefore, exploring the etiological mechanisms of ASD and developing effective therapeutic interventions have become an urgent scientific priority.
Although ASD is strongly influenced by genetic factors, environmental factors such as toxicants, insecticides, infections, and drugs are also suggested to contribute to ASD susceptibility [6]. Exposure to valproic acid (VPA) during pregnancy has been shown to increase the risk of neurodevelopmental disorders in children, with ASD being the most commonly diagnosed [7]. Animal models exposed to VPA during pregnancy can exhibit behavioral characteristics similar to those observed in humans with ASD, making them validated models for studying idiopathic ASD induced by environmental factors [8, 9].
The brain contains various types of cells that form a complex network of connections, contributing to advanced neurological functions [10]. The hippocampus plays an essential role in learning and memory, including the formation of episodic and spatial memories and the spontaneous exploration of new environments [11, 12]. Damage to the hippocampus can cause memory impairment, affecting both memory formation and consolidation [13, 14]. Studies have indicated that interventions targeting the hippocampus can improve memory function [15, 16]. In addition to the characteristic impairment of social functioning, hippocampal-dependent cognitive function is also observed to be impaired in individuals with ASD [17]. Studies have indicated that the hippocampus not only supports memory and spatial reasoning but also plays a role in cognitive mapping that is closely related to social functioning, and ASD patients also display structural changes in the hippocampus [18, 19]. Therefore, the hippocampus is proposed as a valuable brain region associated with the ASD phenotype.
Long non-coding RNAs (lncRNAs) are a class of RNA molecules exceeding 200 bp in length without protein-coding potential [20, 21]. LncRNAs have emerged as novel regulators with important roles in brain developmental processes and the spatial and temporal control of cell fates [22, 23], including the pathogenic processes of ASD [24, 25]. Liu et al. [26] have demonstrated that lncRNA MEG3 exacerbates learning and memory impairment, along with hippocampal neuronal apoptosis, through the recruitment of EP300 to facilitate the expression of CDH2 in rat models of ASD. Jiang et al. [27] have found that lncRNA PVT1 expression is low in the peripheral blood of children with ASD, suggesting it as a promising diagnostic marker for ASD. Similarly, Luo et al. [28] revealed that lncRNA MSNP1AS is upregulated in ASD patients, promoting ASD progression through the regulation of moesin, which influences synapse structure and neuronal apoptosis. These studies highlight the important roles of lncRNAs in the pathogenesis of ASD.
Nuclear paraspeckle assembly transcript 1 (NEAT1) is a lncRNA located at the familial tumor syndrome multiple endocrine neoplasia (MEN) type 1 locus on chromosome 11q13.7 [29]. Prior studies have identified the involvement of NEAT1 in the pathogenesis of various neurodegenerative diseases. For example, NEAT1 is highly expressed in the substantia nigra of patients with Parkinson’s disease [30]. NEAT1 aggravates Aβ-induced neuronal damage by sponging miR-107 in Alzheimer’s disease [31]. Consistently, NEAT1 is shown to be overexpressed in the brains of the R6/2 transgenic Huntington’s disease mouse model [32, 33]. A recent study by Sayad et al. [34] demonstrated that NEAT1 is highly expressed in the peripheral blood of ASD patients. However, the physiological role and molecular mechanism of NEAT1 in ASD pathogenesis remain poorly understood.
In the present work, we explored the functional role of NEAT1 in ASD progression and the potential regulatory mechanisms, with the aim of providing potential molecular targets and research bases for early diagnosis and precision treatment.
Materials and Methods
Animals
Sprague–Dawley (SD) rats (200–250 g) were obtained from the Animal Care Center of the Chenzhou First People’s Hospital. The animals were given adequate food and water and were housed in a quiet environment with a 12-h light and dark cycle at 25 °C room temperature. All experimental procedures were performed under strict regulations for laboratory animal care of the institution and the study was approved by the Animal Ethics Committee of Chenzhou First People’s Hospital.
Model Establishment
The experimental design is shown in Fig. 1A. Adult female and male rats were acclimatized separately for 1 week and then caged together overnight in a 1:1 ratio. The next morning, female rats were tested for fertilization by vaginal pessary or vaginal smear. If pregnant, pregnant rats were housed separately and recorded as the first day of gestation (GD 1). The pregnant rats were randomly divided into the VPA and control groups. The rats in the VPA group were injected intraperitoneally with a single dose of 600 mg/kg of valproic acid (VPA, 50 mg/mL, dissolved in sterile saline) on gestational day 12.5 (GD 12.5), while rats in the control group received the same amount of sterile saline on the same day. The offspring of female rats in the VPA group were labeled as VPA-induced ASD rats, and the offspring of rats in the control group were labeled as control rats. All neonatal rats were weaned after 20 days. A total of 40 neonate rats were selected from each group for different experiments.
VPA-induced developmental delay, learning memory deficits, and enhanced neuronal damage in the rat model of ASD. A The workflow of experimental design of the animal study. GD, gestation day; PND, postnatal day. B Body weight of rats was monitored every week from postnatal days 7 to 35. C The eye-opening scores of rats were recorded from postnatal days 12 to 16. D Hang plate test was performed to assess the muscle strength and motor coordination of rats from postnatal days 7 to 11. E Swimming test was conducted to measure rat motor coordination on postnatal days 9, 11, 13, 15, and 17. F Morris water maze test to evaluate learning and memory from postnatal days 15 to 20. The data shown in B–E is representative of n = 3 experiments, and analysis was performed using two-way ANOVA followed by Bonferroni post hoc test. G TUNEL assay was performed to determine dorsal hippocampal apoptosis in VPA-induced ASD rats or control rats. H ELISA was used to examine SOD activity, GSH, and MDA contents in rat dorsal hippocampus tissues on postnatal day 35. I IHC assay was used to detect 4-HNE and 3-NT levels in rat dorsal hippocampus tissues on postnatal day 35. J ELISA to measure TNF-α, IL-1β, and IL-6 levels in dorsal hippocampal tissues on postnatal day 35. The data shown in F–J is representative of n = 3 experiments, and analysis was performed using an unpaired t-test. *P < 0.05, **P < 0.01, ***P < 0.001
To explore the impact of NEAT1 knockdown on VPA-induced ASD rats, adenoviruses carrying short hairpin RNA targeting NEAT1 (sh-NEAT1) or negative control (sh-NC) were administered into the uterus of pregnant female rats (n = 6 per group) after the appearance of the vaginal plug on GD 1, and then rats received ASD treatment on GD 12.5 according to the procedure described above. The offspring of rats injected with adenoviruses were labeled as VPA-induced ASD rats in the sh-NEAT1 or sh-NC groups, respectively. Ten pups in each group were randomly selected for the behavior test. Finally, the pups were sacrificed with sodium pentobarbital (200 mg/kg) before brain tissues were harvested on postnatal day 35.
Observation of Physiological Indicators
The number of litters of pregnant rats after delivery was recorded, and the developmental indicators, including eye-opening time as well as body weights, were monitored. The body weights of the litter were measured on the 7th, 14th, 21st, 28th, and 35th postnatal days, and the eye-opening scores were measured daily at 17:00 from postnatal days 12 to 16. The scoring criteria for the eye-opening time were as follows as reported previously [26]; both eyes were not opened (0 score), only one eye appeared open (1 score), and both eyes appeared open (2 scores).
