Abstract
Growing evidence reported a strong association between the nitrate ingestion and adverse health consequences in humans, including its detrimental impact on the developing brain. The present study identified miRNAs and proteins in SH-SY5Y human neuroblastoma cells and HMC3 human microglial cells using high-throughput techniques in response to nitrate level most prevalent in the environment (India) as X dose and an exceptionally high nitrate level as 5X dose that can be reached in the near future. Cells were exposed to mixtures of nitrates for 72 h at doses of X and 5X, 320 mg/L and 1600 mg/L, respectively. OpenArray and LCMS analysis revealed maximum deregulation in miRNAs and proteins in cells exposed to 5X dose. Top deregulated miRNAs include miR-34b, miR-34c, miR-155, miR-143, and miR-145. The proteomic profiles of both cell types include proteins that are potential targets of deregulated miRNAs. These miRNAs and their targeted proteins involve in multiple functions, including metabolic processes, mitochondrial functions, autophagy, necroptosis, apoptosis, neuronal disorders, brain development, and homeostasis. Furthermore, measuring mitochondrial bioenergetics in cells exposed to nitrate revealed that a 5X dose causes a significant reduction in oxygen consumption rate (OCR) and other bioenergetic parameters in both cell types. In summary, our studies have demonstrated that a 5X dose of nitrate significantly alters cellular physiology and functions by deregulating several miRNAs and proteins. However, X dose of nitrate has not caused any adverse effects on any cell type.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Neurodegeneration refers to a class of neurological diseases characterized by the slow and gradual deterioration of brain regions that are crucial for cognitive, psychological, behavioral, and motor functions [1]. These deliberating illnesses worsen with age [2] and can appear in hereditary or sporadic forms based on genetic and environmental variables. Protein misfolding/fibrillation, mitochondrial dysfunction, and oxidative stress are special features of various neurodegenerative diseases (NDDs), such as Alzheimer’s disease (AD), Parkinson’s disease (PD), amyotrophic lateral sclerosis (ALS), and Huntington’s disease (HD) [1, 3]. Nitrates are well-known water pollutants that can adversely affect groundwater systems. After pesticides, nitrates are the most prevalent anthropogenic pollutants of surface and groundwater. They are applied to the soil as fertilizers during various agricultural practices. This has become a global concern and several international research communities have raised alarm to supply safe drinking water to the human population [4].
Numerous adverse outcomes have been reported in humans exposed to excess nitrates through drinking water, processed and preserved foods, and fertilizers [5,6,7]. According to Schwendimann et al., the brain development of the offspring born to pregnant mice who consumed water with high concentrations of nitrates (319 mg/L) and total dissolved salts (TDS) was altered. Brain from offsprings born to dams who had been given agricultural water (versus forest control water) was significantly smaller. Furthermore, the motor cortex of the brains of these offspring had significantly more microglia at postnatal day 21 (P21), significantly fewer white matter astrocytes, and a considerable increase in cell death, particularly in the dentate gyrus [8]. Nitrate exposure caused moderate anxiogenic-like behavior and altered the brain metabolomic profile in zebrafish [9]. These neuropathological effects suggest that nitrates may alter brain development. However, the underlying cellular and molecular mechanisms are still need to be well studied.
MicroRNAs (miRNA) are small non-coding RNAs that negatively control post-transcriptional gene expression. A huge number of miRNAs are expressed in the brain, and emerging research suggests that miRNAs are crucial for neuronal functions and brain development [10]. In recent decades, miRNA deregulation is emerging as a contributor to neurodegeneration by influencing most of the mechanisms responsible for NDDs [11]. MiR-34 family, miR-200 family, miR-143/145 cluster, and miR-155 are brain-specific miRNAs and have a crucial role in brain development [12,13,14]. The miR-34 family is involved in the regulation of neuronal maturation (proliferation and differentiation) and aging in neuronal or non-neuronal cells [15], and upregulation of miR-34 is associated with the pathogenesis of AD and metabolic syndrome [16]. The brain has reported overexpression of miR-143/145 cluster after ischemic insult and in multiple sclerosis [17, 18]. Increased miR-199a/b expression was strongly correlated with nitrate tolerance in humans [19]. A key research goal in miRNA biology is to understand the post-transcriptional/translational control of the targets and their effects on biological processes. MiRNA targets are typically identified computationally by seed sequence conservation, which yields numerous candidate targets with little overlap. This lack of linkage and correlation between miRNA and protein levels encourages researchers to employ proteomic-based techniques for identifying, estimating, and confirming miRNA-induced protein alterations.
In the present study, we have studied the effects of nitrate on human SH-SY5Y neuroblastoma cells and human HMC3 microglial cells at X nitrate level (320 mg/L) that is most prevalent in the environment (India) and at 5X (1600 mg/L) which is an exceptionally high nitrate level that appeared occasionally, and might be reached in the future [8, 20]. By employing bi-omics approaches, this study identified miRNAs and proteins deregulated following nitrate exposure in cells. Additionally, the effect of nitrate on cellular bioenergetics has been investigated using the Seahorse extracellular flux analyzer.
Materials and Methods
Chemicals and Reagents
Sodium nitrate (Cat# 221341-550G), potassium nitrate (Cat# P8291-500G), magnesium nitrate (Cat# 63084-500G-F), dithiothreitol (DTT) (Cat# 10,197,777), iodoacetamide (Cat# I1149), D-glucose solution (Cat# G8769), and protease inhibitor (Cat# P-8340) were purchased from Sigma-Aldrich. SH-SY5Y neuroblastoma cells (Cat# SH-SY5Y CRL-2266) and HMC3 microglia cells (Cat# CRL 3304) were purchased from ATCC. Media and reagents including minimum essential media (Cat# 61100–01), F-12 nutrient media (Cat# 21700–075), fetal bovine serum (FBS) (Cat# 10270106), antibiotic/antimycotic solution 100x (Ab/Am) (Cat# 5240062), L-glutamine supplement (Cat# 25030), and Gibco sodium pyruvate (Cat# 11360–070) were procured from Thermo Fisher Scientific. Pierce™ Detergent Removal Spin Columns (Cat# 87,776), Pierce™ C18 Spin Columns (Cat# 89,870), TaqMan Custom RT pool (Cat# A25630), TaqMan™ MicroRNA Reverse Transcription Kit (Cat# 4,366,597), TaqManR Custom OpenArray PreAmp Pool (Cat# 2,874,647), TaqManR PreAmp Master Mix (Cat# 4,384,266), TaqManR OpenArray™ RT-PCR Plates 112 format (Part Number# 4,470,813), TaqManR OpenArrayR Real-Time PCR Master Mix (Cat# 4,462,159), high capacity cDNA RT kit (Cat# 4368813), mirVana miRNA isolation kit (Cat# AM1560), and many other reagents required for quantitative PCR were obtained from Applied Biosystems Thermo Fisher Scientific. Trypsin/Lys-C Mix, LCMS grade (Cat# V5073) was purchased from Promega, CA. XFp cell culture microplates (Part Number# 103023-100), Seahorse XFp extracellular flux cartridge (Part Number# 103022–100), XF calibrant (Part Number# 103059–000), extracellular flux assay kits (Part Number# 103010–100), and Agilent Seahorse XF base medium (Part Number# 102353–100) were procured from Agilent Technologies, India. RapiGest SF Surfactant (Cat# 186,001,861) was procured from Waters, Milford, USA.
Cell Culture and Nitrate Exposure
SH-SY5Y cells were cultured in MEM/Ham F12 Complete Medium (1:1 mixture of MEM medium and Ham’s F12 medium with NEAA) supplemented with 1% antibiotic antimycotic and 10% FBS, whereas HMC3 cells were cultured in MEM medium with 1% Ab/Am and with the supplementation of 10% FBS. Cells were incubated and maintained at 37 °C with 5% CO2 and 95% relative humidity. Cells were exposed to nitrates as a mixture of sodium nitrate, potassium nitrate, and magnesium nitrate (320 mg/L nitrates named as X dose and 1600 mg/L nitrates named as 5X dose) for 72 h.
