Abstract
DJ-1 is a protein with a wide range of functions importantly related to redox regulation in the cell. In humans, dysfunction of the PARK7 gene is associated with neurodegeneration and Parkinson’s disease. Our objective was to establish a novel DJ-1 knockout zebrafish line and to identify early brain proteome changes, which could be linked to later pathology. The CRISPR-Cas9 method was used to target exon 1 of the park7-/- gene to produce a transgenic DJ-1-deficient zebrafish model of Parkinson’s disease. Label-free mass spectrometry was employed to identify altered protein expression in the DJ-1 null brain of early adult animals. The park7−/− line appears to develop normally at young adult and larval stages. With aging however, DJ-1 null fish exhibit lower tyrosine hydroxylase levels, respiratory failure in skeletal muscle, and lower body mass which is especially prevalent among male fish. By proteomic analysis of early adult brains, we determined that less than 5% of the 4091 identified proteins were influenced by the lack of DJ-1. The dysregulated proteins were mainly proteins known to be involved in mitochondrial metabolism, mitophagy, stress response, redox regulation, and inflammation. This dysregulation in protein networks of our novel DJ-1-deficient zebrafish model occurs in the early adult stage preceding a Parkinson’s disease-related phenotype and the reduction of tyrosine hydroxylase level. The identified protein changes provide new mechanistic background for DJ-1 function. The experimental power of zebrafish makes this model a highly valuable tool to understand and modulate cellular signaling leading to neurodegeneration.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
The Parkinson’s disease (PD)-associated protein DJ-1 is a multifunctional oxidative stress response protein ubiquitously expressed in all human cell types [1]. DJ-1 is encoded by the PARK7 gene, and loss-of-function mutations are linked to a rare familial type of early onset parkinsonism [2]. However, also in idiopathic cases of PD, post-mortem analysis shows accumulation of oxidatively modified DJ-1 [3]. Multiple roles and functions have been assigned to DJ-1, including transcriptional and post-transcriptional regulation [4], chaperone activity [5], maintenance of mitochondrial function and motility [6], proteasome activity [7], participation in synaptic vesicle recycling [8], stress response regulation, and anti-oxidative defense [9]. Whether this multifacial function is controlled by a single overarching function is not yet known.
Upon oxidative stress, a highly conserved cysteine (C106) becomes reversibly oxidized converting DJ-1 to an oxidative sensor that translocates a subset of DJ-1 to mitochondria [10]. Oxidative stress may also lead to irreversible oxidative modification of DJ-1, which has been linked to idiopathic PD and Alzheimer’s disease [3]. DJ-1 also seems to be able to elicit its antioxidant function through a C106 oxidation independent pathway involving nuclear factor (erythroid-derived 2)-like 2 (Nrf2)-dependent signaling [7].
DJ-1-deficient mice have been shown to be hypersensitive to oxidative stress-inducing neurotoxicants [11]. Also, alternations in both motor and dopaminergic functions have been observed in DJ-1-deficient mouse and rat models, although loss of nigral neurons and reduced tyrosine hydroxylase levels, which are hallmarks of PD, have not always been detected [12, 13]. The lack of consensus regarding nigrostriatal changes might be related to the variation in experimental animal age.
Deciphering the mechanisms underlying DJ-1 function is particularly appealing since its broad functional regulatory action overlaps with a number of aspects associated to PD pathogenesis, i.e., mitochondrial dysfunction, neuroinflammation, and oxidative/nitrosative stress [14,15,16]. Though traditionally linked with PD, DJ-1 also has implications in several other neurological diseases and disorders which have pathologies underpinned by dysregulation of oxidative stress levels, e.g., multiple sclerosis inflammatory lesions [17], Alzheimer’s disease [3], and amyotrophic lateral sclerosis [18]. Thus, understanding how DJ-1 controls multiple pathways important for neuronal protection may offer new opportunities for disease therapy. Mice treated with the DJ-1-based peptide, ND-13, showed protection from oxidative stress-induced dopaminergic loss in both DJ-1 knockout and control mice, possibly via an Nrf2 activation pathway independent of the full-length DJ-1 protein [19]. More recently, ND-13 was shown to be neuroprotective in a mouse model of focal ischemic injury and restore the absence of response in DJ-1 knockout mice [20]. In contrast with rodent models, zebrafish are particularly suited for drug screening and cell signaling studies due to optical transparency for live imaging, ease of uptake of drug dissolved in water, and efficiency of establishing transgenes [21]. Additionally, well-characterized neuronal circuitries make zebrafish excellent vertebrate models for movement disorders.