Behavioral Tests
Direction Ability Test
An orientation test was performed on pups on days 7–11 after birth. Pups were placed on a smooth 25°-tilting plate with their heads downwards. Rats were observed rotating 180°, and their time spent rotating was recorded and compared. Rats exhibited an innate tendency to spontaneously rotate 180° with their heads up when positioned with their heads down on the tilting plate. The rotation time indicated the development of vestibular sensibility and motor function in rats [35].
Swimming Test
A swimming test was conducted on the 9th, 11th, 13th, 15th, and 17th day after birth to test the coordination capability. The pups were placed individually in a thermostatic water tank at 28 °C for a 5–10 s observation period. Their swimming ability was graded and recorded based on the position of their head, nose, and ears. The scoring criteria [26] were as follows: head and nose both underwater, 0 points; nose underwater, head on top of water, 1 point; head and nose both on top of water, ears underwater, 2 points; nose and head out of water, water level between the two ears, 3 points; nose and head out of water, water level lower than the ears, 4 points.
Morris Water Maze (MWM) Test
The MWM test was performed to assess impairment in learning and memory [36]. The MWM apparatus consisted of a cylindrical bucket with a diameter of 140 cm, a height of 50 cm, and a station with a diameter of 7 cm. The bucket was filled with water (24 ± 2 °C) to a depth of 30 cm. The water surface of the pool was divided into 4 quadrants by the two mutually perpendicular diameters of the bucket, labeled as quadrants I, II, III, and IV. In the center of the II-IV quadrant, a black circular platform with a diameter of 10 cm was consistently positioned 2 cm below the water’s surface in all trials. The rats were given a maximum of 60 s (cut-off time) to find the hidden platform and then permitted to stay on it for 30 s. If the rat was unable to identify the hidden platform within 60 s, it was gently directed to it and permitted to remain there for 30 s. Each rat underwent three trials every day for 5 days in a row. The time taken to locate the platform served as the escape latency and was used as an index of learning [36, 37].
Immunohistochemistry (IHC) Assay
The brain tissues were fixed in 4% paraformaldehyde solution overnight and then rinsed with running water for 24 h. Next, the brain tissues were dehydrated with gradient alcohol, cleared with xylene, embedded in paraffin, and then cut into 4–6-µm-thick sections. The sections were then deparaffinized with xylene I and II solution for 10 min, incubated with 3% H2O2 solution for 25 min, and then incubated with anti-4-hydroxynonenal (4-HNE) or 3-nitrotyrosine (3-NT) antibodies overnight at 4 °C. Next morning, the sections were washed with PBS and then added horseradish peroxidase-labeled secondary antibody, and the sections were incubated at room temperature for 1 h. After washing with PBS, diaminobenzidine (DAB) chromogenic solution was added dropwise, and the sections were incubated at room temperature for 10 min. Thereafter, the sections were immersed in hematoxylin, followed by ethanol dehydration, cleared with xylene, mounted with neutral resin, and then observed under a light microscope (DMI8, Leica). A total of 5 visual fields were selected in each section, and the percentage of positive staining was measured using lmageJ software.
Terminal Deoxynucleotidyl Transferase-Mediated dUTP Nick End Labeling (TUNEL) Detection Assay
The apoptosis was assessed using the TUNEL apoptosis detection kit (C1091, Beyotime, Shanghai, China) in accordance with the manufacturer’s guidelines. The xylene-soaked paraffin sections were dehydrated in graded ethanol, and then, 50 µL of TUNEL reaction mixture was applied and incubated for 1 h at 37 °C. Thereafter, DAPI was added for 10 min to stain the nucleus. The sections were then sealed, and the apoptosis was observed by fluorescence microscope in rat hippocampal tissues. Finally, 5 visual fields from each section were randomly chosen to calculate the TUNEL-positive cells using ImageJ image analysis software, and the apoptosis rate was calculated.
Isolation and Transfection of Primary Neuronal Cells
Hippocampal neuronal cells were isolated by referring to the method as previously reported [26]. Briefly, primary neurons were extracted from five hippocampal tissues of rats in the ASD group. Then, 0.25% trypsin was added to digest hippocampal tissues at 37 °C for 25 min, and the digestion was terminated with 20% fetal bovine serum (FBS). Subsequently, the digested tissues were filtered through a 200-mesh filter and centrifuged at 1000 r/min for 5 min, and the supernatant was discarded. The pelleted cells were resuspended in DMEM/F12 medium containing 10% FBS and incubated at 37 °C in 5% CO2. After 6 h, the medium was replaced with Neurobasal medium containing 10% FBS, 2% B27, and 1% L-glutamine. The medium was changed every 3 days in half volume.
For the transfection, neurons were inoculated into 6-well plates and transfection was performed according to the Lipofectamine 2000 (ThermoFisher Scientific, USA) following the manufacturer’s protocol. Hippocampal neuronal cells were transfected with sh-NC, sh-UBE3A, sh-YY1, and sh-NEAT1 vectors. Also, hippocampal neuronal cells were transfected with oe-vector, oe-UBE3A, oe-YY1, and oe-NEAT1 overexpression plasmids, respectively. The cells were then cultured for 48 h at 37 °C with 5% CO2. After 48 h of transfection, cells were collected and used for various assays.
Cell Viability Assay
Cell viability was assessed by CCK-8 assay. Briefly, the transfected hippocampal neuronal cells were inoculated into 96-well plates and cultured for 24 h. Then, 10 µL of CCK⁃8 working solution was added to the wells and incubated at 37 °C for 2 h. Finally, the cell viability was calculated by using a microplate reader to read the optical density (OD) value at 450 nm.
Flow Cytometry
Apoptosis was evaluated by flow cytometry assay. Harvested neuronal cells were fixed with 70% ethanol at 4 ℃ overnight. After rinsing with PBS, cells were resuspended in binding buffer containing propidium iodide (PI) and AnnexinV-FITC solution (Sigma-Aldrich) for 30 min at room temperature in the dark. Cells were then washed and acquired on BD FACS sorter (BD FACSVerse cytometer). The rate of apoptosis was assessed using FlowJo software.
qRT-PCR
Total RNA was extracted from the rat hippocampal tissue homogenates or cells using TRIzol reagent (Yeasen, Shanghai, China), and then reverse transcribed to cDNA using the PrimeScript (Takara). The PCR reaction was performed in triplicate using a 7500 Real-Time PCR System with SYBR Premix Ex Taq II. The PCR amplification was carried out under the conditions of 95 °C for 10 min, 95 °C for 10 s, 58 °C for 30 s, and 72 °C for 30 s for a total of 40 cycles. The relative expression of target genes was calculated by 2−ΔΔCt method.
Western Blot
Total protein was extracted from rat hippocampal tissue homogenates or cells using RIPA buffer containing protease and phosphatase inhibitors (Selleckchem, China) and quantified by BCA kit (Bio-Rad, USA). Following this, 40 µg proteins were separated using 12% SDS-PAGE gel and wet transferred to PVDF membranes. The membranes were then blocked with TBS containing 5% BSA solution for 2 h at room temperature. Thereafter, membranes were incubated with primary antibodies overnight at 4 °C prior. Next morning, the membranes were washed with T-BST and then incubated with horseradish peroxidase-labeled secondary antibody at room temperature for 1 h. Finally, the ECL Plus Reagent (Merck Millipore, Germany) was applied to visualize the protein bands and the ImageJ software was used for quantification.