Cell Viability Assay
Cytotoxicity of nitrate on SH-SY5Y and HMC3 cells was identified using alamarBlue cell viability reagent (Cat# DAL1025). Equal numbers of cells were seeded in each well of 96-well plates and exposed to various concentrations of nitrate-X/10, X/5, X, 5X, and 10X for 24, 48, 72, and 96 h. The culture medium was replaced with fresh medium supplemented with nitrates after every 24 h. AlamarBlue reagent was used in 1/10th ratio of the medium in each well and incubated for 4 h and then fluorescence was measured at a wavelength of 560 nm (excitation) and 590 nm (emission). Blank was used to normalize background fluorescence.
OpenArray Technology Based on Real-Time PCR of miRNA
Total RNA was isolated using the manufacturer’s instructions of the mirVana miRNA isolation kit from Thermo Fisher Scientific in Waltham, Massachusetts. Gel electrophoresis was used to confirm the integrity of the RNA samples. Selected miRNAs important in neural development and neurotoxicity were included in the unique “brain-specific miRNA” OpenArray panel (Part Number# 4,470,813) which was used for miRNA profiling. The OpenArray panel contains primers for 112 miRNAs. For the reverse transcription, a TaqMan Custom RT pool primer of 112 miRNAs was utilized. Thermo Fischer’s TaqMan miRNA Reverse Transcription Kit was used to perform reverse transcription on mature miRNAs. In brief, the reaction mixture includes 100 ng of RNA, 75 units of MultiScribe reverse transcriptase, RT buffer, OpenArray Custom RT primers, RNase inhibitor, MgCl2, and dNTPs, with a maximum reaction volume of 7.5 μL. The reverse transcription process was carried out employing thermal conditions as follows: 40 cycles of 16 °C for 2 min, 42 °C for 1 min, and 50 °C for 1 s. Denaturation at 85 °C for 5 min was used to stop the process. Following that, cDNA was preamplified using a pool of OpenArray preamp primers (Part No. # 4,485,255) and a TaqMan preamp master mixture. The ultimate volume for preamplification was 25 μL, of which 2.5 μL was custom preamp primer pools combined with 12.5 μL of preamp master mix, 2.5 μL cDNA, and additional nuclease-free water. Thermal cycling at 95 °C for 10 min, 55 °C for 2 min, 72 °C for 2 min, and 12 cycles of 95 °C for 15 s and 60 °C for 4 min was employed for preamplification. After cycling, preamplification products were incubated for 10 min at 99.9 °C to inactivate enzymes. The preamplification products were then diluted in 0.1X TE buffer in the ratio of 1:40. The diluted preamplification products were combined with 2X TaqMan OpenArray Real-Time PCR Master Mix (Cat# 4,462,159) and pipetted onto OpenArray plates using an OpenArray AccuFill System from Thermo Fisher Scientific. The preamplification product was then run as directed by the manufacturer on Thermo Fisher Scientific’s QuantStudio 12K Flex RT-PCR machine. Using the Ct method and endogenous control, the relative quantification of miRNA expression was calculated from the raw data using the ExpressionSuite software. There were three biological replicates used in each experiment. The significant changes were determined by performing the Student’s t-test using the ExpressionSuite OpenArray analysis software. The p-value ≤ 0.05 is statistically significant.
Label-Free Quantitative LC-MS/MS Proteomics
Label-free quantification has been performed as described in our earlier lab studies [21,22,23]. In brief, the protein was isolated from SH-SY5Y and HMC3 cells and quantified using Pierce bicinchoninic acid (BCA) protein assay kit (Catalog 23,227). Samples were then cleaned by Pierce™ Detergent Removal Spin Columns (Cat#87,776). Total 100 μg protein from each sample was mixed with 0.1% (v/v) RapiGest SF surfactant (Cat#186,001,861) for solubility and samples were reduced by adding 5 mM dithiothreitol (DTT) and incubated for 30 min at 60 °C. Furthermore, 15 mM iodoacetamide (IAA) was added, and samples were incubated for 30 min at room temperature (RT) in dark for alkylation. The protein samples were digested by using the enzyme Trypsin/Lys-C (Cat# V5071) and incubated overnight at 37 °C in a water bath. The digestion reaction was quenched by mixing 0.5% formic acid (FA) and incubated for 60 min at 37 °C. Finally, the digested samples were centrifuged at 20,000 g for 10 min at 4 °C and the supernatant containing peptides was transferred to a fresh vial. The peptide samples were purified and concentrated by Pierce C18 Spin Columns (Cat# 89,870), as described in manufacturer’s protocol. The purified tryptic peptides were lyophilized and used for proteomic analysis.
The peptide identification was achieved by nanoLC (EASY-nLC 1200; Thermo Fisher) coupled with Q Exactive mass spectrometer (Q Exactive; Thermo Fisher) through a nano-electrospray ionization source (Thermo Fisher Scientific). The lyophilized peptide samples were reconstituted in 0.1% FA and loaded on a reversed-phase column. The mobile phase used for the elution contains solvents A (1% v/v acetonitrile and 0.1% v/v FA in water) and solvent B (0.1% v/v FA and 80% v/v acetonitrile in water). The loaded peptides in solvent A were eluted by applying a linear gradient of solvent B for 140 min at a flow rate of 300 nL/min. The gradient began at 2% B to 5% B in 5 min and then increased linearly to 35% B in 120 min and finally increased to 90% for 15 min. A voltage of 2300 V was applied for electrospray ionization and the heated capillary temperature was set at 300 °C. Data-dependent acquisition (DDA) was employed for peptide identification. Full MS had a resolution of 70,000, a scan range of 350-1700 m/z, and automatic gain control (AGC) of 1 × 106 with a 60-ms maximum injection time. For dd-MS2, a resolution of 17,500 and AGC was 5 × 104 with a maximum injection duration of 100 ms. The MS data was obtained using the top 15 most abundant precursors and dynamic exclusion was set to 20 s.
Identification and Quantification of Proteins
All proteomics data were analyzed using the Proteome Discoverer software (version 2.4, Thermo Fisher, USA). Peptide lists were then searched against the web-based Uniprot database of Homo sapiens (SwissProtTaxID 9606) with the cysteine carbamidomethylation as fixed modification while methionine oxidation and N-terminal acetylation as static modification. A 1% false discovery rate (FDR) was used for peptide identification.
Metabolic Flux Analysis Using Seahorse XFp for Bioenergetics
Using the Seahorse XFp analyzer (Agilent Technologies, CA), oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were assessed with protocol as described in our previous lab study [21, 23]. SH-SY5Y and HMC3 cells were seeded in Seahorse XFp cell culture mini plates and exposed to nitrate at both X and 5X doses and were then proceeded for real-time metabolic flux analysis. On the day of the test, cells were washed with unbuffered Seahorse XF base assay medium (10 mM glucose, 1 mM sodium pyruvate, 2 mM glutamine, pH 7.4). Prior to the start of the test, cells were cultured at 37 °C for 1 h in a CO2-free incubator. The effect of nitrate on the electron transport chain was assessed by adding oligomycin A (a complex V inhibitor), FCCP (an uncoupler), and rotenone (a complex I inhibitor)/antimycin (a complex III inhibitor) in the order recommended by the manufacturer. The manufacturer’s instructions were followed when using the Cell Mito Stress Test kit, with ports A and B holding oligomycin, carbonyl cyanide-p-trifluoromethoxy phenylhydrazone (FCCP), and port C holding a mixture of rotenone and antimycin. The Seahorse XFp experiment was carried out using triplicate readings, and the outcomes were examined using the Seahorse XFp Wave software.
Statistical Analysis
There were three replicates of each experiment. Data are represented as mean ± standard error mean (SEM). GraphPad Prism 7.0 (GraphPad Software, CA) was used to statistically analyze the dataset using the student’s t-test, and Proteome Discoverer 2.4 was used to analyze the proteomic data. A p-value less than or equal to 0.05 (typically ≤ 0.05) was chosen as statistically significant.