We have generated a DJ-1-deficient zebrafish line using clustered regularly spaced palindromic repeat-associated protein 9 (CRISPR-Cas9) technology to introduce a 5-base pair deletion into exon 1 of the zebrafish park7 gene. Here, we have identified the early protein dysregulation pattern of the DJ-1 null brain occurring prior to tyrosine hydroxylase loss or phenotypic change. Our line should therefore be a highly valuable model for pinpointing possible links, sequence, and importance in the complexity of cellular events leading to PD and other neurological pathologies and to identify future drug targets.
Materials and Methods
Animal Maintenance
All the animals used in experimentation were housed at the Zebrafish Facility located in the Department of Biological Sciences at the University of Bergen. The facility is run according to the European Convention for the Protection of Vertebrate Animals used for Experimental and Other Scientific Purposes. Adult zebrafish were maintained at 26–28 °C with a 14/10 light cycle and were fed twice daily. Embryos were maintained at 28 °C and raised in E3 buffer (5 mM NaCl, 0.17 mM KCl, and 0.33 mM MgSO4) until 14 days post-fertilization (dpf). Establishment of the park7−/− (KO) line and euthanization of adult fish were approved by the Norwegian National Animal Research Authority at Mattilsynet (FOTS ID8039 and ID14039).
Guide RNA and Cas9 Preparation
Target selection for the guide RNA (gRNA) was performed using CHOPCHOPv1 [22]. The 20-base pair (bp) region 5′-GGTGGATGTGATGCGCAGAG-3′ with the PAM site CGG was chosen, no off-targets were identified in CHOPCHOPv1 with an allowance for up to 2 bp mismatches. The gRNA was synthesized with a non-cloning PCR based method as previously described [23]. The resulting product was analyzed on 2% agarose gel and cleaned with the QIAquick PCR purification kit (Qiagen P/N 28106). The PCR product was extracted once with phenol:chloroform:isoamyl alcohol (25:24:1) and ethanol precipitation of the DNA followed by in vitro transcription with MEGAshortscript T7 kit (Ambion P/NAM1354). The RNA product was purified with mirVana (Ambion P/N 1560) diluted and stored at − 80 °C.
The nCas9 mRNA was prepared from the pCS2-nCas9n (generously received from Max Suster Addgene No. 47929) by first linearizing with NotI digestion (New England Biolabs) and purifying with phenol/chloroform DNA extraction. Ambion’s mMESSAGE mMACHINE SP6 kit (P/NAM1340) was used for in vitro transcription of the mRNA.
CRISPR and Line Generation
Tübingen AB wild-type (WT) embryos were collected from natural matings and cell injected at the single cell embryo stage with 1.76 nl of the CRISPR injection mix containing: 100 ng/μl, park7 gRNA, 150 ng/μl nCas9n mRNA, 50 ng/μl eGFP mRNA, and 0.05% phenol red. At 1 dpf, unsuccessful injections determined by lack of eGFP expression were removed and the remaining embryos were grown to adulthood. The injected F0 generation was individually mated with Spotty Wild Type in order to screen the progeny by the PCR restriction digest, and founder fish were identified. Founder fish (F0) were outcrossed with WTs to obtain the F1 generation, and the stable mutations were sequenced using Topo TA cloning (Invitrogen P/N 450641) followed by sequencing at the Sequencing Facility based at the University of Bergen (BigDye v3.1 ThermoFisher) according to the facility protocol.
The F2 fish were established by outcrossing two individual F1 fish containing the same 5-bp deletion in exon 1 with WT fish, and the progeny were combined. The population was expanded from F2 by sibling matings.