Oxidative Stress Marker Measurement
Glutathione (GSH), malondialdehyde (MDA) contents, and superoxide dismutase (SOD) activity in rat brain tissues or cell supernatants were measured using relevant commercially available kits. Briefly, the samples were carefully washed three times with pre-cooled PBS, and then lysate was added and fully lysed on ice for 20 min. Thereafter, the samples were centrifuged at 12,000 × g for 10 min at 4 °C, and the supernatants were collected. The GSH, MDA, and SOD levels were measured following the manufacturer’s protocols (Nanjing Jiancheng Bioengineering Institute). Finally, the optical density (OD) of the samples was measured at 450 nm using an automated microplate reader (SpectraMax® M5).
Cellular reactive oxygen species (ROS) contents were monitored using a DCFH-DA fluorescent probe. The harvested cells were incubated with 2 µmol/L DCFH-DA dye for 30 min, and the fluorescence intensity of ROS was observed under a fluorescence microscope.
Inflammation Factor Detection
Supernatants from rat brain tissue or cell cultures were collected and stored at − 20 °C. IL-1β, TNF-α, and IL-6 levels were quantified using commercially available ELISA kits (ThermoFisher Scientific, Waltham, MA, USA) following the manufacturer’s protocols.
RNA Binding Protein Immunoprecipitation (RIP) and Chromatin Immunoprecipitation (ChIP) Assays
For RIP assay, cells underwent precooled PBS washing, lysis with a matched amount of lysate in an ice bath for 5 min, and then centrifuged at 4 °C for 10 min to obtain the supernatants. One part of the cell supernatant was taken out as control (input), and the other part was mixed with antibodies for co-precipitation. For each co-precipitation reaction system, 50 µL of magnetic beads were washed and resuspended in 100 µL of RIP washing buffer. The antibodies used for RIP were YY1 antibody, and rabbit IgG was used as a negative control. Each system was added 5 µg of antibody, mixed well, and processed at room temperature for 30 min. The magnetic bead-antibody complexes were then re-suspended in RIP washing buffer, followed by the addition of 100 μL cell extract and an overnight incubation at 4 °C. The harvested magnetic bead-protein complexes were digested with proteinase K for RNA extraction and subsequent qRT-PCR assay.
For ChIP assay, chromatin was processed into 200–800 bp fragments by sonication. ChIP experimental steps were performed according to the EZ-ChIP kit (Millipore, USA).
Subcellular Localization
Using a Cytoplasmic and Nuclear RNA Purification Kit (Norgen Biotek, USA), cytoplasmic and nuclear extracts were separated. The cytoplasmic endogenous control employed was B-actin, while the nuclear endogenous control used was U6. The distribution of RNA (U6, Β-actin, and NEAT1) in the nucleus and cytoplasm of hippocampal neuronal cells was measured using RT-qPCR.
Dual Luciferase Reporter Gene Assay
The pGL3-NC, pGL3-UBE3A-promoter, vector, and YY1 overexpression plasmids were co-transfected into neuronal cells, which were labeled as pGL3-NC + vector group, pGL3 + YY1 group, pGL3-UBE3A-promoter + vector group, and pGL3-UBE3A-promoter + YY1 group, respectively. Following a 48-h incubation period, luciferase activities were assayed utilizing the dual luciferase reporter system according to the manufacturer’s instructions.
Statistical Analysis
All statistical analyses were performed using GraphPad Prism 9 software, and data are presented as mean ± SD from at least three independent experiments. Statistical analysis between two groups was performed utilizing the unpaired Student’s t-test, and comparisons among multiple groups were performed using one-way ANOVA with the Tukey-Kremer post hoc test or two-way ANOVA with the Bonferroni post hoc test. A statistical difference was considered at P < 0.05.
Results
VPA-Induced Developmental Delay, Learning and Memory Deficits, and Neuronal Injury in Rat Model of ASD
Firstly, we monitored the body weight of rats on postnatal days 7, 14, 21, 28, and 35, and the results are shown in Fig. 1B. Both groups of littermates gained weight rapidly with time, but the weight of VPA-induced ASD rats was markedly lower compared to the control group rats on days 28 (P = 0.023) and 35 (P = 0.015). Meanwhile, the eye-opening of the littermates was observed and recorded on days 12–16 after birth. The eye-opening scores of both groups of rats showed an increasing trend with time (Fig. 1C), whereas on day 12, the eyes of both groups of rats remained closed. The eye-opening scores of the VPA-induced ASD rats were remarkably lower compared to the control rats on day 13 (P = 0.03) and day 14 (P = 0.012), meaning that fewer of the VPA-induced ASD rats opened their eyes. On day 16, both eyes were fully open in both groups of rats. The results of the orientation tendency test revealed that VPA-induced ASD rats took more time to complete the flip than the normal control rats on days 7 (P = 0.006), 8 (P = 0.002), 9 (P = 0.004), and 10 (P = 0.011) (Fig. 1D). Moreover, the swimming ability of VPA-induced ASD rats was remarkably reduced compared to the control rats on day 9 (P = 0.006), day 11 (P = 0.008), and day 15 (P = 0.046) (Fig. 1E), indicating that VPA-induced ASD rats had poorer motor coordination response. Similarly, the escape latency of VPA-induced ASD rats was longer relative to control rats on day 1 (P = 0.048), day 3 (P = 0.017), day 4 (P = 0.046), and day 5 (P = 0.006) (Fig. 1F), indicating the aggravation of learning and memory injury. Furthermore, apoptotic cells were remarkably increased in the hippocampal tissues of VPA-induced ASD rats compared to the control rats (P = 0.0002) (Fig. 1G). VPA exposure markedly decreased SOD activity (P = 0.0001) as well as reduced GSH content (P < 0.0001) in the hippocampal tissues, whereas it increased MDA content (P < 0.0001) (Fig. 1H) and 4-HNE (P = 0.0001) and 3-NT levels (P < 0.0001) (Fig. 1I). Meanwhile, VPA exposure notably increased TNF-α (P < 0.0001), IL-1β (P < 0.0001), and IL-6 (P < 0.0001) levels in rat hippocampal tissues (Fig. 1J). The above results indicated that neuronal apoptosis, oxidative stress, and inflammatory responses were aggravated in VPA-induced ASD rats.
NEAT1 Knockdown Suppressed Hippocampal Neuronal Cell Injury, Oxidative Stress, and Inflammation Response
Next, we evaluated the expression level of NEAT1, and we found that NEAT1 levels were notably elevated in hippocampal tissues of VPA-induced AD rats (P < 0.0001) (Fig. 2A). As NEAT1 was highly expressed in VPA-induced ASD rats, we hypothesize that NEAT1 may contribute to ASD pathogenesis. To explore the functional role of NEAT1, we conducted loss-and-gain-of-function assays. For this purpose, sh-NC, sh-NEAT1, over-expressed empty vector (oe-vector), or over-expressed NEAT1 (oe-NEAT1) vectors were transfected into primary hippocampal neurons and confirmed the transfection efficiency using qRT-PCR (P < 0.01) (Fig. 2B). Functional assays demonstrated increased cell viability (P = 0.013) as well as decreased apoptosis (P = 0.008) in hippocampal neurons after NEAT1 knockdown, whereas NEAT1 overexpression decreased cell viability (P = 0.002) and promoted apoptosis (P = 0.012) (Fig. 2C, D). Furthermore, NEAT1 inhibition markedly decreased cellular ROS (P = 0.007) and MDA (P = 0.002) levels while increasing SOD (P = 0.028) and GSH contents (P = 0.0001) (Fig. 2E, F). Similarly, NEAT1 inhibition significantly decreased TNF-α (P = 0.019), IL-1β (P = 0.017), and IL-6 (P = 0.015) levels (Fig. 2G). In contrast, NEAT1 overexpression showed the opposite effects, with elevating ROS (P = 0.003) and MDA (P = 0.008) levels and reduced SOD (P = 0.038) and GSH (P = 0.002) contents, as well as elevated TNF-α (P = 0.0005), IL-1β (P = 0.007), and IL-6 (P = 0.003) levels. These findings suggest that NEAT1 inhibition results in ameliorating hippocampal neuronal cell injury, oxidative stress, and inflammation response in ASD.