Results
Changes in Cellular Morphology of SH-SY5Y and HMC3 Cells
Under normal conditions, SH-SY5Y cells grow in cluster form and possess a short branch neurite phenotype with a mixed population of adherent and suspension cells, while HMC3 cells (human microglia) possess only an adherent phenotype. Upon exposure to X dose of nitrate for 72 h, both SH-SY5Y and HMC3 cells showed no significant changes in their morphological aspect while on exposure to 5X dose both SH-SY5Y and HMC3 cells show dramatic changes in their morphology as indicated in Figure 1 with single-sided red arrow.
Phase contrast images of SH-SY5Y and HMC3 cells exposed to nitrates. A-C SH-SY5Y cells: control (A), X dose (B), and 5X dose (C). D-F HMC3 cells: control (D), X dose (E), and 5X dose (F). Cells were exposed to nitrate for 72 h and red arrow in SH-SY5Y cells indicates clusters of cells, whereas in HMC3 cells, this indicates the number of detached cells. Scale bar 100 μm
Assessment of Cell Viability and Cytotoxicity
The toxic potency of nitrate was assessed using alamarBlue assay. SH-SY5Y and HMC3 cells were exposed to different concentrations of nitrate, such as X/10, X/5, X, 5X, and 10X with their respective controls. In SH-SY5Y cells, the cell viability has no significant changes up to X dose of nitrate following 24, 48, 72, and 96 h exposure to nitrate. However, the cell viability reached 70% at a 5X dose of nitrate for 72 h in SH-SY5Y cells (Supplementary Figure 1B). HMC3 cells showed a significant decrease in the percentage of viability at 5X with increasing concentrations of nitrate after 72 h of exposure (Supplementary Figure 1A). However, alteration in cell viability of HMC3 cells at X dose was non-significant.
Identification of Nitrate-Induced miRNAs in Neuronal and Microglial Cells Using Real-Time PCR-Based OpenArray
SH-SY5Y and HMC3 cells were exposed to identified X and 5X concentrations of nitrate for 72 h. Nitrate-regulated miRNAs were identified by a real-time system using a custom “brain-specific miRNA array” containing 112 miRNAs in an OpenArray plate. The expression profile of deregulated miRNAs was shown on a volcano plot drawn between log2 fold change with a minimum 1.5 fold change boundary and p-value less than or equal to 0.05. Results of a customized brain-specific OpenArray panel with 112 miRNAs indicate the differential regulation of miRNAs in different groups following nitrate exposure, as listed in Figure 2. The deregulation of miRNAs was more profound in the case of SH-SY5Y cells. In SH-SY5Y cells, the expression of miR-27b and miR-155 was significantly increased and decreased, respectively at both X and 5X doses of nitrate. MiRNAs such as miR-34c (+7.664 fold), miR-34b (+4.017 fold), miR-194 (+2.7 fold), and miR-200c (+2.524 fold) are the top upregulated miRNAs identified in SH-SY5Y cells exposed to 5X dose of nitrate while miR-213 (+2.141) and miR-363 (+2.03 fold) showed significant upregulation at X dose exposure. Furthermore, miR-145 and miR-143 are commonly upregulated following exposure to both X and 5X doses of nitrate in HMC3 cells, while miR-199a-3p, miR-213, miR-210, miR-34a, miR-9, and miR-339-5p were found to be significantly downregulated upon exposure with 5X dose of nitrate in HMC3 cells. The list of miRNAs that are commonly deregulated in both groups is shown in Figure 3 for both SH-SY5Y and HMC3 cells.
Expression profiling of miRNAs by real-time PCR-based OpenArray in HMC3 and SH-SY5Y cells exposed to nitrates. A-B Volcano plot and list of miRNAs deregulated in HMC3 cells exposed to X dose of nitrate. C-D Volcano plot and list miRNAs deregulated in HMC3 cells exposed to 5X dose of nitrates. E-F Volcano plot and list of miRNAs deregulated in SH-SY5Y cells exposed to X dose of nitrates. G-H Volcano plot and list of miRNAs deregulated in SH-SY5Y cells exposed to 5X dose of nitrates. The volcano plot is drawn between fold change versus p-value. The additional two vertical lines illustrate the cut-off limit (1.5 fold) for downregulated and upregulated miRNA expression, whereas the single horizontal line in the volcano plot depicts the p-value of the t-test, with a threshold set at 0.05. All red dots above the vertical lines represent significantly upregulated miRNAs, while all green dots represent significantly downregulated miRNAs. The lists of significantly deregulated miRNAs in each group display relative quantification of miRNA expression (RQ). Three biological replicates were used in each experiment
Commonly deregulated miRNAs in different groups. A Venn diagram of SH-SY5Y cells exposed to X dose and 5X dose of nitrates with two commonly deregulated miRNAs. B Venn diagram of HMC3 cells exposed to X dose and 5X dose of nitrates with two commonly deregulated miRNAs. C Venn diagram of both cell types exposed to 5X dose of nitrate showed 6 commonly deregulated miRNAs. Venn diagrams were generated by an online tool Venny 2.0
Changes in Proteome Profile Due to Nitrate Exposure to SH-SY5Y and HMC3 Cells
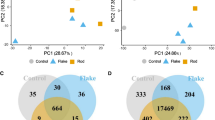
SH-SY5Y and HMC3 cells were exposed to nitrate for 72 h to identify the differentially expressed proteins (DEPs) by LC/MS-MS. A volcano plot analysis of SH-SY5Y and HMC3 cells was performed to identify DEPs by setting fold change threshold boundary equal to 2 and p-value less than 0.05 (Figure 4A-D). In SH-SY5Y cells, we found a total of 6305 proteins in which 218 proteins (117 downregulated and 101 upregulated) were significantly deregulated in the X dose group, while a total of 484 proteins (229 downregulated and 255 upregulated) were significantly deregulated in 5X dose group. On the other hand, we identified a total of 5752 proteins in nitrate-exposed HMC3 cells with 491 proteins (219 downregulated and 272 upregulated) significantly altered in the X dose group while 623 proteins (280 downregulated and 343 upregulated) significantly deregulated in 5X dose group. Furthermore, in order to improve accuracy, we followed a tight selection standard that included proteins with unique peptides greater than one. Finally, we found 47 proteins significantly deregulated in the X dose group while 181 proteins were significantly deregulated in the 5X dose group in SH-SY5Y cells. Moreover, Venn diagram analysis showed that 7 common proteins (PURA, H2BC18, ADI1, ASAH1, LSM2, NAB2, and MRPL54) were significantly deregulated in both X and 5X dose group of SH-SY5Y cells (Figure 5A). In HMC3 cells, 163 proteins in the X dose group and 277 proteins in the 5X dose group were significantly deregulated after applying the selection criteria of unique peptide more than one. Additionally, Venn diagram analysis showed that 44 deregulated proteins (as mentioned in Table 1) were common in both X dose and 5X dose groups of HMC3 cells (Figure 5B). A detailed list of significantly deregulated proteins in SH-SY5Y and HMC3 cells is provided in Supplementary Tables 2-9.