Genotype Screening
The 23-bp gRNA sequence 5′-GGTGGATGTGATGCGCAGAGCGG-3′ including the PAM site (underlined) contained a BsrBI restriction site (bolded). Loss of this restriction site was used to screen both 3 dpf embryos (F0, F1) and tail fin clips (F2) for mutations. DNA was obtained from tissues as previously described [24]. Screening was performed using the Expand High Fidelity PCR System (Roche P/N 04738) to amplify a 254-bp region of exon 1 using primers flanking the target region Fwd 5′-GGAGATGGAGACTGTAATCCC-3′ and Rev 5′-CTGACCACCATAACGCTGC-3′ and an annealing temperature of 58 °C. The PCR fragment was digested at 37 °C overnight with BsrBI (New England Biolabs) and run on a 2% agarose gel.
cDNA Analysis
For larval RNA extraction, up to 20 WT and F3 knockout (KO) larvae were killed and stored in 1 ml of Trizol overnight at − 80 °C. Total RNA extraction was performed using the RNeasy Plus Universal kit (Qiagen 73404) according to manufacturer’s instructions. First-strand RNA was produced from 1 μg of total DNase-treated RNA with the oligo(dT)16 primer and SuperScript IV reverse transcriptase (Invitrogen 18090010). Reverse transcription was performed at 55 °C for 10 min and inactivated by incubation at 80 °C for 10 min. Samples were treated with RNase H, aliquoted, and stored at − 80 °C.
In order to validate the knockout at the level of transcription, the gene was amplified from cDNA using Expand High Fidelity PCR System and the primers Fwd 5′-GCATTGCAGACACGCACAGG-3′ and Rev 5′-TCAGACTGACAGAGCGGTGC-3′ with an annealing temperature of 60 °C for 30 cycles. Gel bands were purified with the UltraClean 15 DNA purification kit (Mo-Bio 12100-300) and sequenced at the Sequencing Facility as previously described.
Dissection of Adult Fish
Adult male WT and KO zebrafish were euthanized in ice water slurry and skeletal muscles, and brains (removing the optical nerves and eyes) were dissected out, rinsed in PBS, snap frozen in liquid nitrogen, and stored at − 80 °C.
In Gel Complex I Activity Assay
Whole larvae 5 dpf and muscle tissue from male 12- and 16-month-old DJ-1 KO and WT fish were homogenized with a Dounce homogenizer (40 strokes) in 3 mM EDTA, 250 mM sucrose, and 100 mM HEPES at pH 7.5 homogenization buffer. Samples were centrifuged at 600×g for 10 min at 4 °C to pellet cell debris, followed by 7000×g for 10 min to isolate mitochondria. Mitochondria were washed once with homogenization buffer. Samples (50 μg) were prepared using 4 g/g digitonin/protein ratio and 1% G250 sample additive [25] and separated on a 4–16% or 3–12% native PAGE Bis-Tris Gel System (Life Technologies, BN1002BOX, BN1001BOX) together with clear cathode buffer. The gel ran for approximately 2 h on ice.
The gel was transferred to ice cold water before adding the substrate for complex I activity assay, 2 mM Tris/HCl (pH 7.4), 0.1 mg/ml NADH (Sigma No. N8129), and 2.5 mg/ml nitrotetrazolulium blue (ThermoFisher scientific No. J60230) and stained for 10 min on a shaking platform in room temperature. The reaction was stopped by adding 10% acetic acid. The gel was imaged with BioRad ChemiDoc XRS+ and stained with Imperial Protein Stain (Pierce) for loading control.