NEAT1 knockdown attenuated hippocampal neuronal injury. A qRT-PCR was used to detect NEAT1 mRNA levels in the hippocampal tissues of VPA-induced ASD and normal control rats on postnatal day 35. B qRT-PCR was performed to examine the transfection efficiency of NEAT1 knockdown/overexpression in primary neuronal cells isolated from VPA-induced ASD rats. C CCK-8 assay was conducted to assess the cell viability of primary hippocampal neurons after culturing for 24 h in the indicated groups. D Flow cytometry was performed to evaluate cell apoptosis of primary hippocampal neurons in indicated groups. E DCFH-DA fluorescent probe assay was conducted to measure cellular ROS levels in primary hippocampal neurons after indicated transfection. Commercially available kits were used to measure F SOD activity, GSH, and MDA contents in the primary hippocampal neuronal cells. ELISA was used to quantify the levels of G TNF-α, IL-1β, and IL-6 in the supernatants of cultured neuronal cells. Each experiment was repeated at least three times (n = 3), and data was analyzed using one-way ANOVA with the Tukey-Kremer post hoc test. *P < 0.05, **P < 0.01, ***P < 0.001
NEAT1 Knockdown Ameliorated Learning Memory Deficits and Neuronal Injury in VPA-Induced ASD Rats
To further confirm the functional role of NEAT1 in ASD pathogenesis, we injected adenovirus carrying sh-NC or sh-NEAT1 into gestational rats. On postnatal day 13, some sh-NEAT1 rats started their eyes opening (P = 0.04), whereas sh-NC rats did not yet; by days 14 and 15, sh-NEAT1 rats consistently displayed more eye-opening tendency compared to sh-NC rats (P = 0.01); and by day 16, both sh-NC and sh-NEAT1 rats’ eyes remained open (Fig. 3A). Similarly, sh-NEAT1 rats took remarkably less time to complete the flip and had enhanced swimming ability, indicating improved motor coordination response (P < 0.05) (Fig. 3B, C). Furthermore, sh-NEAT1 rats experienced a remarkable reduction in escape latency on day 4 (P = 0.034) and day 5 (P = 0.006) (Fig. 3D), indicating improved spatial learning and memory deficits. In addition, apoptosis levels were notably reduced in the hippocampal tissues of sh-NEAT1 rats compared to the sh-NC group rats (P = 0.005) (Fig. 3E). Moreover, NEAT1 knockdown markedly enhanced SOD (P = 0.004) and GSH (P = 0.002) contents and lowered MDA (P = 0.003) levels (Fig. 3F). Subsequently, significantly reduced pro-inflammatory cytokine levels such as TNF-α (P = 0.003), IL-1β (P < 0.001), and IL-6 (P = 0.006) were measured upon NEAT1 knockdown when compared to the sh-NC group (Fig. 3G). These results confirm that targeting NEAT1 can ameliorate learning memory deficits and neuronal injury in VPA-induced ASD rats.
NEAT1 knockdown ameliorated learning memory deficits, neuronal apoptosis, oxidative stress, and inflammatory responses in VPA-induced ASD rats. A Recording of eye-opening scores from postnatal days 12 to 16 in each group of rats. B Hang plate test was used to assess muscle strength and motor coordination from postnatal days 7 to 11. C Swimming test was performed to measure rat motor coordination on postnatal days 9, 11, 13, 15, and 17. D Morris water maze test was conducted to evaluate learning and memory from postnatal days 15 to 20. The data shown in A–D is representative of n = 3 experiments, and analysis was performed using two-way ANOVA followed by Bonferroni post hoc test. E TUNEL assay was used to examine apoptosis in hippocampal tissue on postnatal day 35. F Commercially available kits were used to detect SOD activity, GSH, and MDA contents in rat hippocampal tissue on postnatal day 35. G TNF-α, IL-1β, and IL-6 levels in rat hippocampal tissue on postnatal day 35 were measured using ELISA kits. Each experiment was repeated at least three times (n = 3), and data was analyzed using an unpaired t-test. *P < 0.05, **P < 0.01, ***P < 0.001
UBE3A Was Regulated by NEAT1 to Promote Hippocampal Neuronal Injury
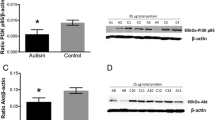
Previously, it has been reported that UBE3A expression is high in ASD patients [38, 39]; therefore, we hypothesized that NEAT1 may mediate its function via regulating UBE3A expression. To this end, we performed qRT-PCR and western blot quantification of UBE3A in the hippocampal tissues of control and VPA-induced ASD rats. We found that the expression levels of UBE3A mRNA and protein in the hippocampal tissues of VPA-induced ASD rats were markedly enhanced compared to the control group rats (P < 0.001) (Fig. 4A, B). Then, we analyzed the correlation between NEAT1 and UBE3A expression in hippocampal tissues by GEPIA database and found a remarkable positive correlation between them (Fig. 4C). Biological assays further confirmed that NEAT1 knockdown markedly decreased UBE3A mRNA (P = 0.007) and protein (P = 0.001) expression levels, while NEAT1 overexpression greatly increased UBE3A mRNA (P = 0.017) and protein (P = 0.005) expression (Fig. 4D, E). This data suggests that NEAT1 regulates UBE3A expression.
UBE3A was highly expressed in hippocampal tissues of VPA-induced ASD rats and regulated by NEAT1. A qRT-PCR and B western blot assays were performed to detect UBE3A expression levels in hippocampal tissues of VPA-induced ASD and control rats on postnatal day 35. The data is representative of n = 3 experiments, and analysis was performed using an unpaired t-test. C The GEPIA database was used to assess the correlation analysis between NEAT1 and UBE3A in human hippocampal tissues. D qRT-PCR and E western blot were conducted to measure UBE3A expression levels in primary hippocampal neurons of the indicated groups. Each experiment was repeated at least three times (n = 3), and data was analyzed using one-way ANOVA with the Tukey-Kremer post hoc test. *P < 0.05, **P < 0.01, ***P < 0.001
We then explored the impacts of UBE3A knockdown/overexpression upon hippocampal neuronal cell viability, apoptosis, oxidative stress, and inflammation processes through a series of cell function assays. We transfected sh-NC, sh-UBE3A, oe-vector, or oe-UBE3A vectors into primary hippocampal neurons and then checked the transfection efficiency utilizing qRT-PCR (P < 0.01) (Fig. 5A). Neuronal viability was notably enhanced after UBE3A knockdown (P = 0.02) (Fig. 5B), and apoptotic cells (P = 0.008) were markedly reduced (Fig. 5C). In contrast, UBE3A overexpression severely reduced neuronal cell viability (P = 0.006) and induced apoptosis (P = 0.009). Subsequently, UBE3A knockdown notably increased SOD (P = 0.022) activity and GSH (P = 0.006) content, while decreased ROS (P = 0.001) and MDA (P = 0.005) levels (Fig. 5D, E). Furthermore, UBE3A knockdown also decreased TNF-α (P = 0.008), IL-1β (P = 0.006), and IL-6 (P = 0.02) levels (Fig. 5F). In contrast, UBE3A overexpression markedly decreased SOD (P < 0.001) and GSH (P = 0.002) contents, while increased ROS (P = 0.007), MDA (P = 0.002), TNF-α (P = 0.003), IL-1β (P = 0.008), and IL-6 (P < 0.001) levels (Fig. 5D–F). Collectively, this data indicates that NEAT1 regulates UBE3A that in turn participates in hippocampal neuronal injury.
