Abstract
The human KCTD13 gene is located within the 16p11.2 locus and copy number variants of this locus are associated with a high risk for neuropsychiatric diseases including autism spectrum disorder and schizophrenia. Studies in zebrafish point to a role of KCTD13 in proliferation of neural precursor cells which may contribute to macrocephaly in 16p11.2 deletion carriers. KCTD13 is highly expressed in the fetal human brain and in mouse cortical neurons, but its contribution to the development and function of mammalian neurons is not completely understood. In the present study, we deleted the KCTD13 gene in human-induced pluripotent stem cells (iPSCs) using CRISPR/Cas9 nickase. Following neural differentiation of KCTD13 deficient and isogenic control iPSC lines, we detected a moderate but significant inhibition of DNA synthesis and proliferation in KCTD13 deficient human neural precursor cells. KCTD13 deficient cortical neurons derived from iPSCs showed decreased neurite formation and reduced spontaneous network activity. RNA-sequencing and pathway analysis pointed to a role for ERBB signaling in these phenotypic changes. Consistently, activating and inhibiting ERBB kinases rescued and aggravated, respectively, impaired neurite formation. In contrast to findings in non-neuronal human HeLa cells, we did not detect an accumulation of the putative KCTD13/Cullin-3 substrate RhoA, and treatment with inhibitors of RhoA signaling did not rescue decreased neurite formation in human KCTD13 knockout neurons. Taken together, our data provide insight into the role of KCTD13 in neurodevelopmental disorders, and point to ERBB signaling as a potential target for neuropsychiatric disorders associated with KCTD13 deficiency.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Neuropsychiatric disorders including autism spectrum disorder (ASD), schizophrenia (SZ), and bipolar disorder are the third leading cause of disability worldwide and are associated with major personal and social burdens [1]. These disorders are characterized by complex genetics and a high heritability [2]. Numerous risk genes for various neuropsychiatric disorders have been discovered by large-scale genetic studies [3, 4]. However, there is still a need to understand the pathomechanisms caused by these genomic variants for the development of novel therapeutic treatments.
In contrast to common single nucleotide variants (SNVs) detected by genome-wide association studies (GWAS) [5], rare copy number variants (CNVs) confer a high risk to develop neuropsychiatric disorders [6, 7]. One of the most frequent CNVs involved in neuropsychiatric disorders is located in the 16p11.2 locus and harbors 29 genes. Patients carrying 16p11.2 CNVs suffer from intellectual disability, ASD, SZ, and bipolar disorders [8,9,10,11]. In a recent GWAS of CNV burden in SZ, 16p11.2 duplications attained the highest genome-wide significance, and a meta-analysis of 16p11.2 deletions reported an odds ratio of nearly 40 for autism [12, 13].
Potassium-channel-tetramerization-domain-containing 13 (KCTD13), also known as polymerase delta-interacting protein 1 (PDIP1), or BTB/POZ domain-containing adapter for Cullin-3-mediated RhoA degradation protein 1 (BACURD1) [14, 15], is located in the 16p11.2 locus. KCTD13 is one of five genes that has been identified in ASD patients carrying smaller 16p11.2 deletions [16], and KCTD13 is one of four genes in the larger 16p11.2 deletions that harbors brain-critical exons (i.e., high brain expression but low missense mutation burden) [17]. In a recent GWAS of schizophrenia, KCTD13 was functionally assigned to non-coding genetic risk variants by chromatin conformation capture using human brain tissue [18].
In zebrafish, morpholino-mediated knockdown of the KCTD13 orthologue increases both head size and the level of neural progenitor cell proliferation [19]. In human Hela cells, KCTD13/BACURD1 acts a substrate-specific adapter of the Cullin-3 (CUL3) ubiquitin E3 ligase complex, which mediates the ubiquitination and degradation of the small GTPase Ras homolog gene family member (RhoA) [14]. In a cell-free assay, KCTD13/PDIP1 interacts with DNA polymerase delta and with proliferation cell nuclear antigen (PCNA). Interaction increases DNA polymerase activity twofold to threefold, and thus may play a role in DNA replication or cell cycle control during mitosis [15]. Additionally, single cell RNA-sequencing (RNA-seq) revealed a high expression of KCTD13 in excitatory glutamatergic neurons of the mouse cerebral cortex [20]. Based on these findings, we hypothesized that KCTD13 may also modulate proliferation of human neural precursor cells, or RhoA-mediated neurite outgrowth [21, 22] in human post-mitotic cortical neurons.
Disease modeling based on human-induced pluripotent stem cells (iPSCs) has revealed specific pathomechanisms, unknown disease etiologies, and new therapeutic strategies [23, 24]. During the last decade, iPSC lines were generated from patients diagnosed with various neuropsychiatric diseases and differentiated into forebrain neurons. Subsequent structural and functional analyses contributed to a better understanding of the neurodevelopmental pathomechanisms that increase the risk for neuropsychiatric disorders [25,26,27,28,29].
16p11.2 co-expressed and interacting proteins (including KCTD13-Cullin-3) are enriched in the mid-fetal period of the developing human cortex [21]. Moreover, since the transcriptome of human iPSC-derived neurons most closely recapitulates that of human fetal forebrain neurons [30], they represent an ideal cellular system for 16p11.2/KCTD13 disease modeling. In the present study, we used CRISPR/Cas9 nickase to generate KCTD13 deficient human iPSC lines. Following differentiation of the KCTD13 deficient iPSC lines and the isogenic parental control iPSC line into neural progenitor cells (NPCs) and cortical neurons, we detected decreased cell proliferation and impaired neurite formation, respectively. RNA-sequencing and pathway analysis pointed to a role for ERBB signaling in impaired neurite formation, which we confirmed using activators and inhibitors of ERBB kinases.
Materials and Methods
Human iPSC Culture
Human iPSCs were cultured under feeder cell-free conditions in Essential 8 medium (E8, Thermo Fisher Scientific) supplemented with 1:100 Antibiotic-Antimycotic (Thermo Fisher Scientific) on Matrigel-coated (BD Bioscience) 6-well tissue culture plates. Cells were split before reaching 100% confluence using 0.02% ethylenediaminetetraacetic acid (EDTA, Sigma) and replated in E8 supplemented with 10 μM ROCK inhibitor Y-27632 (Tocris). All cell lines were cultured at 37 °C and 5% CO2 in a humidified atmosphere. All iPSC-derived cells were negatively tested for mycoplasma using MycoAlert™ PLUS Mycoplasma Detection Kit (Lonza) according to the manufacturer’s protocol.
Genome Editing Using CRISPR/Cas9 Nickase
The guide RNA pair (sense 5′-gAGTCCCCATGCCTATGTTCC(TGG)-3′ and antisense 5′-gCCAGCTGGCAGTCCTCAATC(AGG)-3′) targeting exon 2 of the KCTD13 gene was identified using the Sanger Institute CRISPR webtool (http://www.sanger.ac.uk/htgt/wge/find_crisprs). The guide RNAs (gRNA) were chosen on the basis of having the lowest combined off-targeting score while targeting as many of the known and predicted transcripts as possible. Analysis of the KCTD13 locus (ENSG00000174943, NC_000016.10 (29906335..29926232)) using both NCBI and Ensembl suggested that in addition to the main isoform (ENST00000568000.5, NM_178863) there existed a smaller 209 residue variant (ENST00000561540.2) and a further five predicted transcript variants X1–X5. The region spanning the end of exon 2 and the 5′ end of intron 2–3 was determined to be the optimal region for CRISPR targeting, since this region contains exonic sequence shared between all of the identified transcripts with the exception of X3. Transcription of the latter predicted variant initiates further downstream and was not targeted, since it lacks the BTB/POZ domain and there is no experimental evidence of expressed protein. The chosen gRNA pair was ID: 1136385458_1136385466 and the individual CRISPRs 1136385458 (0: 1, 1: 0, 2: 1, 3: 12, 4: 115) and 1136385466 (0: 1, 1: 0, 2: 0, 3: 12, 4: 126) (http://www.sanger.ac.uk/htgt/wge/find_crisprs). The sense gRNA has one off-target of two mismatches in an intergenic region and 12 off-targets of three mismatches in intergenic or intronic regions. The antisense gRNA has 12 off-targets of three mismatches in intergenic, intronic, or exonic regions. However, there exists no off-target pairing of these gRNAs closer than 1 kb to one another such that the possibility of off-target DNA cleavage is considered to be negligible. Complementary oligonucleotides with BbsI compatible overhangs were designed, annealed according to the Zhang method [31], and these DNA guide inserts ligated into BbsI-digested target vectors; the antisense guide was cloned into the spCas9 D10A nickase expressing vector pX335 (Addgene Plasmid #42335) and the sense guide into the puromycin selectable plasmid pBABED P U6 (University of Dundee, DU48788) yielding clones DU52808 and DU52809, respectively.
The quality-controlled (Sendai virus clearance, pluripotency, normal karyotype) iPSC line SB Ad3 clone 4, which was reprogrammed from skin fibroblasts of a female, 31 years old, healthy donor (StemBANCC consortium, [32]), was used for genome editing. Cells were treated with 10 μM ROCK inhibitor Y27632 for 2 h before nucleofection. Cells were dissociated into single cells using 0.5 ml Accutase (Merck). Subsequently, 800,000 iPSCs and 5 μg of each plasmid were transferred into the nucleofection solution of the Amaxa human stem cell nucleofector starter kit according to the manufacturer’s instructions (Lonza). Nucleofection was performed using program B-016 on the Amaxa nucleofector device (Lonza). Nucleofected iPSCs were seeded on a Matrigel-coated 6-well plate in medium containing 10 μM ROCK inhibitor Y27632. The next day, 0.5 μg/ml puromycin (Merck) was added for 48 h. Surviving single colonies were picked, transferred into 24-well plates, and expanded for characterization as described below.
Differentiation of iPSCs to NPCs
NPCs were generated using a protocol well-established by the StemBANCC consortium [33], which is based on a previously published method [34, 35] with minor modifications. In brief, neural induction of confluent iPSCs was induced by feeding with medium containing 10 μM SB431542 and 1 μM Dorsomorphin (both Merck) for 8–12 days depending on appearance of a uniform neuroepithelial sheet. Medium consisted of 250 ml Neurobasal™ Medium, 250 ml DMEM/F-12, GlutaMAX™ Supplement, 2.5 ml N-2 Supplement (100X), 5 ml B-27™ Supplement (50X, serum free), 500 μl 2-mercaptoethanol (50 mM), 2.5 ml MEM Non-Essential Amino Acids Solution (100X), 1.25 ml Penicillin-Streptomycin (10,000 U/mL), l-Glutamine (200 mM) (all Thermo Fisher Scientific), 125 μl Insulin solution human and 2.5 ml Sodium pyruvate solution (both Sigma). Afterwards, neuroepithelial sheets were dissociated with 1 mg/ml Dispase (Thermo Fisher Scientific) and replated as clusters on laminin-coated (Sigma) 6-well plates. Neural rosette formation was promoted by feeding with medium containing 20 ng/ml FGF2 (R&D Systems) from day 13 on. Background differentiation was removed by multiple 1 mg/ml Dispase dissociation steps. Cells were fed daily with medium from days 17 to 25. Cells were passaged using Accutase to obtain a single cell solution on day 25 and were subsequently expanded or frozen.