Overview of differentially abundant proteins induced by nitrate exposure in SH-SY5Y and HMC3 cells. Volcano plot of deregulated proteins at X dose (A) and 5X dose (B) of nitrate in SH-SY5Y cells and volcano plot of deregulated proteins at X dose (C) and 5X dose (D) of nitrate in HMC3 cells. The volcano plot is plotted between p-value versus fold change. Two vertical lines in the volcano plot illustrate the cut-off limit (±2 fold) for downregulated and upregulated protein expression, while a horizontal line in the volcano plot depicts the threshold for the p-value of the t-test, set at 0.05
Venn diagram of proteins deregulated at X dose and 5X dose of nitrate in SH-SY5Y and HMC3 cells. Venn diagram representing significantly deregulated proteins in SH-SY5Y cells: 47 and 181 proteins were identified in X dose and 5X dose of nitrate respectively (A) with 7 proteins being common in both groups. Venn diagram depicting significantly deregulated 163 and 277 proteins identified in X dose and 5X dose of nitrate, respectively, in HMC3 cells (B) with 44 proteins being commonly deregulated at both doses
Functional Enrichment and Protein-Protein Interaction (PPI) Network of Nitrate-Regulated Proteins in SH-SY5Y and HMC3 Cells
All significantly deregulated proteins obtained with unique peptides more than one were uploaded on DAVID online pathway search engine tool (https://david.ncifcrf.gov) for Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) analysis. KEGG pathway analysis in SH-SY5Y cells indicated the involvement of significantly deregulated proteins in various biological pathways, such as pathways of neurodegeneration involved in multiple diseases, ALS, focal adhesion, autophagy, and sphingolipid metabolism (Figure 6A) in 5X dose group, while no significant pathway was found in X dose group. In HMC3 cells, proteins deregulated at 5X dose were found to be involved in major significant processes such as pathways of neurodegeneration involved in multiple diseases, ALS, PD, AD, prion disease, HD, protein processing in endoplasmic reticulum, ribosome, oxidative phosphorylation, arginine and proline metabolism, proteasome, arginine biosynthesis, tyrosine metabolism, 2-oxocarboxylic acid metabolism, phenylalanine metabolism, and various types of N-glycan biosynthesis (Figure 6B). However, proteins those were deregulated at X dose in HMC3 cells were implicated in pathways such as endocytosis, Epstein-Barr virus infection, ubiquitin-mediated proteolysis, pyrimidine metabolism, viral myocarditis, p53 signaling pathway, and nucleotide metabolism. A detailed list of pathways is provided in Supplementary Table 14-15. Furthermore, GO analysis was performed to classify the significantly deregulated proteins in biological processes, cellular components, and molecular functions (Supplementary Table 10-13).
The STRING algorithm (https://string-db.org) was used to identify the protein-protein interactions (PPIs) among significantly deregulated proteins in SH-SY5Y and HMC3 cells exposed to nitrate. Results of STRING analysis revealed that the deregulated proteins exhibited strong and complex interactions among them. Deregulated proteins at a 5X dose of nitrate in SH-SY5Y cells were found to be involved in pathways such as metabolic process (including nitrogen metabolism and catabolism), mitochondrial function, autophagy, neurodegeneration, Ras signaling, lysosomal function, neuronal apoptosis, mitophagy, cellular response to stress, and processes involved in neurodegenerative diseases (such as AD, PD, HD, ALS, and prion disease) (Figure 7). Furthermore, the biological pathways obtained from the PPI network analysis of proteins deregulated in HMC3 cells following 5X nitrate exposure were part of pathways like metabolic process (including nitrogen metabolism and catabolism), neurodegeneration, macroautophagy, necroptosis, oxidative phosphorylation, mitochondrial translation, stress response, and events involved in neurodegenerative diseases (such as AD, PD, HD, ALS, and prion disease) (Figure 8). The detail of several other interactive networks identified in SH-SY5Y cells (such as DNA ligation, spliceosomal complex, disease of metabolism, and cellular component disassembly) and in HMC3 cells (such as nitrogen compound transport, cellular response to cytokine stimulus, negative regulation of cell cycle G2-M phase transition, negative regulation of protein silencing, programmed cell death, Wnt signaling pathway, and cell cycle) is provided in Supplementary Figures 2-3.
Differentially Expressed miRNAs and Protein Correlation Studies
In general, miRNAs negatively regulate the expression of their target genes. Therefore, miRNA-protein interaction studies were carried out employing the TargetScan software (https://www.targetscan.org) to find the potential binding sites of identified miRNAs with proteins deregulated in SH-SY5Y and HMC3 cells following exposure to nitrate as depicted in Supplementary Figure 4 . The results of the present study showed that upregulated miRNAs reduce the level of their target proteins and vice versa. The significantly deregulated proteins identified in nitrate-exposed SH-SY5Y and HMC3 cells possess binding sites for the significantly deregulated miRNAs at both X and 5X doses of nitrate in SH-SY5Y cells (viz., miR-27b and miR-155) and HMC3 cells (viz., miR-143 and miR-145) and at 5X doses of SH-SY5Y cells and HMC3 cells (viz., miR-34a, miR-34c, miR-143, miR-200c, miR-210, and miR-339-5P) as shown in Figure 9. In SH-SY5Y cells, miR-27b which was significantly increased upon nitrate exposure showed interaction with a maximum number of downregulated proteins, such as QKI, KCMF1, ZEB2, AK4, KMT2A, and PURA. Interestingly, PURA protein is commonly downregulated in both the X and the 5X groups of SH-SY5Y cells, while the remaining proteins (namely, QKI, KCMF1, ZEB2, AK4, KMT2A) were identified only in the 5X dose group. In addition, miR-155 also showed an interaction with RAPH1 and KANSL1 in the X dose group and SCG2 protein in the 5X dose group of SH-SY5Y cells. Several downregulated proteins such as KIAA0319L, ERLIN1, RAB5C, DPYSL2, NEDD4L, STAM, MBD2, STX16, STK25, IRS1, PKN2, CAMK2D, and EPN1 were predicted as direct targets of miR-145, which was significantly upregulated upon nitrate exposure in HMC3 cells. Among these downregulated proteins, 2 proteins (ERLIN1 and KIAA0319L) were identified in both the X and 5X dose groups, 6 proteins (RAB5C, DPYSL2, NEDD4L, STAM, MBD2, and STX16) were identified only in the X dose group, and 5 proteins (EPN1, CAMK2D, PKN2, IRS1, STK25) were found in the 5X dose group.
TargetScan analysis showing correlation between identified miRNAs and proteins in nitrate-exposed SH-SY5Y and HMC3 cells. Deregulated proteins found in nitrate-exposed SH-SY5Y (A) and HMC3 (B) cells are regulated by altered miRNAs identified in X and 5X dose groups. Proteins targeted by miRNAs commonly deregulated in 5X dose of both SH-SY5Y and HMC3 cells (C). Black line indicates interaction of proteins found in 5X dose group, brown line indicates interaction of proteins identified in X dose group, and blue line shows proteins interaction commonly identified in both X and 5X dose groups. Pink circle illustrates protein identified in SH-SY5Y cells, green circle indicates protein identified in HMC3 cells, and orange circle indicates common protein identified in both SH-SY5Y and HMC3 cells
The target analysis of miRNAs deregulated in the 5X group of both SH-SY5Y and HMC3 cells showed that only 4 miRNAs (viz., miR-34a, miR-34c, miR-143, miR-339-5p) possess binding sites for proteins deregulated following nitrate exposure. VPS37B, a protein that is significantly downregulated only in nitrate-exposed HMC3 cells, is a direct target of miR-34a, which showed significant upregulation following nitrate exposure. ABR (identified at 5X dose of nitrate in SH-SY5Y cells), ANP32B, and AXL (identified at 5X dose of nitrate in HMC3 cells) are predicted targets of miR-34c as illustrated in Figure 9. PDF and NEDD4L are direct targets of miR-339-5p, in which PDF was identified in both cells exposed to the 5X dose of nitrate while NEDD4L was identified only in SH-SY5Y cells exposed to 5X dose of nitrate. In SH-SY5Y, several deregulated proteins such as QKI, CBL, KCMF1, BLMH, and SLC35A4 have predicted binding sites for miR-143 while SLU7 is the only predicted target of miR-143 identified in HMC3 cells.