Dopamine Analysis
Snap frozen brains from 16-month-old F3 DJ-1 KO zebrafish and WT were analyzed for dopamine quantitation, n = 7 for WT and n = 6 for KO. Dopamine (DA) detection was performed following the previously described method, with slight modifications [26, 27]. Zebrafish brain tissues were homogenized with 500 μl of 0.1 N perchloric acid solution containing sodium metabisulphite (0.02%, w/v) and disodium ethylenediaminetetraacetate (Na2EDTA) (0.05%, w/v). Treated samples, after sonication, were centrifuged at 12,000 rpm for 20 min at 4 °C. The supernatant was passed through 0.45 μm membrane filters and analyzed by HPLC, consisting of a Waters 600 pump, a Rheodyne 7725i injector, and an Antec Leyden Decade II detector, operating at + 750 mV. The mobile phase composed of methanol (6%, v/v), sodium acetate (13.61 g/l), n-octyl-sodium sulfate (19 mg/l), and Na2EDTA (13 mg/L), solubilized in Milli-Q water, was acidified until pH 4.1 with glacial acetic acid. The stationary phase was a Luna C18 column (250 × 4.6 mm, 5 μm). The flow-rate was 0.7 ml/min, and the injected volume was 10 μl. Standard solutions of DA (1 mg/ml) were diluted to the desired concentrations to quantify DA amount in each sample [28]. Final DA values were expressed as nanograms per milligram of brain tissue.
Protein Extraction of Adult Brains
The homogenates were prepared by thawing the brain tissue on ice weighing the tissue and suspending in 100 μl homogenization buffer (10 mM K2HPO4, 10 mM KH2PO4, 1 mM EDTA, 0.6% CHAPS, 0.2 mM Na3VO4, 50 mM NaF, and protease cocktail (Roche Diagnostics GmbH: 11836153001)). The brain tissues were disrupted by sonication (4 × 5 s) followed by incubation on ice for 20 min. Samples were pelleted at 15,000×g for 15 min at 4 °C and stored at − 80 °C.
Label-Free Mass Spectrometry
Four individual brains lysates for both WT and F4 DJ-1 KO animals aged 90 dpf were analyzed by mass spectrometry. The protein extracts (20 μg) were digested using the FASP method followed by reduction, alkylation, and peptide up-concentration as described in Frøyset et al. [29].
Approximately, 0.5 μg protein as tryptic peptides dissolved in 2% acetonitrile (ACN) and 0.5% formic acid (FA) was injected into an Ultimate 3000 RSLC system (ThermoFisher Scientific, Sunnyvale, California, USA) connected online to a Q-Exactive HF mass spectrometer (ThermoFisher Scientific, Bremen, Germany) equipped with EASY-spray nano-electrospray ion source (ThermoFisher Scientific). The sample was loaded and desalted on a pre-column (Acclaim PepMap100, 2 cm × 75 μm ID nano-Viper column, packed with 3 μm C18 beads) at a flow rate of 5 μl/min for 5 min with 0.1% TFA.
Peptides were separated during a biphasic ACN gradient from two nanoflow UPLC pumps (flow rate of 200 nl/min) on a 50-cm analytical column (PepMap RSLC, 50 cm × 75 μm ID EASY-spray column, packed with 2 μm C18 beads). Solvents A and B were 0.1% FA (vol/vol) in water and 100% ACN, respectively. The gradient composition was 5% B during trapping (5 min) followed by 5–8% B over 0.5 min, 8–24% B for the next 109.5 min, 24–35% B over 25 min, and 35–90% B over 15 min. Elution of very hydrophobic peptides and conditioning of the column were performed during 15 min isocratic elution with 90% B and 20 min isocratic conditioning with 5% B. Total length of the LC run was 195 min.
The eluting peptides from the LC column were ionized in the electrospray and analyzed by the Q-Exactive HF. The mass spectrometer was operated in the DDA mode (data-dependent acquisition) to automatically switch between full-scan MS and MS/MS acquisition. Instrument control was through Q-Exactive HF Tune 2.8 and Xcalibur 3.1.
Survey full-scan MS spectra (from m/z 375 to 1500) were acquired in the Orbitrap with resolution R = 120,000 at m/z 200, automatic gain control (AGC) target of 3E6 and a maximum injection time (IT) of 100 ms. The 15 most-intense eluting peptides above an intensity threshold of 50,000 counts, and charge states 2 to 6, were sequentially isolated to a target value (AGC) of 1E5 and a maximum IT of 110 ms in the C-trap, and isolation width maintained at m/z 1.6 (offset of m/z 0.3), before fragmentation in the higher-energy collision dissociation cell. Fragmentation was performed with a normalized collision energy of 28%, and fragments were detected in the Orbitrap at a resolution of 15,000 at m/z 200, with first mass fixed at m/z 100.