Differentiation of NPCs to Neurons
For neuronal differentiation, confluent NPCs were fed daily with medium containing 10 μM DAPT (STEMCELL Technologies), 50 μM cAMP (Sigma), 20 ng/ml BDNF, and 20 ng/ml GDNF (both Peprotech) for 7 days and re-seeded (Day 0) on plates coated with 0.07% polyethyleneimine (PEI) in borate buffer and 1:100 laminin (both Sigma) for neuronal maturation. DAPT was excluded from maturation medium and cells were fed by 50% medium change three times per week. Neurons derived from iPSCs were assayed on the days of maturation indicated in the “Results” section.
DNA Extraction
QuickExtract DNA Extraction Solution was used for the DNA extraction according to the manufacturer’s protocol (Epicenter). Briefly, a 6-well plate with confluent iPSCs was washed twice with PBS. Thereafter, 0.5 ml QuickExtract solution was added per well. The cells were scraped off the plate, vortexed for 15 s, and transferred to a 1.5 ml tube. The sample was incubated at 65 °C for 6 min, vortexed for 15 s, and incubated at 98 °C for 2 min. DNA was stored at − 80 °C.
T7 Endonuclease Assay
EnGen Mutation Detection Kit (NEB) was used according to the manufacturer’s instructions. The Tm calculator tool (https://www.neb.com/) was used for calculating the melting temperatures of primers. The genomic sequences containing the gRNA target sites were amplified using 5’-GTGGCACAGGTTTGAGACAGATCA-3’ as forward primer, and 5’-GCTGTTACACTCGCACCTGA-3’ as reverse primer. The amplified products were loaded on an E-Gel™ General Purpose Agarose Gels, 1.2% (Thermo Fisher Scientific) and run on an E-Gel iBase™ Power System device with 2 min of PRE-RUN and the program 1E-Gel 0.8–2% for 26 min.
DNA Sequencing
The genomic DNA fragment including the gRNAs target sites was amplified using HotStart Q5 Polymerase (NEB), and the primers described above. DNA sequencing was performed by Sequiserve (Vaterstetten, Germany).
Karyotyping
DNA was extracted from iPSC clones using Qiagen AllPrep DNA/RNA Micro Kits according to the manufacturer’s protocol (Qiagen, Hilden, Germany). DNA samples were sent to Life & Brain Genomics (Bonn, Germany) for karyotype analysis using the Illumina BeadArray Technology (HumanOmni2.5Exome-8 BeadChip v1.3, Illumina, San Diego, CA, USA). Genotypes were analyzed using GenomeStudio V2.0.2. For copy number analysis, the CNV-Partition algorithm version 3.2 (Illumina, San Diego, CA, USA) was applied. Copy number variants were reported, if larger than 350,000 base pairs.
Quantitative RT-PCR
RNA was isolated using RNeasy Mini Kit (Qiagen). Subsequently, cDNA synthesis was performed with VILO Super Script Master mix (Thermo Fisher Scientific) according to the manufacturer’s protocol. The cDNA was amplified in triplicates in a 384-well plate on a Quantstudio 6 device (Thermo Fisher Scientific) using QuantiFast Probe RT-PCR Master Mix (Qiagen) and TaqMan probes (Supplementary Table 1).
Immunocytochemistry
Cells were fixed in 4% paraformaldehyde (Alfa) and permeabilized in 0.1% Triton X-100. Afterwards, cells were blocked in 5% normal goat serum (NGS, Vector) for 2 h, followed by overnight incubation with the primary antibodies in 10% fetal calf serum (FCS, Thermo Fisher Scientific) (Supplementary Table 2). Subsequently, secondary antibodies (all Thermo Fisher Scientific) were applied in 5% FCS for 2 h. Cells were stained with a 1:15,000 dilution of Hoechst 33342 solution (1 mg/ml, Thermo Fisher Scientific) to visualize nuclei.
BrdU Labeling
5-Bromo-2′-deoxy-uridine (BrdU) Labeling and Detection Kit I (Roche) was used according to the manufacturer’s protocol. Cells were counted using a Neubauer counting chamber and Trypan blue solution (0.04%, Sigma) to stain dead cells. 100,000 NPCs per well were seeded on 96-well plates. On the next day, cells were incubated for 1 h with BrdU labeling agent, followed by fixation and incubation with anti-BrdU antibody. Nuclei were counterstained with Hoechst 33342 dye.
EdU Labeling
Briefly, 40-μm cell strainers (Falcon) were used to remove cell clumps, and 100,000 NPCs per well were seeded on 96-well plates. Click-iT™ 5-Ethynyl-2’-Deoxyuridin (EdU) Alexa Fluor™ 488 Imaging Kit was used according to the manufacturer’s protocol (Thermo Fisher Scientific) on the next day. Additionally, NPCs were immunostained for Nestin as described above.
Digital Image Analysis
All fluorescence images were taken from 96-well high-content imaging plates (Greiner) using 20× (49 visual fields per well) or 63× (125 visual fields per well) water objectives and the Opera Phenix imaging system (PerkinElmer). Image analyses were performed using either Columbus or Acapella Studio 4.1 software (PerkinElmer) with in-house customer scripts as described in details elsewhere [36]. The results were transferred to GraphPad Prism 7 (GraphPad Software) for statistical analysis.
The building block toolbox of Columbus was used for simple image analysis, e.g., quantification of the percentage of BrdU/EdU, Nestin, or VGLUT1/VGAT immunostained cells. The “Find Nuclei” building block was used to detect Hoechst stained nuclei. Size and intensity of Hoechst signal were used to discard condensed nuclei (dead cells). Next step was to detect the intensity of different channels (e.g., 488 nm for BrdU) to determine the number of positive cells of this population (e.g., % BrdU-positive cells of living cells). Cytoplasm was determined by MAP2 staining with the “Find Cytoplasm” building block. MAP2-positive neurons were determined by setting a threshold for MAP2 intensity. VGAT- or VGLUT1-positive neurons were determined by counting all MAP2-positive neurons displaying VGLUT1 or VGAT immunofluorescence in the cytoplasm above a certain threshold.
Acapella Studio 4.1 software was used to design scripts that extract the following read-out parameters. For the definition of MAP2-positive cells, the nuclei mask was determined by applying the nucleus detection algorithm “C” to the HOECHST staining. MAP2-positive neurons were determined by defining a 15–18 μm reference ring region around the nucleus and calculating MAP2 immunofluorescence intensity values for the cytosol region (mean intensity) and for the reference region (10% quantile intensity = 10% darkest pixel). This quantile calculation became necessary, since the neurons form roots (extensions) crossing the reference region. In case of applying simply the mean intensity for the reference region, different values would result for neurons with different number of roots. To score for MAP2-positive neurons the ratio of “MAP2 mean intensity (cytosol region)”/“MAP2 10% quantile intensity (ring region)” had to exceed a factor of 40. The number of MAP2+ neurons with roots was determined by all MAP2+ cells expressing at least one extension (root) from the neuron body. The mean neurite length was calculated by summing up the length of all MAP2 extensions (determined by the Acapella neurite detection module using a signal to background of 1.5) normalized to the number of all MAP2+ cells. Number of roots per MAP2+ roots was determined by dividing the total number of roots by total number of MAP2+ with roots.
Pre- and post-synaptic spots were determined by discriminating spots with a 2.0 (pre) and 1.4 (post) fold higher mean intensity than the local background (calculated by the ring region at a distance of 0.4–0.6 μm around the spot). The spot search mask was defined as the MAP2 detected dendrite network including 1.0 μm beyond the MAP2 mask. In case pre- and post-synaptic spots overlap with at least one pixel (0.2 μm × 0.2 μm), they were counted as overlapping spots (synapses). This value was finally normalized to the length (in μm) of the neurite network.
Phalloidin staining was used to determine F-actin labeled growth cone areas and intensities. Phalloidin labeled cones located at MAP2-positive extensions had to express a higher Phalloidin intensity than the Phalloidin reference ring at a distance of 0.6–1.2 μm around cones. The growth cone area per neuron was calculated by summing up all growth cone areas and dividing by the number of neurons. The growth cone intensity per neuron was calculated by summing up all growth cone pixel intensities and dividing by the number of neurons.
DNA Histograms
NPCs were filtrated using 40 μm cell strainers to remove cell clumps, and seeded on 6-well plates at a density of 2 × 106 cells per well. Medium was completely changed on the next day. On day 3, cells were dissociated using Accutase (5 min at 37 °C), pelleted by centrifugation, resuspended in PBS, and counted. Cells were transferred into 2 ml tubes (1 × 106 cells per tube). The Propidium Iodide Flow Cytometry Kit (Abcam) was used following the manufacturer’s protocol. Cell suspensions were measured with a MACSQuant flow cytometer (Miltenyi Biotech) and analyzed with FlowJo software (Tree Star).
Neurite Outgrowth Assay
After 7 days of neuronal differentiation, cells were mechanically lifted, filtrated using a 40 μm cell strainer, and re-seeded at a density of 25,000 cells per well on PEI and laminin-coated 96-well plates. Cells were incubated for 1 h at 37 °C. Subsequently, non-adherent cells were gently removed by changing the medium, and adherent cells were fixed with 4% PFA. Nuclei of adherent cells were stained with Hoechst dye and counted. The neurites were visualized by immunostaining for MAP2. Growth cones were detected by Phalloidin staining of actin filaments (Thermo Fisher Scientific) [37]. Image analysis was performed using Acapella software as described above. For rescue experiments, iPSC-derived neurons were treated with the compounds/proteins listed in Supplementary Table 3 after 7 days of differentiation. Neurons were treated 4 h before mechanical detachment and 4 h after subsequent re-seeding of the neurons to ensure inhibition/activation of the signaling pathway of interest throughout the experimental procedure (8 h in total). Neurite outgrowth was analyzed as described above.
Multi-electrode Array Analysis
Neurons were plated at a density of 100,000 cells per well on 24-well multi-electrode array (MEA) plates (MultiChannel Systems, MCS) coated with 0.07% PEI and laminin. Recordings were performed using Multiwell Screen software (MCS) using a bandwidth filter from 100 Hz to 5 kHz cutoff frequencies and at a sampling rate of 20 kHz. The plates were recorded for 10 min at 37 °C after 5 min equilibration. Analysis was performed using Multiwell Analyzer software (MCS). A spike was counted when the extracellularly recorded signal exceeded a threshold of five times the standard deviation of the baseline noise level [38]. Electrodes were considered active, if spike rate ≥ 0.01 Hz. The ERBB3/4 ligand Neuregulin-1b peptide (50 nM) was added during each 50% medium exchange three times per week.
RhoA Activity Assay
The RhoA G-LISA Activation Assay Kit (Colorimetric format, Cytoskeleton) was used to determine the levels of active (GTP-bound) RhoA according to manufacturer’s protocol.