UBE3A mitigated hippocampal neuronal injury. A qRT-PCR was used to detect the transfection efficiency of sh-UBE3A or oe-UBE3A. B CCK-8 assay was performed to assess the cell viability of primary hippocampal neurons after UBE3A knockdown or overexpression. C Flow cytometry was used to evaluate cell apoptosis of primary hippocampal neurons in indicated groups. D DCFH-DA fluorescent probe was used to examine cellular ROS levels in primary hippocampal neurons after transfection in the indicated groups. E Commercially available kits were used to measure SOD activity, GSH, and MDA contents in primary hippocampal neurons in indicated groups. F TNF-α, IL-1β, and IL-6 levels in the supernatants of primary hippocampal neurons in indicated groups were measured using ELISA kits. Each experiment was repeated at least three times (n = 3), and data was analyzed using one-way ANOVA with the Tukey-Kremer post hoc test. *P < 0.05, **P < 0.01, ***P < 0.001
NEAT1 Recruited YY1 to Regulate UBE3A Expression
To investigate the potential molecular mechanism of NEAT1 in ASD, we predicted the subcellular localization of NEAT1 by LncATLAS and lncLocator databases and discovered that NEAT1 was predominantly localized in the nucleus (Fig. 6A, B). We also confirmed this by cellular nucleoplasmic separation assay (Fig. 6C). Therefore, we hypothesized that NEAT1 might regulate UBE3A expression by recruiting a certain transcription factor and thereby function in ASD development. RPISeq and hTFtarget databases predicted a potential interaction between NEAT1 and the transcription factor YY1 (Fig. 6D, E). Furthermore, the GEPIA database indicated a strong positive correlation between UBE3A and YY1 (Fig. 6F). Subsequently, RIP (P = 0.004) as well as RNA pull-down assays further supported the binding between NEAT1 and YY1 (Fig. 6G, H).
NEAT1 bound to transcription factor YY1. A LncATLAS and B lncLocator databases were used to predict the subcellular localization of NEAT1. C qRT-PCR was used to detect NEAT1 mRNA abundance in isolated nuclei and cytoplasm fractions. D RPISeq database was used to predict the interactions between NEAT1 and YY1. E hTFtarget database was used to predict the potential regulation of UBE3A and transcription factor YY1. F The GEPIA database was used to predict the expression correlation between UBE3A and YY1 in the human hippocampus using Spearman correlation analysis. G RIP assay was conducted to determine the binding between NEAT1 and YY1 in primary hippocampal neurons. H RNA pull-down assay was performed to determine the interaction between NEAT1 and YY1 in primary hippocampal neurons. Each biological experiment was repeated at least three times (n = 3). **P < 0.01
To clarify the regulatory pattern between NEAT1-YY1-UBE3A, we conducted rescue experiments. YY1 knockdown remarkably decreased UBE3A mRNA and protein expression levels (P < 0.001), while YY1 overexpression significantly increased UBE3A mRNA and protein expression levels (P = 0.002) (Fig. 7A, B), suggesting the interaction between YY1 and UBE3A. To confirm that YY1 bound to the UBE3A promoter region, a ChIP assay was performed, which demonstrated the presence of UBE3A promoter enrichment in the YY1 antibody immunoprecipitated DNA complexes compared to the control (P < 0.001) (Fig. 7C), confirming that YY1 binds to the UBE3A promoter region. Subsequently, the results of dual luciferase reporter gene assay displayed that luciferase activity was markedly elevated after YY1 overexpression (P < 0.001), while there was no notable change in luciferase activity in the pGL3-UBE3A-promoter + vector group (P = 0.969) (Fig. 7D). In addition, knockdown of NEAT1 notably suppressed the enrichment of YY1 at the UBE3A promoter site, while overexpressing NEAT1 markedly enhanced the enrichment of YY1 (P < 0.001) (Fig. 7E). Taken together, these findings reveal that NEAT1 recruits YY1 to regulate UBE3A expression.
NEAT1 bound to YY1 to enhance UBE3A expression. A qRT-PCR and B western blot assays were conducted to detect UBE3A expression levels in rat primary hippocampal neurons with YY1 knockdown or overexpression. C ChIP assay was used to examine the binding ability of YY1 to the UBE3A promoter in primary hippocampal neurons. D Dual luciferase reporter gene assay was performed to test UBE3A promoter activity in primary hippocampal neurons. E ChIP assay was used to measure the relative UBE3A promoter enrichment level under NEAT1 knockdown/overexpression in primary hippocampal neurons. Each experiment was repeated at least three times (n = 3), and data was analyzed using either an unpaired t-test or two-way ANOVA followed by a Bonferroni post hoc test. **P < 0.01, ***P < 0.001
NEAT1 Induced Hippocampal Neuronal Cell Injury via UBE3A
Finally, we verified the regulatory function of the NEAT1-UBE3A axis in the hippocampal neuronal cell injury by conducting rescue experiments. We co-transfected primary hippocampal neuron cells with either sh-NC, sh-NEAT1 alone, or sh-NEAT1 together with UBE3A overexpressing vector. Data demonstrated that sh-NEAT1 induced an increase in cell viability (P < 0.001), which was partially suppressed by UBE3A overexpression (P = 0.002) (Fig. 8A). We also found that sh-NEAT1 showed an inhibitory effect on apoptosis (P = 0.003), which was restored by overexpressing UBE3A (P = 0.002) (Fig. 8B). Similarly, UBE3A overexpression reversed sh-NEAT1-induced increase in SOD (P = 0.031) and GSH (P < 0.001) contents, as well as reductions in ROS (P < 0.001), MDA (P < 0.001), TNF-α (P = 0.006), IL-1β (P = 0.004), and IL-6 (P = 0.015) levels (Fig. 8C–E). Collectively, these results indicate that NEAT1 mediates hippocampal neuronal cell injury via regulating UBE3A expression.