One MS/MS spectrum of a precursor mass was allowed before dynamic exclusion for 20 s with “exclude isotopes” on. Lock-mass internal calibration (m/z 445.12003) was enabled.
The spray and ion-source parameters were as follows. Ion spray voltage = 1800 V, no sheath and auxiliary gas flow, and capillary temperature = 275 °C.
Processing of Mass Spectrometry Data
The raw files were searched in MaxQuant (v 1.6.0.16) using Danio rerio a combined UniProtKB database (downloaded: 8 June 2018, 56,929 entries). The same search parameters as previously published [29] were used including fixed modifications of carbamidomethyl (C) and variable modifications of oxidation (M), acetyl (protein N-term), phospho (ST), and phospho (STY). A maximum of two missed trypsin cleavages were allowed with 20 ppm tolerance for precursors and 0.5 Da fragment ion mass tolerance. The false-discovery rate (FDR) was set to 1%. In order to be used for quantification, only proteins identified with at least two unique + razor peptides were considered. The data was further analyzed with Perseus (v 1.6.1.3). The protein list was reduced by removing proteins only identified by site reverse hit and removing potential contaminants. WT only and KO only lists were prepared by keeping IDs that had at least three valid values uniquely in the WT or the KO samples. Additionally, a list of regulated proteins was prepared by keeping proteins which had at least three valid values in both WT and KO samples. The LFQ intensity values were log2 transformed and considered significant if they passed either a one-sample t test (WT and KO only) or two-sample t test (proteins found in both samples) such that the p value 0.05 is used for truncation and S0 = 2. In Perseus, p values were given as the log10 value and the difference between KO and WT was specified as −log2 difference in Perseus. Fold change was calculated as the ratio of −log2 difference of KO over WT.
Bioinformatic Analysis
All significant proteins with a 1.5-fold regulation were combined with the proteins found unique to KO or unique to WT to make up a list of total regulated proteins. Of the 99 proteins, 93 could be mapped with the STRING version 10.5 database [30].
To determine which proteins were considered to localize to the mitochondria, we first had to convert our list of 99 proteins to human orthologues. The OrthoRetriever v1.2 tool was used to convert zebrafish gene symbols to human gene names, which could then be use in further analysis [31]. Out of the 99 genes symbols input into OrthoRetriever, a total of 73 human genes were found. The list of 73 genes could then be used in MitoMiner 4.0 with the gene → IMPI mitochondrial score template with threshold > 0.8 strongly indicative of mitochondrial localization [32]. A total of 71 out of the 73 genes were identified with the program and assigned scores. The same list of 73 human orthologues was compared with known stress granule protein database lists to determine which proteins associate with stress granule [33, 34].
Western Blotting
For mass spectrometry validation, three adult brains were pooled to make protein homogenates from male 4-month-old F5 DJ-1 KO and WT fish. For analysis on aged samples, homogenates were prepared from individual brain samples of 16-month-old zebrafish F3 DJ-1 KO and WT. Samples for Western blotting were separated by SDS-PAGE and transferred to PVDF membranes using 14 V overnight at 4 °C. Membranes were blocked in 1% BSA at room temperature for an hour. Membranes were incubated with the appropriate primary antibody for 1 h at room temperature: anti-DJ-1 1:3000 (NovusBiologicals NB300-270), anti-β-actin 1:5000 (Sigma-Aldrich A2228), anti-GFAP 1:1000 (NSJ Bioreagents F52537), anti-ABCE1 1:10,000 (NB400-116), anti-GSTA1/A2/A3/A4/A5 1:400 (BolserBio PB9627), and anti-TH 1:10,000 (ImmunoStar 22941). Primary antibody was followed by the appropriate secondary antibody incubation for 1 h at room temperature. The membranes were washed and developed using Super Signal West Pico Plus chemiluminescent substrate (ThermoScientific 34577) and imaged with the BioRad ChemiDoc XRS+.