Cell Lysis
Cells were lysed with cell lysis buffer (Cell Signaling Technology) supplemented with 1:100 protease inhibitor cocktail and phosphatase inhibitor cocktail 2 (both Sigma). Samples were incubated on ice for 15–20 min. Subsequently, cell debris was pelleted by centrifugation at 21,100 ×g for 10 min at 4 °C, and supernatants were used for further analysis.
Extraction of Nuclear and Cytoplasmic Proteins
Protein extraction was performed using the NE-PER Nuclear Cytoplasmic Extraction Reagent kit according to the manufacturer’s protocol, (Thermo Fisher Scientific). Briefly, cells of a 6-well plate were washed twice with PBS and immediately put on ice. The cells were lysed with 200 μl of cytoplasmic extraction reagent I supplemented with 1:100 protease inhibitor cocktail and phosphatase inhibitor cocktail 2 (both Sigma). Cytoplasmic and nuclear fraction were separated by centrifugation at 21,100 ×g and used for Western blotting.
Protein Concentration Assay
Protein concentration was determined using BioRad Assay Dye Reagent and a microplate reader Wallac Victor (Perkin Elmer). Briefly, 10 μl of BSA protein standards and 10 μl protein samples (diluted 1:10 in ddH2O) were added to a 96-well plate. Two hundred microliters of BioRad Assay Dye Reagent was added and incubated for 5 min in the dark. Plate was measured and protein concentration was calculated using Excel and GraphPad Prism.
Western Blot Analysis
Proteins were loaded on NuPAGE Novex 4–12% Bis-Tris Gels (LifeTechnologies). Gel electrophoresis was performed for 45 min at 200 V in 3-(N-morpholino)propanesulfonic acid (MOPS) buffer (Invitrogen). Next, proteins were transferred to nitrocellulose membranes using Tris-Glycine Buffer (BioRad) supplemented with 20% methanol at 100 V for 90 min. Membranes were stained with MemCode Reversible Protein Stain (Pierce) as control for equal protein loading. Afterwards, membranes were blocked with 5% milk in TBS-T for 3 h, and incubated in primary antibodies overnight at 4°C in 5% BSA blocking buffer (see Supplementary Table 2). Secondary antibodies were used in 2% BSA blocking buffer for 3 h, followed by chemiluminescence detection with ECL Western Lightning Plus (Perkin Elmer) on a ChemiDoc Imager (Biorad). Densitometry analysis was performed with ImageQuantTL software (GE).
RNA Extraction for RNA-Sequencing
NPCs (n = 3 wells per clone) and early neurons (n = 5 wells per clone) derived from iPSCs were lysed using RLT buffer supplemented with 1% beta-mercaptoethanol after 1 week of differentiation. The Ambion Magmax™-96 total RNA isolation kit (Thermo Fisher Scientific) was used according to the manufacturer’s instructions for total RNA extraction. RNA quality and concentration were assessed using a Biotek Reader (Take3 Plate, UV) and a Fragment Analyzer using a Standard sensitivity total RNA Kit (Agilent Technologies). All RNA samples had RNA integrity numbers (RIN) > 7.
Illumina Library Preparation and Sequencing
TrueSeq RNA Sample Prep Kit v2-Set B (Illumina Inc.) was used to produce a 300 bp fragment including adapters in average size from 200 ng of total RNA input. The adapter indices were applied for the normalization and pooling of 12 individual libraries before sequencing. Subsequently, clustering of pooled libraries was done on the cBot Instrument using the TruSeq SR Cluster Kit v3-cBot – HS (Illumina Inc.). Followed by sequencing as 74 bp, single reads and 7 bases index read using the TruSeq SBS Kit HS-v3 (50-cycle) on an Illumina HiSeq4000 instrument (Illumina Inc).
Bioinformatics Analysis of RNA-Sequencing Data
RNA-seq reads were aligned to the human genome using the STAR Aligner v2.5.2a with their corresponding Ensembl 84 reference genome (http://www.ensembl.org). Sequenced read quality was checked with FastQC v0.11.2 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and alignment quality metrics were calculated using the RNASeQC v1.18. Following read alignment, duplication rates of the RNA-Seq samples were computed with bamUtil v1.u.11 to mark duplicate reads and the dupRadar v1.4 Bioconductor R package for assessment. The gene expression profiles were quantified using Cufflinks software version 2.2.1 to get the Reads Per Kilobase of transcript per million mapped reads (RPKM) as well as read counts from the feature counts software package. The matrix of read counts and the design file were imported to R, normalization factors calculated using trimmed mean of M-values (TMM) and subsequently voom normalized, before subjected to downstream descriptive statistics analysis. Transcripts showing RPKM values > 5 in at least one group were considered in the final analyses [39]. The Benjamini-Hochberg’s method was used to correct for multiple testing, and only protein-coding genes with adjusted p value < 0.01, independent of magnitude of change, were considered as differentially expressed and used in the subsequent analyses.
Ingenuity Pathway Analysis (IPA)
IPA (version 01–7) was used for pathway enrichment analysis in Ingenuity knowledge base (http://www.qiagen.com/ingenuity). Genes (transcripts) with an adjusted p value < 0.01 and an expression level > 5 RPKM were included in the analysis.
Statistical Analysis
GraphPad Prism 7 was used for all statistical analysis. Data are from at least three independent experiments and three different cell batches unless indicated otherwise. Error bars represent mean ± standard error of the mean (SEM). Statistical testing was performed using unpaired t test (two-tailed) or one-way ANOVA followed by Dunn’s multiple comparisons test, if more than two groups were compared. Results were considered statistically significant, if p value < 0.05.
Results
Generation of Human Isogenic KCTD13 Knockout iPSC Lines
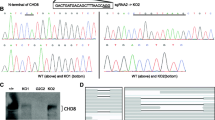
Since several studies point to a critical role of KCTD13 in neuropsychiatric disorders induced by 16p11.2 deletions [16, 17], we investigated the consequences of KCTD13 deficiency in human iPSC and iPSC-derived neurons. We used CRISPR/Cas9 genome modification, which enables the generation of isogenic mutant iPSC lines, thereby reducing genetic background variability [40, 41]. Analysis of the KCTD13 locus (ENSG00000174943, NC_000016.10 (29,906,335...29926232)) highlighted exon 2 as the farthest upstream common coding exon shared by all known transcript variants. A human iPSC line, generated and characterized by the StemBancc consortium, was nucleofected with a plasmid vector encoding CRISPR/Cas9 nickase (Cas9n) and antisense gRNA targeting the KCTD13 locus at exon 2, and a second plasmid vector encoding sense gRNA and an antibiotic resistance gene (Fig. 1a). The Cas9n double-nicking approach was chosen to increase target specificity and reduce off-target effects as shown by others [42]. After puromycin selection, iPSC clones carrying CRISPR/Cas9n-mediated insertions/deletions were identified by T7 endonuclease assay (Fig. 1c). By genomic DNA sequencing, we detected a five-base-pair (bp) deletion in one allele (at bp position 3632–3636 of the human KCTD13 gene ENSG00000174943) of a heterozygous KCTD13 KO (KCTD13+/−) iPSC clone (Fig. 1b). We also detected a homozygous KCTD13 KO (KCTD13−/−) clone carrying two different deletions on each allele (at bp position 3569–3631 and 3621–3683, respectively). All deletions affect the BTB/POZ domain of KCTD13 protein which is required for its interaction with Cullin-3 [14, 43]. Quantitative real-time polymerase chain reaction (qRT-PCR) confirmed a corresponding decrease in KCTD13 mRNA expression in the heterozygous and homozygous KO neurons (Fig. 1d). Consistently, KCTD13 protein levels were reduced to 45 ± 3.8% (mean ± SEM) in the heterozygous KO neurons and to 11 ± 1.5% in the homozygous KO neurons as shown by Western blotting (Fig. 1e) (data from four independent experiments). Karyotyping of iPSC lines detected no larger chromosomal aberrations.
Generation and validation of KCTD13 knockout (KO) by CRISPR/Cas9 nickase in human iPSCs. a Gene targeting strategy using Cas9 nickase and gRNA sequences targeting the KCTD13 gene. Exons are shown as black rectangles. Guide RNA sequences are labeled in red. b Data from DNA sequencing showing deletions in heterozygous (KCTD13+/−) KO iPSC clones (5 bp in one allele) and in homozygous (KCTD13−/−) KO iPSC clones (63 bp in both alleles). The gRNA target sequences are labeled in red. c Identification of CRISPR/Cas9 nickase induced insertions/deletions in the KCTD13 gene by T7 endonuclease 1 cleavage assay. Lane 1: genomic DNA from WT control iPSC line; lane 2: Cas9/gRNA-transfected homozygous KO iPSC clone; lane 4: heterozygous KO iPSC clone; lane 5 (M): DNA Ladder (0.1–10.0 kb, NEB). d qRT-PCR analysis demonstrating decreased KCTD13 mRNA expression in KCTD13 KO iPSC-derived neurons compared to WT neurons. Data from 2 independent experiments; One-way ANOVA followed by Dunnett's multiple comparison test; ****p < 0.0001. e Representative Western blot image from iPSC-derived neurons showing a corresponding decrease in KCTD13 protein levels. Total protein stain confirms similar protein loading (10 μg protein each)
KCTD13 Deficiency Does Not Alter Cell Identity or Differentiation Efficiency of iPSCs, NPCs, and Cortical Neurons
Since KCTD13 is expressed in human iPSCs [44], we investigated a potential role in stemness. Expression of cell surface protein TRA1-60 and nuclear proteins NANOG, OCT4, and SOX2 was assessed in iPSC cultures by immunostaining and high-content digital image analysis. Both immunofluorescence intensity and number of iPSCs immunopositive for these stemness markers did not differ between KCTD13 KO lines and the isogenic control cell line (KCTD13+/+) (Fig. 2a, b). These findings were confirmed using qRT–PCR analysis of NANOG, OCT4, and SOX2 which revealed similar mRNA expression levels (Supplementary Fig. S1a–c). Our results suggest that KCTD13 deficiency does not affect stemness of human iPSCs. Similar findings have been reported from iPSCs derived from 16p11.2 deletion carriers [44]. Expression levels of KCTD13 mRNA are high in the human fetal brain [17]. Therefore, we investigated the effects of KCTD13 deficiency on iPSC differentiation along the neural lineage. KCTD13 KO iPSC lines and the isogenic control line were differentiated into NPCs and into cortical neurons using a well-established small molecule-based protocol [34]. Differentiation of iPSCs into NPCs resulted in a similar percentage of Nestin+ cells (an early neural progenitor marker) in KCTD13 KO lines compared to isogenic controls (Fig. 2c, d). Moreover, we found no differences in cell adhesion or cell viability (ratio of condensed to non-condensed nuclei) in KCTD13 KO NPC clones (Fig. 2e, f).