Effect of Nitrate Exposure on Cellular Bioenergetics and Mitochondrial Functions in SH-SY5Y and HMC3 Cells
The Seahorse XFp analyzer was used to investigate the effect of nitrate exposure on mitochondrial respiration in SH-SY5Y and HMC3 cells. Our results revealed that nitrate dramatically affects cellular bioenergetics in both cell types. In both SH-SY5Y and HMC3 cells, we found that exposure to a 5X dose of nitrate showed a dramatic reduction in oxygen consumption rate (OCR), a sign of dysfunctional oxidative phosphorylation (OXPHOS). However, the X dose could not induce such significant changes in both the cells (Figure 10 A). Furthermore, the extracellular acidification rate (ECAR) was also decreased considerably at 5X dose, but changes in ECAR at X dose were not so prominent (Figure 10 B). In SH-SY5Y and HMC3 cells, the level of changes in basal respiration at X dose was found to be non-significant. However, at a 5X dose exposure, SH-SY5Y cells showed a dramatic reduction in basal respiration rate compared to control, while no changes in basal respiration were observed at both X and 5X doses in HMC3 cells. A similar pattern of changes in maximal respiration and proton leak was observed in both cell lines, with the results showing no signification alteration at X dose, whereas a notable decrease in maximal respiration and proton leak was observed in both SH-SY5Y and HMC3 cells exposed to 5X dose of nitrate. A distinct pattern in the alterations in ATP synthesis in response to nitrate exposure was seen in SH-SY5Y cells compared to HMC3 cells. A marked decrease in ATP production was observed in SH-SY5Y cells following exposure to 5X dose of nitrate, whereas the level of ATP production showed no significant changes in HMC3 cells at both X and 5X doses of nitrate. Furthermore, there was no significant change in spare respiratory capacity in both cell types exposed to X dose of nitrates. The change in spare respiratory capacity in SH-SY5Y cells at 5X dose was non-significant (Figure 10 C). However, spare respiratory capacity was found to be significantly reduced at 5X dose in HMC3 cells.
Mitochondrial function and cellular bioenergetics factors were assessed in SH-SY5Y and HMC3 cells. OCR (A) and ECAR (B) pattern of SH-SY5Y and HMC3 cells exposed to nitrates. At the times shown in the figure, injections of oligomycin (1 μM) [ATP synthase inhibitor], FCCP (1 μM) [uncoupler], and rotenone/antimycin C (0.5 μM) [complex I/complex III inhibitor] were delivered to cells. Changes in bioenergetic parameters like basal respiration, maximal respiration, spare respiratory capacity (%), proton leak, and ATP production in both cell lines (C) are shown as mean ± SEM of the three representative experiments. P* < 0.05, P** < 0.01, P*** < 0.001 versus control
Discussion
Environmental factors have been identified as significant contributors to neural dysfunction-associated disorders. The majority of neurological illnesses are mostly caused by prenatal or postnatal exposure to environmental toxins [24]. Some experimental and clinical studies have also reported the adverse effect of nitrates on health by targeting various physiological and metabolic functions in the body [8, 25,26,27]. Epidemiological studies have shown that high levels of nitrate can be toxic and its chronic exposure may affect brain development [8, 27]. Studies from our laboratory and elsewhere have provided evidence that miRNA plays an important role in neuronal development and neurodegeneration [12, 15, 28,29,30,31]. Indeed, increasing literature has also suggested the involvement of miRNAs and proteins in regulating nitrate-induced toxicity [19, 32]. However, the role of miRNAs and proteins in regulating nitrate-induced toxicity to the brain is yet to be elucidated. The present study aimed to identify critical miRNAs and associated proteins regulating nitrate-induced toxicity in neuronal and microglial cells employing high-throughput technology. Furthermore, the effect of nitrate on the bioenergetic function of SH-SY5Y and HMC3 cells has also been explored. Our results demonstrate that nitrate exposure has an adverse impact on brain functions by affecting the neurons and microglia at a molecular level, as indicated by alterations in the morphological features of cells. Earlier studies reported that offsprings born to pregnant and lactating mice fed with nitrate-contaminated water from heavily farmed terrain exhibit neurodevelopmental issues, as indicated by the loss of neurons and astrocytes, rise in microglial cells, and cell death [8]. Several studies demonstrated the adverse effect of nitrate/nitrite on the pathophysiology of the brain leading to anxiety-like behavior, whereas other studies revealed no significant changes in the level of epinephrine, nor-epinephrine, or dopamine as well as no alteration in electrical activity following infusion of sodium nitrite/nitrate [9, 33]. Various types of cancers have been caused by exposure to nitrate/nitrite in drinking water; however, brain tumors showed no significant association with nitrate from well water [34].
The present study identified several miRNAs and associated proteins deregulated following both X dose and 5X dose in both SH-SY5Y and HMC3 cells. The high dose of nitrate caused a more profound deregulation in miRNA expression in both neuronal and microglial cells, with more miRNAs deregulated in neuronal cells. Top significant deregulated miRNAs identified following exposure to nitrate include miR-213, miR-363, miR-27b, miR-146a, and miR-203 at X dose and miR-34c, miR-34b, miR-194, miR-200c, miR-34a, miR-55, miR-7, and others at 5X dose in SH-SY5Y cells. Significant deregulation was observed in miR-143 and miR-145 at X dose and miR-145, miR-200c, miR-34c, and miR-296 at 5X dose in HMC3 cells. Several experimental studies have reported the role of miR-34 family in various biological functions such as cellular senescence, apoptosis, cell cycle, immune homeostasis, stress response, signal transduction, and organ development [35, 36]. Acidic nuclear phosphoprotein 32 family member B (ANP32B) is an important protein involved in the extracellular vesicle secretion in neuronal cells and has also been implicated in several neurodegenerative diseases [37]. Target analysis showed that miR-34c negatively regulated ANP32B protein expression by binding to the 3′UTR of ANP32B mRNA. LCMS analysis revealed that 47 and 181 proteins in SH-SY5Y cells and 163 and 277 proteins in HMC3 cells, at X and 5X dose respectively, were deregulated following nitrate exposure. KEGG enrichment analysis showed that proteins deregulated in both cells following exposure to a 5X dose of nitrate are involved in neurodegeneration and associated disorders. Furthermore, deregulated proteins identified in SH-SY5Y cells at X dose of nitrate showed no association with relevant pathways. However, proteins altered at X dose in HMC3 cells are found to be involved in pathways such as endocytosis, ubiquitin-mediated proteolysis, pyrimidine metabolism, p53 signaling pathway, and nucleotide metabolism.