NEAT1 regulated hippocampal neuronal cell viability, apoptosis, oxidative stress, and inflammatory response through UBE3A. Hippocampal neuronal cells were divided into three groups: sh-NC, sh-NEAT1, and sh-NEAT1 + UBE3A. A CCK-8 assay was used to assess the cell viability of primary hippocampal neurons after indicated transfection. B Flow cytometry was performed to measure the apoptosis rate of primary hippocampal neurons in indicated groups. C DCFH-DA fluorescent probe assay was performed to detect cellular ROS levels of primary hippocampal neurons in indicated groups. D Commercially available kits were used to measure SOD activity, GSH, and MDA contents of primary hippocampal neurons in indicated groups. ELISA was used to measure E TNF-α, IL-1β, and IL-6 levels in the cell supernatants of primary hippocampal neurons in indicated groups. Each experiment was repeated at least three times (n = 3), and data was analyzed using one-way ANOVA with Tukey-Kremer post hoc. *P < 0.05, **P < 0.01, ***P < 0.001
Discussion
As a class of widespread neurodevelopmental disorders, the incidence of ASD is increasing annually, attracting growing attention and research from the neuroscience field in recent years. The etiology of ASD is uncertain, and there are no effective treatment options; therefore, research on the etiology and pathogenesis of ASD is particularly essential. ASD is generally regarded as a disorder stemming from neurodevelopmental abnormalities that lead to brain dysfunction. With the advances in CT and MRI technologies, many studies have identified certain central nervous system developmental features that are unique to children with ASD, such as overgrowth of the cortex and limbic system in the early stages of neurodevelopment, followed by slower or even stagnant growth in later stages [40, 41]. These disturbances in neurodevelopment and the resulting abnormalities in neural circuits may play an important role in the eventual development of ASD [42].
VPA is an antiepileptic drug, and mothers who take large amounts of VPA prenatally have greatly increased chances of their offspring developing ASD [8]. Mid-pregnancy intraperitoneal injection of VPA in pregnant rats interferes with the neurodevelopment of the offspring and has become a widely used classical animal model of ASD. Its offspring rats showed autism-related neurobehavioral manifestations such as social impairment, repetitive stereotyped behaviors, and interest preference disorder. Altered neurotransmitters and protein expression, abnormalities in synaptic number, and dendritic spine density in the brain of offspring rats were also observed [43, 44]. Previous studies have mainly focused on VPA-induced alterations in brain regions, such as the prefrontal cortex and cerebellum, in rat models of ASD [45,46,47]. Consistent with these studies, we also found poor development, impaired motor coordination, and defects in spatial learning and memory in VPA-induced ASD rats. A previous study has shown that individuals with autism often have restricted physical activity, delayed motor abilities, and reduced physical fitness [48]. Additionally, we explored hippocampal apoptosis in rats from both the control and ASD groups, demonstrating that the number of apoptotic cells increased in the hippocampal tissues of VPA-induced ASD rats compared to control rats. This finding indicates that the neurotoxicity of VPA is associated with apoptotic neurodegeneration in the developing rat hippocampus.
Inflammatory and immune system disorders are apparently involved in the etiopathogenesis of ASD [49]. Analysis of pro-inflammatory indicators has revealed increased inflammatory activity in ASD children. Previous studies have reported elevated levels of pro-inflammatory cytokines such as IL-1β, IL-6, IL-17, and TNF-α in the plasma of ASD patients [49,50,51]. Similarly, ASD is characterized by elevated levels of ROS or free radicals and a decrease in the antioxidant enzymes/substances levels [52, 53]. Accordingly, in our study, we found significantly increased levels of pro-inflammatory cytokines such as IL-1β, IL-6, and TNF-α in the VPA-induced ASD rats. Likewise, we also found prominently high 4-HNE, 3-NT, ROS, and MDA levels while reduced SOD and GSH contents in VPA-induced ASD rats, thus confirming the existence of a relationship between oxidative stress, neuroinflammation in the hippocampus, and ASD in neonate rats [54, 55].
NEAT1, a long non-coding RNA (lncRNA), is reported to be highly expressed and exerts an essential role in neurological diseases such as sepsis-related encephalopathy [56], Alzheimer’s disease [57], Parkinson’s disease [58], and ischemic stroke [59]. In this study, we successfully established a VPA-induced ASD model and discovered that NEAT1 was notably hyper-expressed in ASD, consistent with the findings of Sayad et al. [34]. The role of NEAT1 in ASD was determined for the first time. We injected a NEAT1 knockdown adenovirus into pregnant rats to obtain NEAT1 knockdown VPA-induced ASD rats. Our results revealed that down-regulated NEAT1 ameliorated developmental delays and learning and memory deficits and attenuated oxidative stress and inflammation in the hippocampus of VPA-induced ASD rats. Furthermore, a series of cell function assays demonstrated that sh-NEAT1 enhanced the viability of hippocampal neurons and effectively reduced apoptosis, oxidative stress, and inflammatory responses. Interestingly, the knockdown of NEAT1 has been reported to decrease cell viability and promote oxidative stress in Parkinson’s disease [30]. This difference may be due to the different rodent models used and the potential dual roles of NEAT1 in regulating oxidative stress under specific contexts. Notably, NEAT1 displayed a predominantly nuclear distribution, which is in agreement with previous reports [60, 61].
LncRNAs are reported to function in cellular and developmental biology by recruiting transcription factors to regulate downstream target genes [62]. For instance, LINC01133 is shown to resist ferroptosis in pancreatic cancer cells by the recruitment of FUS to enhance FSP1 mRNA stability [63]. LINC02454 positively regulates HMGA2 transcription by promoting CREB1 phosphorylation and nuclear translocation, thereby facilitating the malignant progression of thyroid cancer [64]. LncRNA NKILA exacerbates neuronal injury and oxidative stress in Alzheimer’s disease through FOXA1-mediated TNFAIP1 transcription [65]. Based on these findings, we speculated that NEAT1 might mediate its function by recruiting transcription factors. A previous study conducted by Rahman et al. [66] identified 1567 differentially expressed genes in brain tissues using RNA-Seq data from ASD patients and healthy controls in the GSE64018 and GSE30573 datasets and identified several transcription factors, including Yin-Yang 1 (YY1). YY1 is a versatile transcription factor that is ubiquitously expressed in various cells of the nervous system, especially neurons, and can act as both a transcriptional activator and repressor depending on its chromatin structure, modifications, and target genes [67]. YY1 dysregulation has been linked to several neurodegenerative conditions, including Alzheimer’s disease, Parkinson’s disease, and Rett syndrome [68,69,70,71]. A recent study reported de novo harmful single-nucleotide variations (SNVs) in YY1 in 59 Chinese families with a child suffering from ASD [72]. Consistent with these studies, our work also found that NEAT1 mediates its role by recruiting YY1.
An examination of RNA-Seq datasets from human ASD brain samples revealed that YY1 plays a key role in regulating differentially expressed genes in ASD [73]. Accordingly, we found that NEAT1 interacted with YY1 to regulate UBE3A expression in hippocampal neurons. We also observed that UBE3A expression was high in ASD neonates and that UBE3A suppressed cell viability, induced apoptosis, and enhanced oxidative stress and inflammation. Previously, it has been reported that UBE3A deficiency causes neurodevelopmental disorders such as Angelman syndrome [74], while UBE3A overexpression is associated with an increased risk of ASD [75]. This suggests that UBE3A must be expressed at precise levels to ensure proper neuronal function and may provide novel therapeutic targets for ASD.
Conclusions
In conclusion, we demonstrated that NEAT1 is upregulated in hippocampal tissues of VPA-induced ASD rats. NEAT1 promotes ASD development by recruiting YY1 to facilitate UBE3A expression, which in turn induces apoptosis, oxidative stress, and inflammatory processes in hippocampal neurons. This finding provides a potential new direction for developing therapeutic strategies to treat ASD.