KCTD13 deficiency does not affect cell identity or differentiation efficiency of iPSCs, NPCs or neurons. a Representative confocal images of immunostainings for stemness markers (OCT4, TRA-1-60) in iPSC cultures. Scale bar, 50 μm. b High content image analysis revealed similar mean immunofluorescence intensity of stemness-associated markers (TRA-1-60, NANOG, SOX2, OCT4). Total n = 3–12 wells per iPSC line. c Confocal images of immunostaining for the NPC marker Nestin and nuclear Hoechst 33342 labeling. d, e Analysis of Nestin+ cells and cell viability (number of non-condensed Hoechst+ nuclei) revealed no differences between KCTD13 KO and isogenic control NPCs. Total n = 26–27 wells per NPC line. f KCTD13 deficiency did not alter percentage of adherent NPCs. Total n = 21 wells per NPC line. g Representative confocal images of immunostainings of iPSC-derived neurons for the pan-neuronal marker MAP2 (red) and the GABAergic neuron marker VGAT (green). Scale bar, 50 μm. h High content image analysis revealed similar percentage of MAP2+ neurons per Hoechst-labeled nuclei in cultures of KCTD13 KO lines and controls. Total n = 28–35 wells per neuron line. i, j KCTD13 deficiency did not significantly change percentage of VGAT+ (i) or VGLUT1+ (j) neurons per MAP2+ neurons. Total n = 15–17 wells per neuron line. All data are from at least three independent experiments and three different cell batches unless indicated otherwise
After 21 days of maturation, cortical neurons differentiated from WT and KCTD13 KO iPSC were immunostained for MAP2 Microtubule-associated protein 2 (MAP2), a pan-neuronal marker, for vesicular GABA transporter (VGAT), a marker for GABAergic neurons, and for vesicular glutamate transporter (VGLUT), a marker for glutamatergic neurons (Fig. 2g). The percentage of MAP2+ neurons did not differ between the genotypes (Fig. 2h). About 10–20% of the MAP2+ cells were VGAT+ GABAergic neurons (Fig. 2i), and 75–85% were VGLUT+ glutamatergic neurons (Fig. 2j). High content image analysis did not detect a significant difference between the genotypes.
NPCs Differentiated from KCTD13 Knockout iPSCs Exhibit a Moderate Decrease in Cell Proliferation
Dysregulation of early developmental pathways affecting proliferation and apoptosis in NPCs may contribute to macrocephaly in 16p11.2 deletion carriers [10, 45]. Examples include knockdown of the KCTD13 orthologue in zebrafish which led to increased brain size, and knockdown of KCTD13 in the embryonic mouse brain which led to enhanced cell proliferation [19]. To examine the effect of KCTD13 KO on human NPC proliferation, we analyzed the incorporation of 5-bromo-2′-deoxyuridine (BrdU) into newly replicated DNA, which is widely used to assess DNA replication in proliferating cells [46]. The percentage of BrdU+ NPCs was significantly reduced in KCTD13 KO clones, whereas cell viability (ratio of condensed to non-condensed nuclei) was unchanged (Fig. 3a and Supplementary Fig. S2a, b). Double staining for the early neuronal marker Nestin and incorporated 5-ethynyl-2-deoxyuridine (EdU) confirmed a significant reduction of proliferation of KCTD13 deficient, Nestin+ NPCs (Fig. 3b, c and Supplementary Fig. S2c). Distribution of NPCs within the cell cycle was analyzed using flow cytometry after DNA staining with propidium iodide (Fig. 3d). Propidium iodide intercalates into double-stranded DNA producing a fluorescent signal. Signal intensity depends on the amount of DNA present in the cell [47]. The percentage of cells in the S phase was significantly lower in the KCTD13 KO NPCs (Fig. 3e), whereas no significant changes were observed in the percentage of cells in the G0/G1 or G2/M phase of the cell cycle, and in the percentage of cells within the gate (Supplementary Fig. S2d–f). Consistently, mRNA expression of multiple cell cycle–associated genes significantly declined in KCTD13 KO NPCs (Fig. 3f). Furthermore, Ingenuity pathway analysis (IPA) of our RNA-seq data indicated deregulated pathways relevant to cell cycle regulation and DNA repair/replication (Supplementary Fig. S2g ). In a biochemical assay, recombinant KCTD13 interacted directly with PCNA and polymerase delta, and stimulated DNA polymerase activity twofold to threefold [15]. Therefore, we investigated the localization of KCTD13 and PCNA in our iPSC-derived NPCs. We first confirmed the separation of the cytoplasmic and nuclear protein fraction by immunoblotting c-FOS, a nuclear transcription factor. Next, we showed that KCTD13 and PCNA are co-localized both in the cytoplasmic and nuclear protein fraction (Fig. 3g). Unfortunately, we could not co-immunoprecipitate KCTD13 interacting proteins from human iPSC-derived NPCs or neurons, since all commercially available antibodies against KCTD13 did not immunoprecipitate endogenous KCTD13 from mouse brain lysates or KCTD13 overexpressed in non-neuronal HEK293 cells (data not shown). Moreover, non-specific protein bands were detected by the anti-KCTD13 antibody on Western blots (Fig. 3g) precluding its use in immunocytochemistry.
Decreased cell proliferation and DNA synthesis in KCTD13 KO iPSC-derived NPCs. a KCTD13 KO NPCs exhibit a decreased number of BrdU+ cells relative to isogenic WT NPCs. Total n = 29 wells per NPC line; one-way ANOVA followed by Dunnett's multiple comparison test; ****p = 0.0001. b Representative confocal images of EdU/Nestin double stainings of NPCs. Scale bar, 50 μm. c High content image analysis revealed a significant reduction in EdU+/Nestin+ double-positive NPCs in KCTD13 KO cultures. Total n = 28–29 wells per NPC line; one-way ANOVA followed by Dunnett's multiple comparison test; ****p < 0.0001. d Top: Flow cytometry graph showing gated cell population. Bottom: Representative DNA histogram showing cell cycle phases in red and percentage of cells in a specific phase. e Reduced percentage of KCTD13 KO NPCs in S-phase (DNA synthesis). Total n = 28–31 measurements per NPC line from five independent experiments; One-way ANOVA by Dunnett's multiple comparison test; **p = 0.0027; ***p = 0.0001. f Significant decline in mRNA expression (RPKM values) of numerous cell cycle associated genes in KCTD13 KO NPCs compared to WT with a minimum p value of < 0.05 (for detailed statistics see Supplementary Table 4). g Detection of c-FOS protein in the nuclear (N) but not in the cytoplasmic (C) protein fraction. KCTD13 and PCNA were found in both protein fractions of WT NPCs. Please note a non-specific protein band at 25 kDa detected by the anti-KCTD13 antibody precluding its use in immunocytochemistry
KCTD13-deficient Cortical Neurons Show Impairment in Early Neurite Formation
Neurite outgrowth is a critical step in the formation of neural network connections in the developing mammalian central nervous system [48]. Since RhoA is involved in early neurite formation [49], and since KCTD13/Cullin-3 regulates RhoA degradation in non-neuronal HeLa cells [14], we hypothesized that knockout of KCTD13 may affect neurite outgrowth in our iPSC-derived neurons in a RhoA-dependent manner. For detailed morphological analysis, we replated iPSC-derived neurons after 7 days of neuronal differentiation (Day 0). We combined MAP2 staining with the Acapella Studio software to detect MAP2+ neurons, neurite length, and neurite roots by high content image analysis (Fig. 4a). Percentage of MAP2+ neurons, viability of neurons (ratio of condensed to non-condensed nuclei), and neuronal cell adhesion was unchanged following KCTD13 KO (Fig. 4b–d). More importantly, percentage of MAP2+ neurons with roots, and number of roots per MAP2+ neurons were significantly reduced in the KCTD13 KO clones compared to the isogenic controls (Fig. 4e, f). Additionally, mean neurite length was significantly shorter in the KCTD13 KO neurons compared to control neurons (Fig. 4g).
Impaired early neurite formation in KCTD13 deficient iPSC-derived neurons. a Representative confocal images of MAP2 immunostaining of postmitotic neurons after reseeding. Scale bar, 50 μm. b–d High content image analysis revealed no changes in cell viability (number of non-condensed Hoechst+ nuclei) (b), in percentage of adherent cells (c), and in percentage of MAP2+ neurons (d) in KCTD13 KO versus isogenic WT cultures. e–g KCTD13 KO neurons exhibit a significant decrease in percentage of MAP2+ neurons with roots (e), in number of roots per MAP2+ neuron (f), and in mean neurite length (g). Total n = 28–31 wells per genotype; one-way ANOVA followed by Dunnett's multiple comparison test; ****p < 0.001. Data are from at least three independent experiments and three different cell batches unless indicated otherwise
Inhibition of RhoA Does Not Rescue Impaired Neurite Formation
To detect a potential accumulation of active (GTP-bound) RhoA and total RhoA protein in KCTD13 deficient neurons, we used RhoA GLISA and Western blotting, respectively. Both methods detected similar levels of RhoA in whole cell lysates from KCTD13 KO neurons and isogenic controls (Fig. 5a, b). Since changes in RhoA and F-actin might be restricted to the neuronal growth cone (neurite compartment) in KCTD13 KO neurons and not detectable by whole cell analyses, we performed two additional experiments. Treatment of KCTD13 KO iPSC-derived neurons with the small molecule RhoA inhibitor Rhosin (50 μM, 8 h) did not rescue impaired neurite formation, although Rhosin at this concentration reversed synaptic deficits in slices from KCTD13 KO mice [50]. In our human iPSC-derived neuron cultures, however, Rhosin treatment caused a decrease in neurite length, in the percentage of MAP2+ neurons with roots, and in the number of roots per MAP2+ neurons (Fig. 5c and Supplementary Fig. S3a, b). Consistently, treatment with an inhibitor (Y-27632, 10 μM, 8 h) of Rho kinase, a downstream effector of RhoA, resulted in a similar decrease in neurite length and in number of roots (data not shown). Staining for F-actin using the fluorescent dye Phalloidin and high-content image analysis indicated a reduction of F-actin area and fluorescence intensity per growth cone in KCTD13 KO neurons (Fig. 5d–f).
Analysis of the role of RhoA in iPSC-derived KCTD13 KO neurons. a Active RhoA protein levels were unchanged in KCTD13 KO iPSC-derived neurons as measured by GLISA (absorbance at 490 nm). Total n = 5 wells per neuron line. b Representative Western blot image showing similar total RhoA protein levels. Total protein stain (MemCode) confirms similar protein loading. c Reduced neurite formation in KCTD13 KO neurons was not rescued, but further impaired by treatment with the RhoA inhibitor Rhosin (50 μM). Total n = 14–18 wells per neuron line. Phenotypic changes: One-way ANOVA followed by Dunnett's multiple comparison test; **p = 0.002 (KCTD13+/+ vs. KCTD13+/−), **p = 0.0017 (KCTD13+/+ vs. KCTD13−/−). Treatment effect: Unpaired t test: **p = 0.0091, ***p = 0.0009, ****p < 0.0001. d Representative confocal images of (filamentous) F-actin staining by Phalloidin in neuronal growth cones. Scale bar, 50 μm. e, f Both area and intensity of F-actin staining per growth cone were reduced in KCTD13 KO neurons. 1 pixel = 0.6 μm × 0.6 μm. Total n = 28 wells; one-way ANOVA followed by Dunnett's multiple comparison test; ****p < 0.0001
ERBB Signaling Contributes to Impaired Neurite Formation in KCTD13 KO Neurons
For an unbiased detection of potential alternative mechanisms that might underlie impaired neurite formation in KCTD13 KO neurons, we performed Ingenuity pathway analysis (IPA) using RNA-seq data from all genotypes. Principal component analysis of differential gene expression data showed a clear separation of homozygous KCTD13 KO neurons from heterozygous KO neurons and isogenic WT controls (Fig. 6a). In addition, neurological diseases and psychological disorders were indicated as top-ranking diseases (Fig. 6b). More importantly, IPA pointed to ERBB2, TP53, and VEGF as most significant upstream regulators in KCTD13 KO iPSC-derived neurons (Fig. 6c). A critical role for pro-apoptotic TP53 seemed unlikely, since KCTD13 deficiency did not affect cell viability (Figs. 2e and 4b). Treatment with recombinant VEGF did not rescue impaired neurite formation in KCTD13 KO neurons (data not shown) excluding a central role for VEGF in this phenotypic alteration.