Results
Generation of park7 −/− Zebrafish Line
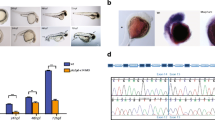
Using the CRISPR-Cas9 method, a targeted mutation was introduced into the first exon of the zebrafish park7 gene (Fig. 1a). At the F0 generation, a variety of mosaic mutations including deletions of between 2 and 7 base pairs or silent point mutations were idetified. Loss of the BsrBI restriction site overlapping with the PAM site was used to identify successful mutagenesis. At the F1 generation, a stable 5-base pair deletion, occurring at residue 27, resulting in a frameshift and premature stop codon after 11 nonsense amino acids was identified for expansion of the line. By the F2 generation, DNA fingerprint utilizing the loss of the BsrBI site was used to identify the genotype of individual fish from caudal fin clips (Fig. 1b). The knockout was validated at the level of transcription by producing a cDNA from the mRNA of larvae and amplifying the park7 gene (Fig. 1c). The band for park7−/− was still visible but fainter compared with the WT possibly due to nonsense-mediated decay of the mRNA. Furthermore, sequencing of the cDNA showed no alternative splicing and confirmed a premature stop codon at the level of transcription. At the level of the protein expression, Western blotting of larval lysates using a DJ-1 antibody showed total absence of the protein in the park7−/−/− sample (Fig. 1d).
Generation of DJ-1 knockout line based on CRISPR-Cas9 method. a CRISPR-Cas9 was used to target a 20-base pair (bp) region of exon 1 in the zebrafish park7 gene. The 3′ end of the target region and protospacer adjacent motif (PAM) overlapped with a unique BsrBI restriction site allowing for screening of mutations using restriction digestion. Fish containing a 5-bp deletion conferring a frame shift and early stop codon were crossed to expand the line. b DNA restriction pattern analysis of F2 generation using DNA extracted from the caudal fin. Undig, undigested. c Transcriptional analysis amplifying the park7 mRNA from cDNA produced from F3 park7−/− and wild-type embryos. d Protein level validation of the knockout using Western blotting of 4 dpf larval protein lysate and probing with anti-DJ-1. β-Actin is shown as a loading control. KO, park7−/−; WT, wild type
Late Adult Stage DJ-1-Deficient Animals Have Reduced Tyrosine Hydroxylase Levels, Are Underweight, and Show a Decrease in Mitochondrial Complex I Activity in Skeletal Muscle
The park7−/− line was raised through the adult F4 generation. No anomalies in larval development (Fig. 2a) or gross anatomy in young adult stages (not shown) were observed. The adult fish was viable and able to reproduce, but as the fish aged beyond 3 months, there was a trend toward smaller size compared with age-matched WT fish (Fig. 2a). By 16 months, the adults, especially males, had a significantly lower body mass compared with WT (Fig. 2a, b). This encouraged us to determine if any change in basal metabolism had been altered in the KO fish. As reduced mitochondrial complex I activity, both in the brain areas and peripheral tissues [6] has been reported in PD patients, we isolated mitochondria from muscle of adult fish and measured complex I activity. In the 16-month-KO adult fish, complex I activity level was below detectable levels in muscle tissue (Fig. 2d). At the 12-month stage, the complex I activity in the KO was measurable but significantly lower than in WT (Fig. 2c). On the other hand, complex I activity measured from mitochondria isolated from whole larvae did not show any difference between KO and WT (Fig. 2d). Thus, there seems to be a progressive loss of muscle mitochondrial complex I activity in the KO animals compared with WT.