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
References
Genovese A, Butler MG (2020) Clinical assessment, genetics, and treatment approaches in autism spectrum disorder (ASD). Int J Mol Sci 21(13):4726. https://doi.org/10.3390/ijms21134726
Gillberg C (2010) The ESSENCE in child psychiatry: early symptomatic syndromes eliciting neurodevelopmental clinical examinations. Res Dev Disabil 31(6):1543–1551
Moreno-De-Luca A et al (2013) Developmental brain dysfunction: revival and expansion of old concepts based on new genetic evidence. Lancet Neurol 12(4):406–414
Maenner MJ et al (2021) Prevalence and characteristics of autism spectrum disorder among children aged 8 years - autism and developmental disabilities monitoring network, 11 sites, United States, 2018. MMWR Surveill Summ 70(11):1–16
Solomon C (2020) Autism and employment: implications for employers and adults with ASD. J Autism Dev Disord 50(11):4209–4217
Wang L, Wang B, Wu C, Wang J, Sun M (2023) Autism spectrum disorder: neurodevelopmental risk factors, biological mechanism, and precision therapy. Int J Mol Sci 24(3):1819. https://doi.org/10.3390/ijms24031819
Bromley RL et al (2013) The prevalence of neurodevelopmental disorders in children prenatally exposed to antiepileptic drugs. J Neurol Neurosurg Psychiatry 84(6):637–643
Nicolini C, Fahnestock M (2018) The valproic acid-induced rodent model of autism. Exp Neurol 299(Pt A):217–227
Kuo HY, Liu FC (2018) Molecular pathology and pharmacological treatment of autism spectrum disorder-like phenotypes using rodent models. Front Cell Neurosci 12:422
Lodato S, Arlotta P (2015) Generating neuronal diversity in the mammalian cerebral cortex. Annu Rev Cell Dev Biol 31:699–720
Knierim JJ (2015) The hippocampus. Curr Biol 25(23):R1116–R1121
Rolls ET, Wirth S (2018) Spatial representations in the primate hippocampus, and their functions in memory and navigation. Prog Neurobiol 171:90–113
Opitz B (2014) Memory function and the hippocampus. Front Neurol Neurosci 34:51–59
Sheldon S, Levine B (2016) The role of the hippocampus in memory and mental construction. Ann N Y Acad Sci 1369(1):76–92
Ayabe T et al (2019) The lacto-tetrapeptide gly-Thr-Trp-Tyr, β-Lactolin, improves spatial memory functions via dopamine release and D1 receptor activation in the hippocampus. Nutrients 11(10):2469. https://doi.org/10.3390/nu11102469
Hampson RE et al (2013) Facilitation of memory encoding in primate hippocampus by a neuroprosthesis that promotes task-specific neural firing. J Neural Eng 10(6):066013
Banker SM et al (2021) Hippocampal contributions to social and cognitive deficits in autism spectrum disorder. Trends Neurosci 44(10):793–807
Behrens TEJ et al (2018) What is a cognitive map? Organizing knowledge for flexible behavior. Neuron 100(2):490–509
Reinhardt VP et al (2020) Understanding hippocampal development in young children with autism spectrum disorder. J Am Acad Child Adolesc Psychiatry 59(9):1069–1079
Statello L et al (2021) Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol 22(2):96–118
Mattick JS et al (2023) Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat Rev Mol Cell Biol 24(6):430–447
Chodroff RA et al (2010) Long noncoding RNA genes: conservation of sequence and brain expression among diverse amniotes. Genome Biol 11(7):R72
Oe S, Kimura T, Yamada H (2019) Regulatory non-coding RNAs in nervous system development and disease. Front Biosci (Landmark Ed) 24(7):1203–1240
Tang J, Yu Y, Yang W (2017) Long noncoding RNA and its contribution to autism spectrum disorders. CNS Neurosci Ther 23(8):645–656
Ghafouri-Fard S et al (2022) Emerging role of non-coding RNAs in autism spectrum disorder. J Mol Neurosci 72(2):201–216
Liu X et al (2022) LncRNA MEG3 activates CDH2 expression by recruitment of EP300 in valproic acid-induced autism spectrum disorder. Neurosci Lett 783:136726
Jiang M, Chen G (2023) Investigation of LncRNA PVT1 and MiR-21-5p expression as promising novel biomarkers for autism spectrum disorder. J Mol Neurosci 73(9–10):865-873. https://doi.org/10.1007/s12031-023-02161-8
Luo T et al (2020) The autism-related lncRNA MSNP1AS regulates moesin protein to influence the RhoA, Rac1, and PI3K/Akt pathways and regulate the structure and survival of neurons. Autism Res 13(12):2073–2082
Dong P et al (2018) Long non-coding rNA NEAT1: a novel target for diagnosis and therapy in human tumors. Front Genet 9:471
Simchovitz A et al (2019) NEAT1 is overexpressed in Parkinson’s disease substantia nigra and confers drug-inducible neuroprotection from oxidative stress. FASEB J 33(10):11223–11234
Ke S et al (2019) Long noncoding RNA NEAT1 aggravates Aβ-induced neuronal damage by targeting miR-107 in Alzheimer’s disease. Yonsei Med J 60(7):640–650
Sunwoo J-S et al (2017) Altered expression of the long noncoding RNA NEAT1 in Huntington’s disease. Mol Neurobiol 54:1577–1586
Chanda K et al (2018) Altered levels of long ncRNAs Meg3 and Neat1 in cell and animal models of Huntington’s disease. RNA Biol 15(10):1348–1363
Sayad A et al (2019) Aberrant expression of long non-coding RNAs in peripheral blood of autistic patients. J Mol Neurosci 67(2):276–281
Yao W, Huang J, He H (2019) Over-expressed LOC101927196 suppressed oxidative stress levels and neuron cell proliferation in a rat model of autism through disrupting the Wnt signaling pathway by targeting FZD3. Cell Signal 62:109328
Ishola IO, Balogun AO, Adeyemi OO (2020) Novel potential of metformin on valproic acid-induced autism spectrum disorder in rats: involvement of antioxidant defence system. Fundam Clin Pharmacol 34(6):650–661
Ishola IO, Adamson FM, Adeyemi OO (2017) Ameliorative effect of kolaviron, a biflavonoid complex from Garcinia kola seeds against scopolamine-induced memory impairment in rats: role of antioxidant defense system. Metab Brain Dis 32(1):235–245
Xu X et al (2018) Excessive UBE3A dosage impairs retinoic acid signaling and synaptic plasticity in autism spectrum disorders. Cell Res 28(1):48–68
Roy B et al (2024) UBE3A: The role in autism spectrum disorders (ASDs) and a potential candidate for biomarker studies and designing therapeutic strategies. Diseases 12(1):7
Girault JB, Piven J (2020) The neurodevelopment of autism from infancy through toddlerhood. Neuroimaging Clin N Am 30(1):97–114
Beopoulos A, Géa M, Fasano A, Iris F (2022) Autism spectrum disorders pathogenesis: toward a comprehensive model based on neuroanatomic and neurodevelopment considerations. Front Neurosci 16:988735. https://doi.org/10.3389/fnins.2022.988735
Courchesne E, Gazestani VH, Lewis NE (2020) Prenatal origins of ASD: the when, what, and how of ASD development. Trends Neurosci 43(5):326–342
Traetta ME et al (2021) Hippocampal neurons isolated from rats subjected to the valproic acid model mimic in vivo synaptic pattern: evidence of neuronal priming during early development in autism spectrum disorders. Mol Autism 12(1):23