Phenocopy and rescue, respectively, of impaired neurite formation by modulation of ERBB signaling in iPSC-derived neurons. a Principal component analysis based on RNA sequencing data showed a clear separation of homozygous KCTD13 KO neurons from heterozygous KO and WT neurons. Mutant and WT NPCs cluster together. b Ingenuity pathway analysis of RNA sequencing data revealed top 5 diseases in KCTD13 KO neurons. c Top 3 upstream regulators identified by Ingenuity pathway analysis points to a significant role of ERBB signaling in homozygous KCTD13 KO neurons. d Phenocopy of decreased neurite formation in WT neurons and further impairment in KO neurons by treatment with the specific ERBB family inhibitor Lapatinib (10 μM). Total n = 59–64 wells per neuron line; ****p < 0.0001; unpaired t test. e Rescue of decreased neurite formation in KCTD13 KO neurons by treatment with the specific ERBB3/4 ligand NRG1b (50 nM). Total n = 63–90 wells per neuron line from 3 to 4 independent experiments. Phenotypic changes: One-way ANOVA followed by Dunnett's multiple comparison test; **p = 0.0045, ****p = 0.0001. Treatment effect: Unpaired t test: *p = 0.0147, ****p < 0.0001
On the mRNA level, expression of ERBB2 was highest compared to the other ERBB family members in iPSC-derived neurons. On the protein level however, expression of ERBB2 was lower compared to ERBB4, which showed a trend towards a decrease in homozygous KCTD13 KO neurons (Supplementary Fig. S4a, b). To assess the role of ERBB signaling, we first tested the effect of the specific ERBB family inhibitor Lapatinib [51] on neurite formation. Interestingly, Lapatinib treatment (10 μM, 8 h) phenocopied the effect of KCTD13 KO in WT neurons and further reduced neurite length in KCTD13 KO neurons (Fig. 6d). The reversible inhibitor Tucatinib, which shows a higher selectivity for ERBB2 and a higher potency compared to Lapatinib [52] did not alter neurite formation (1 μM, 8 h) in WT and KO neurons (Supplementary Fig. S4c). For activation of the ERBB signaling cascade, we used the ERBB3/4 ligand Neuregulin-1b (NRG1b), which has already been shown to increase neurite length in rat hippocampal and motor neurons in culture [53,54,55]. Treatment with the EGF-like domain of NRG1b (50 nM, 8 h) reversed the deficit in neurite formation in both homozygous and heterozygous KCTD13 KO neurons (Fig. 6e; Supplementary Fig. S4d). To assess a potential contribution of ERBB1 signaling, we also tested the specific ERBB1 ligand epidermal growth factor (EGF) [56], which has been demonstrated to increase neurite outgrowth in mouse cortical neuron cultures [57]. Treatment with EGF at different concentrations, however, impaired neurite formation in human iPSC-derived cortical neurons (Supplementary Fig. S4e).
To confirm these findings in additional iPSC lines, we assessed neurite formation and the effect of NRG1b treatment in neuron cultures derived from an isogenic subclone of our wildtype parental iPSC line (KCTD13+/+_2) and from one additional homozygous KCTD13 KO iPSC clone (KCTD13−/−_2). By qRT-PCR we detected a decrease in KCTD13 mRNA expression to 4.5 ± 0.5% and by Western blotting a decline in KCTD13 protein levels to 12 ± 2.6% (mean ± SEM) in the KCTD13−/−_2 line compared to the isogenic wildtype line. Sequencing of genomic DNA revealed a 254 bp deletion in one allele (at bp position 3428–3681 of the human KCTD13 gene) and a 51 bp insertion in the other allele (position 3664–3681). Both a significant impairment in neurite formation and a rescue by single NRG1b treatment in homozygous KCTD13 KO neurons derived from KCTD13−/−_2 iPSCs could be confirmed in these experiments (Supplementary Fig. S5).
Impaired Neurite Elongation and Decreased Neuronal Network Activity in Mature KCTD13 KO Neurons
There is growing evidence for deficits in synaptic connection and function in many neuropsychiatric disorders [58,59,60]. Consequently, we assessed whether impaired neurite elongation was detectable in mature KCTD13 KO neurons (21 days of maturation). As shown in Fig. 7a, mean neurite length was significantly reduced in a gene dose-dependent manner in KCTD13 deficient iPSC-derived neurons. Next, we measured the number of synaptic puncta labeled by the presynaptic marker Synapsin 1/2 (SYN1/2) and the postsynaptic marker postsynaptic density protein 95 (PSD95). We focused on puncta which showed an overlap of the pre- and post-synaptic markers and which localize close to MAP2+ dendrites. High-content image analysis revealed no significant differences in synapse density per micrometer neurite between the genotypes (Fig. 7b). As a functional assay for neuronal network activity, we performed recordings on 24-well multi-electrode array (MEA) plates. We detected a significant, gene dose-dependent decreased spike rate in KCTD13 deficient neurons, whereas the mean number of active electrodes per well was unchanged (Fig. 7c–e). Unexpectedly, repeated administration of NRG1b (50 nM) to KCTD13 deficient neurons cultured on MEA plates did not rescue decreased neuronal electrical activity (Supplementary Fig. S6).
Deficits in neurite elongation and in spontaneous network activity in mature neurons derived from KCTD13 KO iPSCs. a Mean neurite length showed a gene dosage dependent decrease in KCTD13 deficient neurons after 21 days of maturation. Total n = 32–36 wells per neuron line; one-way ANOVA followed by Dunnett's multiple comparison test; *p = 0.031, ****p < 0.0001. b Left: Representative confocal images of immunostaining for presynaptic SYN1/2 (green) and postsynaptic PSD95 (red) in iPSC-derived MAP2+ (blue) neurons after 21 days of maturation. Right: High content image analysis of overlapping PSD95+/SYN1/2+ puncta revealed no significant differences in synapse density per μm neurite between genotypes. Total n = 16–18 wells per neuron line. Scale bar, 20 μm. c Multi-electrode array analysis revealed similar number of active electrodes per well in KCTD13 KO versus WT neuron cultures. d Mean spike rate showed a gene dosage dependent decrease in KCTD13 deficient neurons after 21 days of maturation. Total n = 39–41 wells per neuron line; One-way ANOVA followed by Dunnett's multiple comparison test; *p = 0.0111, ****p = 0.0001. e Representative raster plots of spontaneous neuronal activity on 24-well multi-electrode array plates (one representative well containing 12 electrodes each)
Discussion
KCTD13, one of 29 genes located in the 16p11.2 locus, is considered an important contributor to neuropsychiatric diseases associated with 16p11.2 CNVs [17, 19], however, its molecular function in human brain development is poorly understood. A knockdown screen in zebrafish pointed to KCTD13 as a major driver for increased brain size associated with 16p11.2 CNV deletions, whereas more recent studies revealed no alterations in brain size and proliferation of NPCs in KCTD13 deficient zebrafish and mice, respectively [19, 50, 61]. In the present study, we used CRISPR/Cas9 nickase to generate KCTD13 deficient human iPSCs and isogenic controls, in order to identify structural and functional alterations during differentiation into human cortical neurons. Unexpectedly, our results demonstrate that heterozygous and homozygous knockout of KCTD13 moderately decreases proliferation of human NPCs in culture. We detected decreased proliferation of KCTD13 deficient NPCs in independent experiments using different read-outs. Importantly, viability, adhesion, and differentiation of KCTD13 KO NPCs were not affected. Our data from RNA-seq indicated decreased expression of many cell cycle-regulating genes in KCTD13 KO NPCs. Moreover, pathway analysis revealed that numerous biological processes associated with cell cycle and DNA replication were dysregulated. In a cell-free assay, binding of recombinant KCTD13 stimulates activity of DNA polymerase delta, and thus may play a role in DNA synthesis and cell cycle control during mitosis [15]. Consistently, we detected a moderate (≤ 18%) decrease in DNA synthesis in KCTD13 deficient NPCs, which may slow cell cycle progression. Interestingly, crossing KCTD13-deficient mice with genetic interactors identified in zebrafish lead to a reduction in brain volume in double mutant mice pointing to complex interactions of genes within the 16p11.2 locus [61].
Mitochondrial dysfunction has been reported in various neurodegenerative/neuropsychiatric disorders including Alzheimer’s disease, Parkinson’s disease, schizophrenia, and bipolar disorder [62]. Mitochondria-related gene expression was globally downregulated in post-mortem brains of patients with schizophrenia and bipolar disorders, however, an effect of medication could not be excluded [63]. Interestingly, iPSC-derived dopaminergic neurons from three schizophrenia patients showed a significant downregulation of mitochondrial genes encoding complex 1 subunits (NDUFV1, NDUFV2) and a decrease in mitochondrial membrane potential [64] speaking in favor of cell intrinsic defects. In a recent larger iPSC study (15 schizophrenia lines, 15 healthy control lines) downregulated mRNA expression of mitochondrial complex 1 subunits (MT-ND2, MT-ND4L) and decreased maximal respiration of mitochondria were observed. Importantly, these alterations were detectable in iPSC-derived cortical GABAergic interneurons, but not in cortical glutamatergic neurons pointing to cell-specific pathomechanisms [65]. In our KCTD13 deficient iPSC-derived glutamatergic neurons, we observed a minor (≤ 1.2-fold) increase in mRNA levels of mitochondrial complex 1 subunits (NDUFS4, NDUFS2). We cannot exclude however, that biologically-relevant mitochondrial changes may become detectable under conditions of cellular stress (e.g., depletion of medium supplements containing antioxidants) or following differentiation into other neuronal subtypes. In line with recent studies showing reduced dendrite length in KCTD13 knockout mice and decreased dendritic arborization following KCTD13 knockdown in Drosophila [50, 66], we detected decreased neurite formation/elongation in KCTD13 knockout cortical neurons derived from human iPSCs. Deficits in neurite outgrowth have also been reported in iPSC-derived neurons from patients diagnosed with Timothy syndrome and Fragile X syndrome, respectively [29, 67,68,69] pointing to converging pathomechanisms. By contrast, increased neurite length has been detected in iPSC-derived cortical neurons from 16p11.2 deletion carriers [44]. The latter findings may be explained by the complex genetic interactions that ultimately determine the neurodevelopmental deficits associated with 16p11.2 deletions, as shown by a genetic screen in Drosophila [66]. Consistent with reduced glutamatergic neurotransmission in hippocampal slices from KCTD13 KO mice and in the prefrontal cortex of 16p11.2 deletion mice [50, 70], we observed decreased spontaneous network activity in human KCTD13 KO neurons cultured on MEA plates. Decreased spontaneous neuronal activity of KCTD13 KO neurons may be due to decreased neurite length causing a reduction in the total number of synapses, while synapse density and the ratio of glutamatergic/GABAergic neurons remained unchanged in our iPSC model. Deficits in neurite elongation/pathfinding may ultimately contribute to hypoconnectivity between brain circuits detected by neuroimaging in ASD and schizophrenia patients [71, 72].