Late adult park7−/− zebrafish exhibit slimmer shape, decrease in tyrosine hydroxylase, and reduced muscle mitochondrial complex I activity compared with controls. a Phenotypes of 5 dpf wild-type (WT) and park7−/− (KO) larvae and male adult fish at 16 months. b Body weight difference of aged male WT (n = 12) and KO (n = 13) zebrafish; ***p < 0.001. c Representative image of in-gel activity assay of mitochondrial complex I alone and in supercomplex with complex III and complex IV, and quantitation of complex I alone from mitochondria isolated from muscle of 12-month WT (n = 3) and KO (n = 3) fish; ***p < 0.001. Post-staining of gel with imperial blue was used to show equal loading. d Images of representative in-gel mitochondrial complex I activity in total lysate of 5 dpf larvae and muscle from 16-month WT and KO animals. e Representative Western blot of tyrosine hydroxylase (TH) expression and quantitation in brain lysates from 16-month adults. Ponceau-S was used as loading control; *p < 0.05. f Dopamine levels in 16-month WT (n = 7) and KO (n = 6) zebrafish brain tissue. Data are expressed as the mean ± SEM and analyzed using Student’s two-sample t test
To address the effect of DJ-1 loss on dopaminergic neurons, we measured the level of tyrosine hydroxylase and dopamine in the brains from 16-month adults. As shown in Fig. 2e, there was a significant reduction in tyrosine hydroxylase levels in the KO brains. Although there seemed to be a trend toward reduction, also in dopamine levels, there was a high variation between individuals (Fig. 2f).
Label-Free Proteomic Profiling of DJ-1-Deficient Young Adult Brains
We now wanted to identify DJ-1-dependent protein changes that could be responsible for the phenotype and PD-related neuronal changes of the late-adult DJ-1 KO animals (Fig. 2). Early adults, recently gender differentiated males (3 months), were therefore selected for brain proteome analysis. At this early adult stage, no phenotypic difference between WT and KO could be observed (not shown). Using label-free mass spectrometry, 4091 proteins were identified based on two or more unique peptides with a mass accuracy ≤ 20 ppm and peptide score ≥ 4. The full list of proteins identified can found in Online resource 1. This table also shows the expression level of the identified proteins in the individual samples. To check the correlation between the biological replicates, scatter plots were generated and the Pearson Correlation values were calculated (Fig. 3a, b). A high positive correlation value for each biological group was found from 0.986 to 0.992 and 0.960 to 0.993 and in the knockout and wild-type samples, respectively.
Label-free mass spectrometry-based proteome analysis. a, b Multiscatter plots of correlation between biological replicates. The log-transformed LFQ intensity values are shown for replicates of park7−/− (KO) (a) or wild type (WT) (b). The Pearson correlation values on the individual plots show a high level of reproducibility. c Venn diagram showing the number of proteins identified by mass spectrometry with at least three valid identifications in both groups. Also shown is the total number of proteins with at least three valid values found uniquely in a single group. All proteins have a threshold score above 4 and have been identified by at least two unique peptides. d Volcano plot showing significantly regulated proteins. The protein expression ratio (fold change) of KO/WT (log2 scale) was plotted against the −log10 of the probability calculated using Student’s two-sample t test such that S0 = 2. The dashed lines indicated the applied threshold values (p value < 0.05; fold change ≥ 1.5)
For quantitative analysis, a minimum of two unique peptides was required for further statistical analysis. We restricted the analysis to include only proteins identified in at least three out of four samples in either KO or WT samples resulting in 2900 proteins. Out of 2900 proteins, 18 proteins were only identified in KO and 23 only in WT (Fig. 3c).
To analyze regulated proteins identified in both groups, at least three valid values in each group were required. Common for both groups were 2523 proteins (Fig. 3c). Strikingly, while a large number of proteins were identified in both KO and WT brains, only 58 proteins could be seen as regulated with 1.5-fold change or higher (Fig. 3d). The 58 significantly regulated proteins (fold change ≥ 1.5) when added to the list of KO only (18 proteins) and WT only (23 proteins) were considered regulated proteins (99 proteins) for further bioinformatic analysis.
In Table 1, we have listed proteins that showed the most prominent regulation in DJ-1 KO brains. Tables 2 and 3 list proteins identified only in KO or WT brains, respectively. The absence of identification in one group most possibly reflects a protein level below detection limit. For proteins identified in both control and DJ-1 KO brains, we have restricted the table to include only proteins regulated with fold change ≥ 2.0. From these regulated proteins, we selected glutathione S-transferase (gstal) and ATP-binding cassette, subfamily E, member 1 (abce1) for validation by Western blotting (Fig. 4a, b). In all tables, we have included protein characteristics that based on previous knowledge may be relevant for PD-related pathogenesis. In the case where protein function is given in the UniProt database, no reference is included.