Luhach K et al (2021) Vinpocetine amended prenatal valproic acid induced features of ASD possibly by altering markers of neuronal function, inflammation, and oxidative stress. Autism Res 14(11):2270–2286
Xiong Y et al (2023) Thymol improves autism-like behaviour in VPA-induced ASD rats through the Pin1/p38 MAPK pathway. Int Immunopharmacol 117:109885
Varghese M et al (2017) Autism spectrum disorder: neuropathology and animal models. Acta Neuropathol 134(4):537–566
Wang L et al (2022) Progranulin improves neural development via the PI3K/Akt/GSK-3β pathway in the cerebellum of a VPA-induced rat model of ASD. Transl Psychiatry 12(1):114
Toscano CVA, Carvalho HM, Ferreira JP (2018) Exercise effects for children with autism spectrum disorder: metabolic health, autistic traits, and quality of life. Percept Mot Skills 125(1):126–146
Siniscalco D, Schultz S, Brigida AL, Antonucci N (2018) Inflammation and neuro-immune dysregulations in autism spectrum disorders. Pharmaceuticals (Basel) 11(2):56. https://doi.org/10.3390/ph11020056
Ashwood P et al (2011) Elevated plasma cytokines in autism spectrum disorders provide evidence of immune dysfunction and are associated with impaired behavioral outcome. Brain Behav Immun 25(1):40–45
Inga Jácome MC et al (2016) Peripheral inflammatory markers contributing to comorbidities in autism. Behav Sci 6(4):29
Pangrazzi L, Balasco L, Bozzi Y (2020) Oxidative stress and immune system dysfunction in autism spectrum disorders. Int J Mol Sci 21(9):3293. https://doi.org/10.3390/ijms21093293
Chen L et al (2021) Oxidative stress marker aberrations in children with autism spectrum disorder: a systematic review and meta-analysis of 87 studies (N = 9109). Transl Psychiatry 11(1):15
Saceleanu VM, Toader C, Ples H, Covache-Busuioc RA, Costin HP, Bratu BG, Dumitrascu DI, Bordeianu A, Corlatescu AD, Ciurea AV (2023) Integrative approaches in acute ischemic stroke: from symptom recognition to future innovations. Biomedicines 11(10):2617. https://doi.org/10.3390/biomedicines11102617
Singh R, Kisku A, Kungumaraj H, Nagaraj V, Pal A, Kumar S, Sulakhiya K (2023) Autism spectrum disorders: a recent update on targeting inflammatory pathways with natural anti-inflammatory agents. Biomedicines 11(1):115. https://doi.org/10.3390/biomedicines11010115
Wei XB et al (2022) Exosome-derived lncRNA NEAT1 exacerbates sepsis-associated encephalopathy by promoting ferroptosis through regulating miR-9-5p/TFRC and GOT1 Axis. Mol Neurobiol 59(3):1954–1969
Li Y et al (2023) Targeting lncRNA NEAT1 hampers Alzheimer’s disease progression. Neuroscience 529:88–98
Yan W et al (2018) LncRNA NEAT1 promotes autophagy in MPTP-induced Parkinson’s disease through stabilizing PINK1 protein. Biochem Biophys Res Commun 496(4):1019–1024
Zhou Z et al (2022) LncRNA NEAT1 stabilized Wnt3a via U2AF2 and activated Wnt/β-catenin pathway to alleviate ischemia stroke induced injury. Brain Res 1788:147921
Chen D et al (2023) LncRNA NEAT1 suppresses cellular senescence in hepatocellular carcinoma via KIF11-dependent repression of CDKN2A. Clin Transl Med 13(9):e1418
Jia Y et al (2022) Long non-coding RNA NEAT1 mediated RPRD1B stability facilitates fatty acid metabolism and lymph node metastasis via c-Jun/c-Fos/SREBP1 axis in gastric cancer. J Exp Clin Cancer Res 41(1):287
Hosseini E et al (2019) The importance of long non-coding RNAs in neuropsychiatric disorders. Mol Aspects Med 70:127–140
Wang S et al (2023) LINC01133 can induce acquired ferroptosis resistance by enhancing the FSP1 mRNA stability through forming the LINC01133-FUS-FSP1 complex. Cell Death Dis 14(11):767
Cao Y et al (2023) LINC02454 promotes thyroid carcinoma progression via upregulating HMGA2 through CREB1. Faseb J 37(12):e23288
Zhou Y et al (2023) LncRNA NKILA exacerbates Alzheimer’s disease progression by regulating the FOXA1-mediated transcription of TNFAIP1. Neurochem Res 48(9):2895–2910
Rahman MR, Petralia MC, Ciurleo R, Bramanti A, Fagone P, Shahjaman M, Wu L, Sun Y, Turanli B, Arga KY, Islam MR, Islam T, Nicoletti F (2020) Comprehensive analysis of rna-seq gene expression profiling of brain transcriptomes reveals novel genes, regulators, and pathways in autism spectrum disorder. Brain Sci 10(10):747. https://doi.org/10.3390/brainsci10100747
Verheul TC et al (2020) The why of YY1: mechanisms of transcriptional regulation by Yin Yang 1. Front Cell Dev Biol 8:592164
Vargas DM et al (2018) Alzheimer’s disease master regulators analysis: search for potential molecular targets and drug repositioning candidates. Alzheimer’s Res Ther 10(1):59
Forlani G et al (2010) The MeCP2/YY1 interaction regulates ANT1 expression at 4q35: novel hints for Rett syndrome pathogenesis. Hum Mol Genet 19(16):3114–3123
Tiwari PC, Pal R (2017) The potential role of neuroinflammation and transcription factors in Parkinson disease. Dialogues Clin Neurosci 19(1):71–80
Pabian-Jewuła S, Bragiel-Pieczonka A, Rylski M (2022) Ying Yang 1 engagement in brain pathology. J Neurochem 161(3):236–253
Jiao J et al (2020) Identification of de novo JAK2 and MAPK7 mutations related to autism spectrum disorder using whole-exome sequencing in a Chinese child and adolescent trio-based sample. J Mol Neurosci 70:219–229
Rahman MR et al (2020) Comprehensive analysis of RNA-seq gene expression profiling of brain transcriptomes reveals novel genes, regulators, and pathways in autism spectrum disorder. Brain Sci 10(10):747
Geerts-Haages A et al (2020) A novel UBE3A sequence variant identified in eight related individuals with neurodevelopmental delay, results in a phenotype which does not match the clinical criteria of Angelman syndrome. Mol Genet Genomic Med 8(11):e1481
Wang J et al (2019) UBE3A-mediated PTPA ubiquitination and degradation regulate PP2A activity and dendritic spine morphology. Proc Natl Acad Sci U S A 116(25):12500–12505
Author information
Authors and Affiliations
Contributions
HZ and CH conceived and designed the experiments. CH and LC contributed significantly to the experiments and arranging data. CH and ZL performed data analyses. CH wrote the draft manuscript. HZ revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval
All animal-related procedures were carried out in accordance with the ARRIVE guidelines, and this study was approved by the Ethics Committee of the Chenzhou First People’s Hospital.
Consent for Publication
Not applicable.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
He, C., Zhou, H., Chen, L. et al. NEAT1 Promotes Valproic Acid-Induced Autism Spectrum Disorder by Recruiting YY1 to Regulate UBE3A Transcription. Mol Neurobiol (2024). https://doi.org/10.1007/s12035-024-04309-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12035-024-04309-y