In non-neuronal HeLA cells the KCTD13/Cullin-3 complex ubiquitinates RhoA and thereby promotes degradation of active RhoA by the proteasome [14]. Consistently, Escamilla and coworkers [50] detected a 1.5- to 2-fold increase in total RhoA in the hippocampus of young KCTD13 KO mice and a reversal of synaptic dysfunction by the RhoA inhibitor Rhosin. Interestingly, hippocampal RhoA levels were unchanged in embryonic KCTD13 KO mice in their study. Since human iPSC-derived neurons most closely resemble human fetal neurons [30], unchanged RhoA levels in our iPSC-derived neuron cultures are consistent with the findings in the embryonic mouse brain by Escamilla et al. [50]. A more recent study could not detect an accumulation of RhoA in the hippocampus and cortex of a different KCTD13 KO mouse line and a 16p11.2 deletion mouse model [61]. As already discussed by Arbogast and colleagues [61], the entire KCTD13 locus including potential gene regulatory elements was deleted in the study by Escamilla et al. [50], whereas in the study by Arbogast et al. and in our study only exon 2 was excised, which might explain some of the divergent phenotypes. Notably, the KCTD13 KO mouse line generated by Arbogast and coworkers showed reduced spine density in the hippocampus (but not in the cortex) and memory deficits in several behavioral tests, which appear to be independent of RhoA accumulation similar to our findings in human KCTD13 KO neurons.
By pathway enrichment of differentially expressed genes, we identified ERBB signaling as a potential contributor to the deficit in early neurite formation in human KCTD13 KO neurons. Pharmacological inhibition of ERBB signaling using the ERBB family inhibitor Lapatinib lead to a worsening of the deficit, whereas activation of ERBB3/4 signaling by a single administration of recombinant NRG1b lead to a rescue. The ERBB2 selective inhibitor Tucatinib [52] did not alter neurite formation also arguing for a major role of ERBB3/4 signaling. By contrast, impaired neuronal network activity in KCTD13 KO neurons cultured on MEA plates could not be rescued by repeated administration of NRG1b. Both stimulatory and inhibitory effects of exogenous NRG1b on neurotransmission in ex vivo/in vitro cultures have been reported depending on neuronal subtypes, NRG1b concentrations and timing of administration, and ERBB kinases involved. Moreover, repeated administration of NRG1b to iPSC-derived neurons on MEA plates may modulate not only neurite elongation but also axon myelination and synapse formation between glutamatergic and GABAergic neurons as shown in rodent neurons [73, 74]. In addition, KCTD13/Cullin-3 interacts with several protein substrates [75] which may lead to diverse cellular deficits in KCTD13 deficient neurons that are not influenced by ERBB signaling. Thus, further studies are needed to find the optimal treatment regimen for long-term functional rescue of KCTD13 deficient neurons. Unfortunately, we were unable to co-immunoprecipitate KCTD13 interacting proteins from our human iPSC-derived neurons, since commercially available antibodies against KCTD13 did not immunoprecipitate endogenous KCTD13. Thus, it remains unclear whether the KCTD13/Cullin-3 complex ubiquitinates a component of the NRG/ERBB signaling pathway (similar to RhoA), thereby triggering proteasomal degradation, or whether KCTD13/Cullin-3 and NRG/ERBB pathways act in parallel. Erbin (ERBB2IP, LAP2), a protein that interacts with ERBB2, has been identified as a Cullin-3 binding protein by affinity purification and mass spectrometry [75]. Based on findings in Erbin deficient neurons [76, 77], however, accumulation of Erbin protein in Cullin-3 deficient cells should increase NRG1/ERBB2 signaling. Messenger RNA levels of ERBB/NRG family members and protein levels of ERBB2/4 did not change significantly in our KCTD13 KO neurons.
NRG/ERBB signaling has been shown to stimulate neurite outgrowth of various murine neuron classes in vitro [53, 78, 79]. Numerous studies have revealed complex NRG/ERBB signaling networks that modulate neurotransmission, synaptic plasticity, myelination, and circuit formation in the developing and adult mammalian nervous system. Both common, low effect single nucleotide polymorphisms and rare, high-risk genetic variants in NRG/ERBB family members are strongly associated with psychiatric disorders (reviewed in [73, 74]). Moreover, human studies and mouse models show that an optimal range for NRG/ERBB signaling in the brain is needed for normal neurodevelopment, synaptic function, and cognitive performance [80,81,82]. Behavioral deficits caused by ERBB4 deficiency during development in mutant mice were alleviated by restoring ERBB4 levels in adulthood [83]. Notably, single intraperitoneal injection of NRG1 peptide rescued hippocampal theta oscillations and behavioral deficits in a mouse model of the human 22q11 deletion syndrome [84]. In summary, the aforementioned studies and our findings suggest that modulators of NRG/ERBB signaling may represent a novel therapeutic strategy for neuropsychiatric disorders associated with KCTD13 deficiency.
References
Ustun TB (1999) The global burden of mental disorders. Am J Public Health 89(9):1315–1318
Polderman TJ, Benyamin B, de Leeuw CA, Sullivan PF, van Bochoven A, Visscher PM, Posthuma D (2015) Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat Genet 47(7):702–709. https://doi.org/10.1038/ng.3285
Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, O'Dushlaine C, Chambert K et al (2014) A polygenic burden of rare disruptive mutations in schizophrenia. Nature 506(7487):185–190. https://doi.org/10.1038/nature12975
Wang K, Zhang H, Ma D, Bucan M, Glessner JT, Abrahams BS, Salyakina D, Imielinski M et al (2009) Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature 459(7246):528–533. https://doi.org/10.1038/nature07999
Schizophrenia Working Group of the Psychiatri c Genomics C (2014) Biological insights from 108 schizophrenia-associated genetic loci. Nature 511(7510):421–427. https://doi.org/10.1038/nature13595
International Schizophrenia C (2008) Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature 455(7210):237–241. https://doi.org/10.1038/nature07239
Malhotra D, Sebat J (2012) CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell 148(6):1223–1241. https://doi.org/10.1016/j.cell.2012.02.039
Alenzi FQ, Wyse RK, Altamimi WG (2004) Apoptosis as a tool for therapeutic agents in haematological diseases. Expert Opin Biol Ther 4(3):407–420. https://doi.org/10.1517/14712598.4.3.407
Kumar RA, KaraMohamed S, Sudi J, Conrad DF, Brune C, Badner JA, Gilliam TC, Nowak NJ et al (2008) Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet 17(4):628–638. https://doi.org/10.1093/hmg/ddm376
Shinawi M, Liu P, Kang SH, Shen J, Belmont JW, Scott DA, Probst FJ, Craigen WJ et al (2010) Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J Med Genet 47(5):332–341. https://doi.org/10.1136/jmg.2009.073015
Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H et al (2008) Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med 358(7):667–675. https://doi.org/10.1056/NEJMoa075974
Marshall CR, Howrigan DP, Merico D, Thiruvahindrapuram B, Wu W, Greer DS, Antaki D, Shetty A et al (2017) Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet 49(1):27–35. https://doi.org/10.1038/ng.3725
McCarthy SE, Makarov V, Kirov G, Addington AM, McClellan J, Yoon S, Perkins DO, Dickel DE et al (2009) Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet 41(11):1223–1227. https://doi.org/10.1038/ng.474
Chen Y, Yang Z, Meng M, Zhao Y, Dong N, Yan H, Liu L, Ding M et al (2009) Cullin mediates degradation of RhoA through evolutionarily conserved BTB adaptors to control actin cytoskeleton structure and cell movement. Mol Cell 35(6):841–855. https://doi.org/10.1016/j.molcel.2009.09.004
He H, Tan CK, Downey KM, So AG (2001) A tumor necrosis factor alpha- and interleukin 6-inducible protein that interacts with the small subunit of DNA polymerase delta and proliferating cell nuclear antigen. Proc Natl Acad Sci U S A 98(21):11979–11984. https://doi.org/10.1073/pnas.221452098
Crepel A, Steyaert J, De la Marche W, De Wolf V, Fryns JP, Noens I, Devriendt K, Peeters H (2011) Narrowing the critical deletion region for autism spectrum disorders on 16p11.2. Am J Med Genet B Neuropsychiatr Genet 156(2):243–245. https://doi.org/10.1002/ajmg.b.31163
Uddin M, Tammimies K, Pellecchia G, Alipanahi B, Hu P, Wang Z, Pinto D, Lau L et al (2014) Brain-expressed exons under purifying selection are enriched for de novo mutations in autism spectrum disorder. Nat Genet 46(7):742–747. https://doi.org/10.1038/ng.2980
Li M, Santpere G, Imamura Kawasawa Y, Evgrafov OV, Gulden FO, Pochareddy S, Sunkin SM, Li Z et al (2018) Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science 362(6420). https://doi.org/10.1126/science.aat7615
Golzio C, Willer J, Talkowski ME, Oh EC, Taniguchi Y, Jacquemont S, Reymond A, Sun M et al (2012) KCTD13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11.2 copy number variant. Nature 485(7398):363–367. https://doi.org/10.1038/nature11091
Zeisel A, Hochgerner H, Lonnerberg P, Johnsson A, Memic F, van der Zwan J, Haring M, Braun E et al (2018) Molecular architecture of the mouse nervous system. Cell 174(4):999–1014 e1022. https://doi.org/10.1016/j.cell.2018.06.021
Lin GN, Corominas R, Lemmens I, Yang X, Tavernier J, Hill DE, Vidal M, Sebat J et al (2015) Spatiotemporal 16p11.2 protein network implicates cortical late mid-fetal brain development and KCTD13-Cul3-RhoA pathway in psychiatric diseases. Neuron 85(4):742–754. https://doi.org/10.1016/j.neuron.2015.01.010
Sit ST, Manser E (2011) Rho GTPases and their role in organizing the actin cytoskeleton. J Cell Sci 124 (Pt 5):679–683. doi:https://doi.org/10.1242/jcs.064964