Functional protein-protein interaction network analysis revealed that proteins deregulated at 5X dose of nitrate in SH-SY5Y cells were found to be involved in pathways such as metabolic process (including nitrogen metabolism and catabolism), mitochondrial function, autophagy, neurodegeneration, Ras signaling, lysosomal function, neuronal apoptosis, mitophagy, cellular response to stress, and processes involved in neurodegenerative diseases (such as AD, PD, HD, ALS, and prion disease). Furthermore, the biological pathways obtained from the PPI network analysis of proteins deregulated in HMC3 cells following 5X nitrate exposure were part of pathways like metabolic process (including nitrogen metabolism and catabolism), neurodegeneration, macroautophagy, necroptosis, oxidative phosphorylation, mitochondrial translation, stress response, and events involved in neurodegenerative diseases (such as AD, PD, HD, ALS, and prion disease). Overexpression of miR-27b has been reported to lower the expression of some mitochondrial genes that are essential for promoting mitochondrial biogenesis and decreasing the number of mitochondria [38]. In line with this study, we have also found upregulation of miR-27b at both doses (X and 5X) in SH-SY5Y cells. Interestingly, target analysis showed that several downregulated proteins such as PURA, QK1, KCMF1, ZEB2, AK4, and KMT2A possess a binding site for miR-27b. Pur-alpha (PURA) was deregulated in SH-SY5Y cells at both X and 5X doses. PURA has been reported to be involved in various neuronal disorders and transcriptional control mechanisms [39]. The level of PURA protein was upregulated in the 5X dose group and downregulated in the X dose group in SH-SY5Y cells. Consistent upregulation of PURA was also reported in AD [40]; thus, indicating a high dose of nitrate may have neurotoxic effects on the brain. Zinc finger E-box-binding homeobox 2 (ZEB2) is a substantial contributor to astrogliosis and induction of astrogliosis can enhance neuroprotection and neurological function recovery in stroke and spinal cord injury [41, 42]. The expression of ZEB2 was found to be downregulated in SH-SY5Y cells at 5x dose of nitrate and could be responsible for neuronal death. TargetScan analysis revealed that ZEB2 has the binding site for miR-27b which may be responsible for downregulating ZEB2. MiR-155 is known as the master regulator of inflammation and can be regulated by multiple signaling pathways [43]. Regulatory cytokines such as TGF-β and IL-10 can induce or inhibit the expression of miR-155 [43]. TargetScan analysis revealed that secretogranin II (SCG2) is a direct target of miR-155, which was downregulated in nitrate-exposed SH-SY5Y cells. Earlier research has indicated that SCG2 overexpression in plasma could be used as a candidate biomarker for early neurodevelopmental disorders [44]. SLU7 has been reported to regulate the process of alternate splicing [45]. Alternative splicing is a frequent occurrence in brain tissues and plays a role in all stages of brain development, including cell fate determination, axon guidance, neuronal migration, and synapse formation [46]. Downregulation of SLU7 can affect various processes of brain development. TargetScan analysis revealed that SLU7 which was downregulated in HMC3 cells at both X and 5X doses of nitrate has the binding site for miR-143, which was upregulated in HMC3 cells following nitrate exposure. Previous reports have shown the involvement of miR-143 in the induction of apoptosis via regulating ERK5 pathway and protein kinase C-epsilon (PKCε) [47, 48]. Therefore, decreased expression of SLU7 and subsequent neurodegeneration could be caused by increased miR-143 expression in HMC3 cells. KIAA0319L and ERLIN1 were downregulated in HMC3 cells at both X and 5X doses of nitrate. Gostic et al. in their study found that KIAA0319L, which is a dyslexia susceptible gene, is involved in the development of brain and neuronal migration [49]. A membrane protein called ERLIN1 is associated with a number of neurological diseases and may be the root cause of the early onset of ALS [50, 51]. However, the precise role of identified proteins such as KIAA03119L, ERLIN1, and SLU7 which are putative targets of miR-143/145 is not well understood but these are found to be involved in brain development and altered expression of these proteins may affect various brain developmental processes. Overexpression of miR-143 and miR-145 has been reported to promote cellular apoptosis via regulating several pathways [48, 52,53,54]. Bleomycin hydrolase (BLMH) is also an important protein involved in aging, brain plasticity, learning, and neurodegenerative disorders such as AD [55]. The expression of BLMH was downregulated in SH-SY5Y cells following high-dose exposure of nitrate. CBL is an E3-ubiquitin ligase involved in the inhibition of microglial-mediated neuroinflammation via PI3K/AKT/NFKB pathway [56]. Our results have shown significant downregulation in the expression of CBL protein which may lead to increased inflammation in SH-SY5Y cells. Interestingly, both BLMH and CBL possess binding sites for miR-143 which was significantly upregulated in SH-SY5Y cells following 5X dose of nitrate. So, the increased expression of the miR-143 following nitrate exposure may cause downregulation of BLMH and CBL which have been involved in neurodegeneration.
The mitochondria undergo some of the most significant changes during neuronal development and differentiation. The mitochondria are essential for cellular functions such as energy metabolism, aging, formation of reactive oxygen species, calcium homeostasis, and differentiation [57]. In our study, we have measured OCR in live cells using Seahorse extracellular flux analyzer and observed no significant alteration in bioenergetic parameters when cells were exposed to X dose of nitrate. However, OCR as well as other bioenergetic parameters were dramatically altered in both neuronal and microglial cells at 5X dose. Proliferative cells have been reported to engage in various metabolic activities and thus demand high ATP consumption [58]. During the conversion of ADP to ATP, active mitochondria require oxygen. To satisfy the demands, cells may speed up glycolysis or respiration rate [59]. Therefore, we evaluated the major indices of mitochondrial respiration including OCR, basal respiration, maximal respiration, Spare respiratory capacity, proton leak, ATP synthesis, and extracellular acidification rate (ECAR) in both neuronal and microglial cells. Our results showed that SH-SY5Y cells exposed to 5X dose possess lower OCR as well as ECAR compared to control cells. Furthermore, significant downregulation in basal respiration, maximal respiration, proton leak, and ATP production has been observed in SH-SY5Y cells exposed to a 5X dose of nitrate, whereas a slight reduction in OCR and ECAR was observed with X dose of nitrate. In HMC3 cells, changes observed in the X dose group were non-significant. However, both OCR and ECAR were dramatically reduced in HMC3 cells following exposure to a 5X dose of nitrate. Bioenergetic parameters such as maximal respiration, spare respiratory capacity, and proton leak were significantly reduced in HMC3 cells exposed to 5X dose of nitrate. Mitochondrial bioenergetics analysis suggests that nitrate exposure at higher doses targets mitochondria and drastically alters its functions, which may be linked to various pathological mechanisms underlying nitrate-induced toxicity. However, the most prevalent X dose did not cause any prominent impact on mitochondrial bioenergetics. The disruption of cellular metabolism due to miRNA dysfunction is a common event in neuronal toxicity and cell death. Our results indicate that nitrate exposure to both SH-SY5Y and HMC3 cells significantly increased the expression of miR-143 and miR-145, which are transcribed as a bicistronic unit with common regulatory elements. Studies have shown that miR-143 regulates mitochondrial membrane potential, ATP activity, glycolysis, and apoptosis by targeting protein kinase C-epsilon (PKC-ε), ERK5 pathway, and hexokinase 2 (HK2) in different cells [47, 48, 60] and miR-145 regulates cell viability, oxidative stress, and cellular stability via regulating PI3K/AKT signaling pathway [54, 61]. Therefore, increased expression of miR-143 and miR-145 following nitrate exposure may possibly be the cause of cellular bioenergetic dysfunction, as depicted by our bioenergetics studies in both SH-SY5Y and HMC3 cells. Furthermore, miR-200c, which was significantly upregulated in both SH-SY5Y and HMC3 cells in response to nitrate exposure, has also been reported to specifically target SIRT1 and SIRT2, which are key metabolic sensors in cells, and their activities are intrinsically linked to mitochondrial bioenergetics and cellular metabolism [62, 63]. Another miRNA, miR-34c, which was significantly upregulated in both nitrate-exposed SH-SY5Y and HMC3 cells, has been shown to mediate synaptic and memory deficits by targeting synaptotagmin-1 (SYT1) through the ROS-JNK-p53 pathway [64] which may result in mitochondrial-dependent apoptotic cell death. Overexpression of miR-29 has been reported to suppress mitochondrial functions in MCF-7 and LCC9 cells [65]. An increased level of miR-29b in HMC3 cells following nitrate exposure may possibly contribute to bioenergetic impairment, potentially leading to mitochondrial dysfunction.
In summary, the present study is the first to identify miRNAs and associated proteins at lower and higher doses of nitrate in both neuronal and microglial cells. We found that a higher nitrate dose (5X) deregulated more miRNAs and proteins in both cell types, with more prominent alterations observed in SH-SY5Y cells. Dysfunction of cellular bioenergetics indicated that nitrate has adverse effects on neuronal and microglial cells, targeting mitochondrial functions with more pronounced effects at a higher dose of nitrate. Thus, the X dose, which has been highly prevalent in many locations, did not lead to any adverse repercussions. However, the 5X dose, which has been detected in very few locations or could be reached in the near future, has the potential to cause adverse health outcomes in the brain.
Data Availability
The raw data are available in the repository of the institute on its server, and with corresponding author too.