Colman A, Dreesen O (2009) Pluripotent stem cells and disease modeling. Cell Stem Cell 5(3):244–247. https://doi.org/10.1016/j.stem.2009.08.010
Shi Y, Inoue H, Wu JC, Yamanaka S (2017) Induced pluripotent stem cell technology: a decade of progress. Nat Rev Drug Discov 16(2):115–130. https://doi.org/10.1038/nrd.2016.245
Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S, Li Y, Mu Y et al (2011) Modelling schizophrenia using human induced pluripotent stem cells. Nature 473(7346):221–225. https://doi.org/10.1038/nature09915
Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, Chen G, Gage FH et al (2010) A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell 143(4):527–539. https://doi.org/10.1016/j.cell.2010.10.016
Murai K, Sun G, Ye P, Tian E, Yang S, Cui Q, Sun G, Trinh D et al (2016) The TLX-miR-219 cascade regulates neural stem cell proliferation in neurodevelopment and schizophrenia iPSC model. Nat Commun 7:10965. https://doi.org/10.1038/ncomms10965
Pasca SP, Portmann T, Voineagu I, Yazawa M, Shcheglovitov A, Pasca AM, Cord B, Palmer TD et al (2011) Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nat Med 17(12):1657–1662. https://doi.org/10.1038/nm.2576
Sheridan SD, Theriault KM, Reis SA, Zhou F, Madison JM, Daheron L, Loring JF, Haggarty SJ (2011) Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome. PLoS One 6(10):e26203. https://doi.org/10.1371/journal.pone.0026203
Brennand K, Savas JN, Kim Y, Tran N, Simone A, Hashimoto-Torii K, Beaumont KG, Kim HJ et al (2015) Phenotypic differences in hiPSC NPCs derived from patients with schizophrenia. Mol Psychiatry 20(3):361–368. https://doi.org/10.1038/mp.2014.22
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F (2013) Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8(11):2281–2308. https://doi.org/10.1038/nprot.2013.143
Morrison M, Klein C, Clemann N, Collier DA, Hardy J, Heisserer B, Cader MZ, Graf M et al (2015) StemBANCC: governing access to material and data in a large stem cell research consortium. Stem Cell Rev 11(5):681–687. https://doi.org/10.1007/s12015-015-9599-3
Volpato V, Smith J, Sandor C, Ried JS, Baud A, Handel A, Newey SE, Wessely F et al (2018) Reproducibility of molecular phenotypes after long-term differentiation to human iPSC-derived neurons: a multi-site omics study. Stem Cell Reports 11(4):897–911. https://doi.org/10.1016/j.stemcr.2018.08.013
Shi Y, Kirwan P, Livesey FJ (2012) Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat Protoc 7(10):1836–1846. https://doi.org/10.1038/nprot.2012.116
Shi Y, Kirwan P, Smith J, Robinson HP, Livesey FJ (2012) Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat Neurosci 15(3):477–486, S471. https://doi.org/10.1038/nn.3041
Carty N, Berson N, Tillack K, Thiede C, Scholz D, Kottig K, Sedaghat Y, Gabrysiak C et al (2015) Characterization of HTT inclusion size, location, and timing in the zQ175 mouse model of Huntington's disease: an in vivo high-content imaging study. PLoS One 10(4):e0123527. https://doi.org/10.1371/journal.pone.0123527
San Miguel-Ruiz JE, Letourneau PC (2014) The role of Arp2/3 in growth cone actin dynamics and guidance is substrate dependent. J Neurosci 34(17):5895–5908. https://doi.org/10.1523/JNEUROSCI.0672-14.2014
Heikkila TJ, Yla-Outinen L, Tanskanen JM, Lappalainen RS, Skottman H, Suuronen R, Mikkonen JE, Hyttinen JA et al (2009) Human embryonic stem cell-derived neuronal cells form spontaneously active neuronal networks in vitro. Exp Neurol 218(1):109–116. https://doi.org/10.1016/j.expneurol.2009.04.011
Sollner JF, Leparc G, Hildebrandt T, Klein H, Thomas L, Stupka E, Simon E (2017) An RNA-Seq atlas of gene expression in mouse and rat normal tissues. Sci Data 4:170185. https://doi.org/10.1038/sdata.2017.185
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337(6096):816–821. https://doi.org/10.1126/science.1225829
Kim HS, Bernitz JM, Lee DF, Lemischka IR (2014) Genomic editing tools to model human diseases with isogenic pluripotent stem cells. Stem Cells Dev 23(22):2673–2686. https://doi.org/10.1089/scd.2014.0167
Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A et al (2013) Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154(6):1380–1389. https://doi.org/10.1016/j.cell.2013.08.021
Zhou J, Hu X, Xiong X, Liu X, Liu Y, Ren K, Jiang T, Hu X et al (2005) Cloning of two rat PDIP1 related genes and their interactions with proliferating cell nuclear antigen. J Exp Zool A Comp Exp Biol 303(3):227–240. https://doi.org/10.1002/jez.a.150
Deshpande A, Yadav S, Dao DQ, Wu ZY, Hokanson KC, Cahill MK, Wiita AP, Jan YN et al (2017) Cellular phenotypes in human iPSC-derived neurons from a genetic model of autism spectrum disorder. Cell Rep 21(10):2678–2687. https://doi.org/10.1016/j.celrep.2017.11.037
Groszer M, Erickson R, Scripture-Adams DD, Lesche R, Trumpp A, Zack JA, Kornblum HI, Liu X et al (2001) Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo. Science 294(5549):2186–2189. https://doi.org/10.1126/science.1065518
Darzynkiewicz Z, Juan G (2001) Analysis of DNA content and BrdU incorporation. Curr Protoc Cytom Chapter 7:Unit 7 7. doi:https://doi.org/10.1002/0471142956.cy0707s02
Krishan A (1975) Rapid flow cytofluorometric analysis of mammalian cell cycle by propidium iodide staining. J Cell Biol 66(1):188–193
da Silva JS, Dotti CG (2002) Breaking the neuronal sphere: regulation of the actin cytoskeleton in neuritogenesis. Nat Rev Neurosci 3(9):694–704. https://doi.org/10.1038/nrn918
Kozma R, Sarner S, Ahmed S, Lim L (1997) Rho family GTPases and neuronal growth cone remodelling: relationship between increased complexity induced by Cdc42Hs, Rac1, and acetylcholine and collapse induced by RhoA and lysophosphatidic acid. Mol Cell Biol 17(3):1201–1211
Escamilla CO, Filonova I, Walker AK, Xuan ZX, Holehonnur R, Espinosa F, Liu S, Thyme SB et al (2017) Kctd13 deletion reduces synaptic transmission via increased RhoA. Nature 551(7679):227–231. https://doi.org/10.1038/nature24470
Wood ER, Truesdale AT, McDonald OB, Yuan D, Hassell A, Dickerson SH, Ellis B, Pennisi C et al (2004) A unique structure for epidermal growth factor receptor bound to GW572016 (Lapatinib): relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Res 64(18):6652–6659. https://doi.org/10.1158/0008-5472.CAN-04-1168
Duchnowska R, Loibl S, Jassem J (2018) Tyrosine kinase inhibitors for brain metastases in HER2-positive breast cancer. Cancer Treat Rev 67:71–77. https://doi.org/10.1016/j.ctrv.2018.05.004
Gerecke KM, Wyss JM, Carroll SL (2004) Neuregulin-1beta induces neurite extension and arborization in cultured hippocampal neurons. Mol Cell Neurosci 27(4):379–393. https://doi.org/10.1016/j.mcn.2004.08.001
Modol-Caballero G, Santos D, Navarro X, Herrando-Grabulosa M (2017) Neuregulin 1 reduces motoneuron cell death and promotes neurite growth in an in vitro model of motoneuron degeneration. Front Cell Neurosci 11:431. https://doi.org/10.3389/fncel.2017.00431
Krivosheya D, Tapia L, Levinson JN, Huang K, Kang Y, Hines R, Ting AK, Craig AM et al (2008) ErbB4-neuregulin signaling modulates synapse development and dendritic arborization through distinct mechanisms. J Biol Chem 283(47):32944–32956. https://doi.org/10.1074/jbc.M800073200
Harris RC, Chung E, Coffey RJ (2003) EGF receptor ligands. Exp Cell Res 284(1):2–13
Goldshmit Y, Greenhalgh CJ, Turnley AM (2004) Suppressor of cytokine signalling-2 and epidermal growth factor regulate neurite outgrowth of cortical neurons. Eur J Neurosci 20(9):2260–2266. https://doi.org/10.1111/j.1460-9568.2004.03698.x
Pfeiffer BE, Huber KM (2009) The state of synapses in fragile X syndrome. Neuroscientist 15(5):549–567. https://doi.org/10.1177/1073858409333075
Stephan KE, Baldeweg T, Friston KJ (2006) Synaptic plasticity and dysconnection in schizophrenia. Biol Psychiatry 59(10):929–939. https://doi.org/10.1016/j.biopsych.2005.10.005
Sudhof TC (2008) Neuroligins and neurexins link synaptic function to cognitive disease. Nature 455(7215):903–911. https://doi.org/10.1038/nature07456
Arbogast T, Razaz P, Ellegood J, McKinstry S, Erdin S, Currall B, Aneichyk T, Lerch JP et al (2018) Kctd13-deficient mice display short-term memory impairment and sex-dependent genetic interactions. Hum Mol Genet. https://doi.org/10.1093/hmg/ddy436
Manji H, Kato T, Di Prospero NA, Ness S, Beal MF, Krams M, Chen G (2012) Impaired mitochondrial function in psychiatric disorders. Nat Rev Neurosci 13(5):293–307. https://doi.org/10.1038/nrn3229
Iwamoto K, Bundo M, Kato T (2005) Altered expression of mitochondria-related genes in postmortem brains of patients with bipolar disorder or schizophrenia, as revealed by large-scale DNA microarray analysis. Hum Mol Genet 14(2):241–253. https://doi.org/10.1093/hmg/ddi022
Robicsek O, Karry R, Petit I, Salman-Kesner N, Muller FJ, Klein E, Aberdam D, Ben-Shachar D (2013) Abnormal neuronal differentiation and mitochondrial dysfunction in hair follicle-derived induced pluripotent stem cells of schizophrenia patients. Mol Psychiatry 18(10):1067–1076. https://doi.org/10.1038/mp.2013.67
Ni P, Noh H, Park GH, Shao Z, Guan Y, Park JM, Yu S, Park JS et al (2019) iPSC-derived homogeneous populations of developing schizophrenia cortical interneurons have compromised mitochondrial function. Mol Psychiatry. https://doi.org/10.1038/s41380-019-0423-3
Iyer J, Singh MD, Jensen M, Patel P, Pizzo L, Huber E, Koerselman H, Weiner AT et al (2018) Pervasive genetic interactions modulate neurodevelopmental defects of the autism-associated 16p11.2 deletion in Drosophila melanogaster. Nat Commun 9(1):2548. https://doi.org/10.1038/s41467-018-04882-6
Doers ME, Musser MT, Nichol R, Berndt ER, Baker M, Gomez TM, Zhang SC, Abbeduto L et al (2014) iPSC-derived forebrain neurons from FXS individuals show defects in initial neurite outgrowth. Stem Cells Dev 23(15):1777–1787. https://doi.org/10.1089/scd.2014.0030
Krey JF, Pasca SP, Shcheglovitov A, Yazawa M, Schwemberger R, Rasmusson R, Dolmetsch RE (2013) Timothy syndrome is associated with activity-dependent dendritic retraction in rodent and human neurons. Nat Neurosci 16(2):201–209. https://doi.org/10.1038/nn.3307
Liu J, Koscielska KA, Cao Z, Hulsizer S, Grace N, Mitchell G, Nacey C, Githinji J et al (2012) Signaling defects in iPSC-derived fragile X premutation neurons. Hum Mol Genet 21(17):3795–3805. https://doi.org/10.1093/hmg/dds207
Wang W, Rein B, Zhang F, Tan T, Zhong P, Qin L, Yan Z (2018) Chemogenetic activation of prefrontal cortex rescues synaptic and behavioral deficits in a mouse model of 16p11.2 deletion syndrome. J Neurosci 38(26):5939–5948. https://doi.org/10.1523/JNEUROSCI.0149-18.2018
Di Martino A, Yan CG, Li Q, Denio E, Castellanos FX, Alaerts K, Anderson JS, Assaf M et al (2014) The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol Psychiatry 19(6):659–667. https://doi.org/10.1038/mp.2013.78
Rae CL, Davies G, Garfinkel SN, Gabel MC, Dowell NG, Cercignani M, Seth AK, Greenwood KE et al (2017) Deficits in neurite density underlie white matter structure abnormalities in first-episode psychosis. Biol Psychiatry 82(10):716–725. https://doi.org/10.1016/j.biopsych.2017.02.008
Mei L, Nave KA (2014) Neuregulin-ERBB signaling in the nervous system and neuropsychiatric diseases. Neuron 83(1):27–49. https://doi.org/10.1016/j.neuron.2014.06.007
Mei L, Xiong WC (2008) Neuregulin 1 in neural development, synaptic plasticity and schizophrenia. Nat Rev Neurosci 9(6):437–452. https://doi.org/10.1038/nrn2392
Wimuttisuk W, West M, Davidge B, Yu K, Salomon A, Singer JD (2014) Novel Cul3 binding proteins function to remodel E3 ligase complexes. BMC Cell Biol 15:28. https://doi.org/10.1186/1471-2121-15-28
Arikkath J, Israely I, Tao Y, Mei L, Liu X, Reichardt LF (2008) Erbin controls dendritic morphogenesis by regulating localization of delta-catenin. J Neurosci 28(28):7047–7056. https://doi.org/10.1523/JNEUROSCI.0451-08.2008
Tao Y, Dai P, Liu Y, Marchetto S, Xiong WC, Borg JP, Mei L (2009) Erbin regulates NRG1 signaling and myelination. Proc Natl Acad Sci U S A 106(23):9477–9482. https://doi.org/10.1073/pnas.0901844106
Murphy SP, Bielby-Clarke K (2008) Neuregulin signaling in neurons depends on ErbB4 interaction with PSD-95. Brain Res 1207:32–35. https://doi.org/10.1016/j.brainres.2008.02.063
Rio C, Rieff HI, Qi P, Khurana TS, Corfas G (1997) Neuregulin and erbB receptors play a critical role in neuronal migration. Neuron 19(1):39–50
Agarwal A, Zhang M, Trembak-Duff I, Unterbarnscheidt T, Radyushkin K, Dibaj P, Martins de Souza D, Boretius S et al (2014) Dysregulated expression of neuregulin-1 by cortical pyramidal neurons disrupts synaptic plasticity. Cell Rep 8(4):1130–1145. https://doi.org/10.1016/j.celrep.2014.07.026
Bjarnadottir M, Misner DL, Haverfield-Gross S, Bruun S, Helgason VG, Stefansson H, Sigmundsson A, Firth DR et al (2007) Neuregulin1 (NRG1) signaling through Fyn modulates NMDA receptor phosphorylation: differential synaptic function in NRG1+/− knock-outs compared with wild-type mice. J Neurosci 27(17):4519–4529. https://doi.org/10.1523/JNEUROSCI.4314-06.2007
Li B, Woo RS, Mei L, Malinow R (2007) The neuregulin-1 receptor erbB4 controls glutamatergic synapse maturation and plasticity. Neuron 54(4):583–597. https://doi.org/10.1016/j.neuron.2007.03.028
Wang H, Liu F, Chen W, Sun X, Cui W, Dong Z, Zhao K, Zhang H et al (2018) Genetic recovery of ErbB4 in adulthood partially restores brain functions in null mice. Proc Natl Acad Sci U S A 115(51):13105–13110. https://doi.org/10.1073/pnas.1811287115
Marissal T, Salazar RF, Bertollini C, Mutel S, De Roo M, Rodriguez I, Muller D, Carleton A (2018) Restoring wild-type-like CA1 network dynamics and behavior during adulthood in a mouse model of schizophrenia. Nat Neurosci 21(10):1412–1420. https://doi.org/10.1038/s41593-018-0225-y
Acknowledgments
We would like to thank Dr. Michael Brendel (Boehringer Ingelheim Pharma GmbH & Co. KG) for help with statistical analysis of neurite formation assays, and Dr. Jinwei Zhang (University of Dundee) for validation of anti-KCTD13 antibodies for immunoprecipitation.
Funding
The research leading to these results has received support (iPSC line from a healthy donor) from the Innovative Medicines Initiative Joint Undertaking under grant agreement no. 115439. Resources of which are composed of financial contribution from the European Union’s Seventh Framework Programme (FP7/2007-2013) and EFPIA companies’ in kind contribution. This publication reflects only the author's views and neither the IMI JU nor EFPIA nor the Europeam Commission are liable for any use that may be made of the information therein.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Boehringer Ingelheim Pharma GmbH & Co. KG supported this work only by providing financial support in the form of authors’ salaries (all affiliated with Boehringer Ingelheim Pharma GmbH & Co. KG) and research materials. Data collection and analysis, study design, decision to publish, or preparation of the manuscript for this work were performed independently. The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 65 kb)
ESM 2
Supplementary Fig. S1, related to Fig. 2 Supportive data for unaltered stemness of KCTD13 KO iPSCs. a–c Bar graphs showing similar mRNA expression levels of stemness marker genes (NANOG, SOX2, OCT4) assessed by qRT-PCR. n = 3 replicates each. (PNG 134 kb)
ESM 3
Supplementary Fig. S2, related to Fig. 3 Additional data supporting decreased proliferation of KCTD13 KO NPCs. a Cell viability (number of non-condensed Hoechst+ nuclei) of BrdU+ cells was unchanged in all NPC lines. Total n = 30 wells per NPC line. b Representative confocal images of BrdU (green) and Hoechst 33342 (blue) nuclear stainings in NPC cultures. Scale bar, 50 μm. c Percentage of EdU+ NPCs is decreased in KCTD13 KO cultures. Total n = 28–29 wells per NPC line; One-way ANOVA followed by Dunnett’s multiple comparison test; ****p < 0.0001. d Gated cell population used for cell cycle analysis is similar between genotypes. e Percentage of NPCs in G0/G1 phase and f in G2/M phase are comparable between lines. Total n = 28–31 measurements per NPC line from 5 independent experiments. g Enriched canonical pathways in KCTD13 KO versus WT NPCs as detected by Ingenuity Pathway Analysis based on RNA sequencing data. (PNG 1.20 mb)
ESM 4
Supplementary Fig. S3, related to Fig. 5a Treatment with the RhoA inhibitor Rhosin (50 μM) does not rescue decreased percentage of MAP2+ neurons with roots in KCTD13 KO cultures. Phenotypic changes: One-way ANOVA followed by Dunnett’s multiple comparison test; **p = 0.0013, ***p = 0.0008. Treatment effect: Unpaired t test: **p = 0.0082 (KCTD13+/+ vs. 50 μM Rhosin), **p = 0.0027 (KCTD13−/− vs. 50 μM Rhosin). b Rhosin does not rescue decreased number of roots per MAP2+ neuron with roots. Phenotypic changes: One-way ANOVA followed by Dunnett’s multiple comparison test; ***p = 0.0007 (KCTD13+/+ vs. KCTD13+/- ), ***p = 0.0003 (KCTD13+/+ vs. KCTD13−/−). Treatment effect: Unpaired t test: *p = 0.0244, **p = 0.0019 (KCTD13−/− vs. 50 μM Rhosin), **p = 0.0012 (KCTD13+/+ vs. 50 μM Rhosin). Total n = 14–18 wells per genotype. (PNG 294 kb)
ESM 5
Supplementary Fig. S4, related to Fig. 6 Representative Western blot images showing similar protein levels of total ERBB2 (a) and total ERBB4 (b) in KCTD13 KO versus WT iPSC-derived neurons. c Treatment with the potent specific ERBB2 inhibitor Tucatinib (1 μM) has no significant effect on neurite elongation either in WT or KCTD13 KO neurons. Total n = 16–20 wells per neuron line from 1 experiment. Phenotypic changes: Unpaired t test; ***p = 0.0008. d Treatment with NRG1b peptide also rescues decreased percentage of MAP2+ neurons with roots. Total n = 63–90 wells per neuron line from 3 to 4 independent experiments. Phenotypic changes: One-way ANOVA followed by Dunnett’s multiple comparison test; *p = 0.0446, ****p < 0.0001. Treatment effect: Unpaired t-test: *p = 0.0289, ***p = 0.0003. e Treatment with the ERBB1 ligand EGF (3, 10, 30 nM) impairs neurite elongation in WT and KCTD13 KO neurons. Total n = 16–24 wells per neuron line from 1 experiment. Phenotypic changes: Unpaired t test; ****p < 0.0001. Treatment effect: one-way ANOVA followed by Dunnett’s multiple comparison test, **p = 0.0062, ****p = 0.0001. (PNG 514 kb)
ESM 6
Supplementary Fig. S5, related to Fig. 6e Confirmation of the deficit in neurite formation and its rescue by NRG1b treatment in a second homozygous KCTD13 KO clone (KCTD13−/−_2) compared to an isogenic subclone of the parental wildtype clone (KCTD13+/+_2). Mean neurite length is significantly decreased in homozygous KCTD13 KO neurons derived from iPSCs and is significantly increased by single administration of NRG1b peptide (50 nM). Unpaired t test; **p = 0.002 (KCTD13+/+_2, Vehicle vs. NRG1b), **p = 0.0014 (KCTD13+/+_2 vs. KCTD13−/−_2), ****p < 0.0001. Total n = 21–24 wells per experimental group. (PNG 485 kb)
ESM 7
Supplementary Fig. S6, related to Fig. 7d Analysis of mean spike rate using multi-electrode-arrays revealed a significant decrease in iPSC-derived KCTD13 deficient neuron cultures. Reduced neuronal network activity was not rescued by repeated administration of NRG1b peptide at 50 nM during each medium exchange. Total n = 8 wells per neuron line from 1 experiment; Phenotypic changes: One-way ANOVA followed by Dunnett’s multiple comparison test; ***p = 0.0009 (KCTD13+/+ vs. KCTD13+/- ), ***p = 0.0003 (KCTD13+/+ vs. KCTD13−/−). (PNG 433 kb)
Rights and permissions
About this article
Cite this article
Kizner, V., Naujock, M., Fischer, S. et al. CRISPR/Cas9-mediated Knockout of the Neuropsychiatric Risk Gene KCTD13 Causes Developmental Deficits in Human Cortical Neurons Derived from Induced Pluripotent Stem Cells. Mol Neurobiol 57, 616–634 (2020). https://doi.org/10.1007/s12035-019-01727-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-019-01727-1