References
Tewari D, Sah AN, Bawari S, Nabavi SF, Dehpour AR, Shirooie S, Braidy N, Fiebich BL et al (2021) Role of nitric oxide in neurodegeneration: function, regulation, and inhibition. Curr. Neuropharmacol. 19(2):114–126
Roy B, Lee E, Li T, Rampersaud M (2022) Role of miRNAs in neurodegeneration: from disease cause to tools of biomarker discovery and therapeutics. Genes 13(3):425
Karnati HK, Panigrahi MK, Gutti RK, Greig NH, Tamargo IA (2015) miRNAs: key players in neurodegenerative disorders and epilepsy. J. Alzheimers Dis. 48(3):563–580
Bishayee B, Chatterjee RP, Ruj B, Chakrabortty S, Nayak J (2022) Strategic management of nitrate pollution from contaminated water using viable adsorbents: an economic assessment-based review with possible policy suggestions. J. Environ. Manage. 303:114081
Temkin A, Evans S, Manidis T, Campbell C, Naidenko OV (2019) Exposure-based assessment and economic valuation of adverse birth outcomes and cancer risk due to nitrate in United States drinking water. Environ. Res. 176:108442
Ebdrup NH, Schullehner J, Knudsen UB, Liew Z, Thomsen AML, Lyngsø J, Bay B, Arendt LH et al (2022) Drinking water nitrate and risk of pregnancy loss: a nationwide cohort study. Environ Health : A Global Access Sci Source 21(1):87
Rahman A, Mondal NC, Tiwari KK (2021) Anthropogenic nitrate in groundwater and its health risks in the view of background concentration in a semi arid area of Rajasthan India. Sci. Rep. 11(1):9279
Schwendimann L, Sivaprakasam I, Buvaneshwari S, Gurumurthy GP, Mishra S, Ruiz L, Sekhar M, Fleiss B et al (2021) Agricultural groundwater with high nitrates and dissolved salts given to pregnant mice alters brain development in the offspring. Ecotoxicol. Environ. Saf. 224:112635
García-Jaramillo M, Beaver LM, Truong L, Axton ER, Keller RM, Prater MC, Magnusson KR, Tanguay RL et al (2020) Nitrate and nitrite exposure leads to mild anxiogenic-like behavior and alters brain metabolomic profile in zebrafish. PloS One 15(12):e0240070
Petri R, Malmevik J, Fasching L, Åkerblom M, Jakobsson J (2014) miRNAs in brain development. Exp. Cell Res. 321(1):84–89
De Felice B, Montanino C, Oliva M, Bonavita S, Di Onofrio V, Coppola C (2020) MicroRNA expression signature in mild cognitive impairment due to Alzheimer’s disease. Mol Neurobiol 57(11):4408–4416
Jauhari A, Yadav S (2019) MiR-34 and MiR-200: regulator of cell fate plasticity and neural development. Neuromolecular Med. 21(2):97–109
Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee T-H, Miano JM et al (2009) miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature 460(7256):705–710
Varendi K, Kumar A, Härma M-A, Andressoo J-O (2014) miR-1, miR-10b, miR-155, and miR-191 are novel regulators of BDNF. Cell. Mol. Life Sci. 71(22):4443–4456
Jauhari A, Singh T, Singh P, Parmar D, Yadav S (2018) Regulation of miR-34 family in neuronal development. Mol Neurobiol 55(2):936–945
Bazrgar M, Khodabakhsh P, Prudencio M, Mohagheghi F, Ahmadiani A (2021) The role of microRNA-34 family in Alzheimer’s disease: a potential molecular link between neurodegeneration and metabolic disorders. Pharmacol. Res. 172:105805
Volný O, Kašičková L, Coufalová D, Cimflová P, Novák J (2015) Micrornas in cerebrovascular disease. microRNA. Med Evid 888:155–195
Sharaf-Eldin WE, Kishk NA, Gad YZ, Hassan H, Ali MA, Zaki MS, Mohamed MR, Essawi ML (2017) Extracellular miR-145, miR-223 and miR-326 expression signature allow for differential diagnosis of immune-mediated neuroinflammatory diseases. J Neurol Sci 383:188–198
Bai Y-P, Zhang J-X, Sun Q, Zhou J-P, Luo J-M, He L-F, Lin X-C, Zhu L-P et al (2018) Induction of microRNA-199 by nitric oxide in endothelial cells is required for nitrovasodilator resistance via targeting of prostaglandin I2 synthase. Circulation 138(4):397–411
Central Ground Water Board (CGWB) (2018) Ground water quality in shallow aquifers in India. Annexure VI:129–184. http://cgwb.gov.in/WQ/Ground%20Water%20Book-F.pdf
Srivastava AK, Yadav SS, Mishra S, Yadav SK, Parmar D, Yadav S (2020) A combined microRNA and proteome profiling to investigate the effect of ZnO nanoparticles on neuronal cells. Nanotoxicology 14(6):757–773
Yadav SK, Pandey A, Sarkar S, Yadav SS, Parmar D, Yadav S (2022) Identification of altered blood microRNAs and plasma proteins in a rat model of Parkinson’s disease. Mol. Neurobiol. 59(3):1781–1798
Pandey A, Sarkar S, Yadav SK, Yadav SS, Srikrishna S, Siddiqui MH, Parmar D, Yadav S (2022) Studies on regulation of global protein profile and cellular bioenergetics of differentiating SH-SY5Y cells. Mol. Neurobiol. 59(3):1799–1818
Nabi M, Tabassum N (2022) Role of environmental toxicants on neurodegenerative disorders. Frontiers Toxicol 4:837579
Pasupuleti S, Singha SS, Singha S, Kumar S, Singh R, Dhada I (2022) Groundwater characterization and non-carcinogenic and carcinogenic health risk assessment of nitrate exposure in the Mahanadi River Basin of India. J. Environ. Manage. 319:115746
Mukate S, Bhoominathan S, Solanky V (2022) Assessment of human health risk arising due to fluoride and nitrate in groundwater: a case study of Bhokardan tehsil of Maharashtra. Hum Ecol Risk Assess Int J 28(5–6):594–620
Sv R, Basavaiah S (2022) Assessment of sodium nitrate (NaNO3) effects on the reproductive system, liver, pancreas and kidney of male rats. Toxicol. Ind. Health 38(10):702–711
Jauhari A, Singh T, Singh P, Parmar D, Yadav S (2019) Correction to: Regulation of miR-34 family in neuronal development. Mol Neurobiol 56(11):7909–7910
Pandey A, Jauhari A, Singh T, Singh P, Singh N, Srivastava AK, Khan F, Pant AB et al (2015) Transactivation of P53 by cypermethrin induced miR-200 and apoptosis in neuronal cells. Toxicol. Res. 4(6):1578–1586
Rajman M, Schratt G (2017) MicroRNAs in neural development: from master regulators to fine-tuners. Development 144(13):2310–2322
Godlewski J, Lenart J, Salinska E (2019) MicroRNA in brain pathology: neurodegeneration the other side of the brain cancer. Non-coding RNA 5(1):20
Oghbaei H, Hamidian G, Alipour MR, Alipour S, Keyhanmanesh R (2020) The effect of prolonged dietary sodium nitrate treatment on the hypothalamus-pituitary-gonadal axis and testicular structure and function in streptozotocin-induced diabetic male rats. Food Funct. 11(3):2451–2465
Franko E, Ezra M, Crockett DC, Joly O, Pattinson K (2019) Effect of nitrite on the electroencephalographic activity in the healthy brain. Nitric Oxide 90:47–54
Picetti R, Deeney M, Pastorino S, Miller MR, Shah A, Leon DA, Dangour AD, Green R (2022) Nitrate and nitrite contamination in drinking water and cancer risk: a systematic review with meta-analysis. Environ. Res. 210:112988
Tufekci KU, Alural B, Tarakcioglu E, San T, Genc S (2021) Lithium inhibits oxidative stress-induced neuronal senescence through miR-34a. Mol. Biol. Rep. 48(5):4171–4180
Wang C, Jia Q, Guo X, Li K, Chen W, Shen Q, Xu C, Fu Y (2022) microRNA-34 family: from mechanism to potential applications. Int J Biochem Cell Biol 144:106168
Muraoka S, Jedrychowski MP, Iwahara N, Abdullah M, Onos KD, Keezer KJ, Hu J, Ikezu S et al (2021) Enrichment of neurodegenerative microglia signature in brain-derived extracellular vesicles isolated from Alzheimer’s disease mouse models. J. Proteome Res. 20(3):1733–1743
Shen L, Chen L, Zhang S, Du J, Bai L, Zhang Y, Jiang Y, Li X et al (2016) MicroRNA-27b regulates mitochondria biogenesis in myocytes. PLoS One 11(2):e0148532
Molitor L, Bacher S, Burczyk S, Niessing D (2021) The molecular function of PURA and its implications in neurological diseases. Front. Genet. 12:638217
Patel H, Dobson RJ, Newhouse SJ (2019) A meta-analysis of Alzheimer’s disease brain transcriptomic data. J. Alzheimers Dis. 68(4):1635–1656
Vivinetto AL, Kim I-d, Goldberg DC, Fones L, Brown E, Tarabykin VS, Hill CE, Cho S et al (2020) Zeb2 is a regulator of astrogliosis and functional recovery after CNS injury. Cell Rep. 31(13):107834
Zhao Z, Hu X, Wang J, Wang J, Hou Y, Chen S (2021) Zinc Finger E-Box binding homeobox 2 (ZEB2)-induced astrogliosis protected neuron from pyroptosis in cerebral ischemia and reperfusion injury. Bioengineered 12(2):12917–12930
Mahesh G, Biswas R (2019) MicroRNA-155: a master regulator of inflammation. J. Interferon Cytokine Res. 39(6):321–330
Lim S-H, Sung Y-J, Jo N, Lee N-Y, Kim K-S, Lee DY, Kim N-S, Lee J et al (2021) Nanoplasmonic immunosensor for the detection of SCG2, a candidate serum biomarker for the early diagnosis of neurodevelopmental disorder. Sci. Rep. 11(1):1–11
Alberstein M, Amit M, Vaknin K, O’Donnell A, Farhy C, Lerenthal Y, Shomron N, Shaham O et al (2007) Regulation of transcription of the RNA splicing factor hSlu7 by Elk-1 and Sp1 affects alternative splicing. RNA 13(11):1988–1999
Su C-H, Tarn W-Y (2018) Alternative splicing in neurogenesis and brain development. Front. Mol. Biosci. 5:12
Nakagawa Y, Iinuma M, Naoe T, Nozawa Y, Akao Y (2007) Characterized mechanism of α-mangostin-induced cell death: caspase-independent apoptosis with release of endonuclease-G from mitochondria and increased miR-143 expression in human colorectal cancer DLD-1 cells. Bioorg. Med. Chem. 15(16):5620–5628
Hong H, Tao T, Chen S, Liang C, Qiu Y, Zhou Y, Zhang R (2017) MicroRNA-143 promotes cardiac ischemia-mediated mitochondrial impairment by the inhibition of protein kinase Cepsilon. Basic Res. Cardiol. 112:1–16
Gostic M, Martinelli A, Tucker C, Yang Z, Gasparoli F, Ewart JY, Dholakia K, Sillar KT et al (2019) The dyslexia susceptibility KIAA0319 gene shows a specific expression pattern during zebrafish development supporting a role beyond neuronal migration. J Comp Neurol 527(16):2634–2643
Tunca C, Akçimen F, Coşkun C, Gündoğdu-Eken A, Kocoglu C, Çevik B, Bekircan-Kurt CE, Tan E et al (2018) ERLIN1 mutations cause teenage-onset slowly progressive ALS in a large Turkish pedigree. Eur. J. Hum. Genet. 26(5):745–748
Wright FA, Bonzerato CG, Sliter DA, Wojcikiewicz RJ (2018) The erlin2 T65I mutation inhibits erlin1/2 complex–mediated inositol 1, 4, 5-trisphosphate receptor ubiquitination and phosphatidylinositol 3-phosphate binding. J. Biol. Chem. 293(40):15706–15714
Tang W, Zhang X, Tan W, Gao J, Pan L, Ye X, Chen L, Zheng W (2019) miR-145-5p suppresses breast cancer progression by inhibiting SOX2. J. Surg. Res. 236:278–287
Slattery ML, Mullany LE, Sakoda LC, Wolff RK, Samowitz WS, Herrick JS (2018) Dysregulated genes and miRNAs in the apoptosis pathway in colorectal cancer patients. Apoptosis 23:237–250
Xin Z, Tong Z, Tan J, Liu C (2021) MicroRNA-145-5p aggravates cell apoptosis and oxidative stress in tongue squamous cell carcinoma. Exp. Ther. Med. 21(4):1–8
Suszyńska-Zajczyk J, Łuczak M, Marczak Ł, Jakubowski H (2014) Hyperhomocysteinemia and bleomycin hydrolase modulate the expression of mouse brain proteins involved in neurodegeneration. J. Alzheimers Dis. 40(3):713–726
Dong L, Li YZ, An HT, Wang YL, Chen SH, Qian YJ, Wang K, Zhen JL et al (2016) The E3 ubiquitin ligase c-Cbl inhibits microglia-mediated CNS inflammation by regulating PI 3K/Akt/NF-κB pathway. CNS Neurosci. Ther. 22(8):661–669
Wanet A, Arnould T, Najimi M, Renard P (2015) Connecting mitochondria, metabolism, and stem cell fate. Stem Cells Dev. 24(17):1957–1971
Luengo A, Li Z, Gui DY, Sullivan LB, Zagorulya M, Do BT, Ferreira R, Naamati A et al (2021) Increased demand for NAD+ relative to ATP drives aerobic glycolysis. Mol. Cell 81(4):691–707
Fadaka A, Ajiboye B, Ojo O, Adewale O, Olayide I, Emuowhochere R (2017) Biology of glucose metabolization in cancer cells. J Oncol Sci 3(2):45–51
Fang R, Xiao T, Fang Z, Sun Y, Li F, Gao Y, Feng Y, Li L et al (2012) MicroRNA-143 (miR-143) regulates cancer glycolysis via targeting hexokinase 2 gene. J. Biol. Chem. 287(27):23227–23235
Zhao J, Zhou K, Ma L, Zhang H (2020) MicroRNA-145 overexpression inhibits neuroblastoma tumorigenesis in vitro and in vivo. Bioengineered 11(1):219–228
Griffiths B, Xu L, Sun X, Greer M, Murray I, Stary C (2022) Inhibition of microRNA-200c preserves astrocyte sirtuin-1 and mitofusin-2, and protects against hippocampal neurodegeneration following global cerebral ischemia in mice. Front Mol Neurosci 15:1014751
Cha Y, Han M-J, Cha H-J, Zoldan J, Burkart A, Jung JH, Jang Y, Kim C-H et al (2017) Metabolic control of primed human pluripotent stem cell fate and function by the miR-200c–SIRT2 axis. Nat. Cell Biol. 19(5):445–456
Shi Z, Zhang K, Zhou H, Jiang L, Xie B, Wang R, Xia W, Yin Y et al (2020) Increased miR-34c mediates synaptic deficits by targeting synaptotagmin 1 through ROS-JNK-p53 pathway in Alzheimer’s disease. Aging Cell 19(3):e13125
Muluhngwi P, Alizadeh-Rad N, Vittitow SL, Kalbfleisch TS, Klinge CM (2017) The miR-29 transcriptome in endocrine-sensitive and resistant breast cancer cells. Sci. Rep. 7(1):5205
Acknowledgements
Ms Saumya Mishra is grateful to University Grant Commission (UGC), New Delhi, for providing a fellowship. Ms Deepshikha Srivastava is acknowledged for extending technical support in the Central Instrumentation Facility for LC-MS/MS during proteomics studies.
Funding
The research was funded by DST-Science and Engineering Research Board (DST-SERB), New Delhi (grant sanction no. EMR/2017/000889).
Author information
Authors and Affiliations
Contributions
AB Pant has conceptualized the program, designed the protocols, and monitored throughout the course of experimentations. Saumya Mishra performed the experiments, data analysis, and prepared the manuscript. Sana Sarkar has helped in the data analysis and editing of the manuscript. Anuj Pandey assisted in carrying out the cellular bioenergetic experiments, and data analysis. Sanjeev Kumar Yadav assisted in carrying out the OpenArray experiments. Renu Negi assisted in the in vitro experiments. Sanjay Yadav provided the intellectual inputs and reviewed the manuscript.
Corresponding author
Ethics declarations
Ethical Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mishra, S., Sarkar, S., Pandey, A. et al. Crosstalk Between miRNA and Protein Expression Profiles in Nitrate-Exposed Brain Cells. Mol Neurobiol 60, 3855–3872 (2023). https://doi.org/10.1007/s12035-023-03316-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-023-03316-9