Verification of mass spectrometry data. a, c Protein lysates from brains of wild-type and F4 park7−/− zebrafish were separated by SDS-PAGE, Western blotted, and probed with indicated antibodies. Ponceau-S was used as loading control. a, b Representative Western blots of DJ-1-dependent regulation of ACBE1 (a) and GSTα (b) from 4-month-old pooled zebrafish brain lysates. Asterisk indicates GSTα band. n = 3. c Tyrosine hydroxylase (TH) and glial fibrillary acidic protein (GFAP) expression in 3-month-old zebrafish samples used for mass spectrometry analysis. KO, park7−/−; WT, wild type
In all, prominent changes in the brain proteome of DJ-1 KO animals were already observable at an early adult age. However, at this stage the tyrosine hydroxylase (th) (Acc. No. B3DJW5, Online resource 1 levels, as a marker for dopaminergic cell death, did not differ from controls. Neither did we observe any compensatory up-regulation of the antioxidant proteins, superoxide dismutase [Cu–Zn] (SOD1) or peroxiredoxins. Glial fibrillary acidic protein (gfap), a marker for astrocyte activation and inflammation, showed a large variation between individual DJ-1 KO brains and no significant change (Fig. 4c; Acc. No. B2GP50, Online resource 1).
DJ-1-Deficient Brain Protein Profile Reveals Changes in Clusters of Proteins Belonging to the Mitochondrial Electron Transport Chain, Translational Control, Stress Response, Glutathione metabolism, and Inflammation
To gain an overall understanding of relationship between proteins affected by the loss of DJ-1, we used the protein-protein interaction data analysis tool STRING: functional protein networks [30] to obtain a network map of the differentially expressed proteins. The protein-protein interactions predicted by STRING are based on direct (physical) and indirect (functional) associations, of which data are extracted from a large number of different databases. It should be noted that since the zebrafish proteome has yet to be completely annotated, our bioinformatics analysis in many cases relies on existing knowledge of the human or mammalian orthologues. A total of 93 proteins out of the 99 regulated proteins were mapped by either gene name or UniProt accession number.
As shown in Fig. 5, the most prominent cluster consisted of proteins belonging to the mitochondrial electron transport chain, which were up-regulated (blue circles) in the DJ-1 KO brains. Another cluster included down-regulated (red circles) proteins associated to stress response and vesicular trafficking. In addition, proteins known to regulate inflammatory response were down-regulated in DJ-1-deficient brains. The absence of DJ-1 also influenced proteins related to glutathione metabolism and translational regulation/stress granules.
STRING protein-protein interaction network analysis of 93 proteins regulated in the DJ-1-deficient brain. Interaction network was built from the list of significant proteins via two-sample t test (p < 0.05) with three valid values in both wild-type (WT) and park7−/− (KO) samples with a fold change ≥ 1.5 and those proteins only found uniquely in either WT or KO samples. A total of 99 proteins were used for STRING analysis, 93 of which could be mapped in the diagram. Up-regulation and down-regulation are indicated by node color blue and red, respectively, and evidence type is indicated by a line color. A few of the mapped proteins with Ensembl or NCBI gene IDs were changed to the appropriate gene name or mammalian homolog
DJ-1 Loss Affects Proteins with Strong Support of Mitochondrial or Stress Granule Localization
The STRING analysis indicated a DJ-1-dependent regulation associated to specific intracellular localizations: mitochondria and stress granules. This encouraged us to perform a comparison between existing mitochondria and stress granule databases and the mammalian orthologues of our regulated proteins. These are databases containing proteins with strong support of either mitochondrial (MitoMiner 4.0) or stress granule [33, 34] localization. This analysis showed that as many as 45% of the regulated proteins were either known or predicted to have a mitochondrial localization (Fig. 6a). Similarly, also approximately 45% of the regulated proteins are proteins suggested to be involved in stress granule regulation (Fig. 6b).
Differential expressed proteins highlights proteins associated to mitochondria and stress granules The list of 99 regulated proteins was converted to human orthologues for analysis resulting in a list of 73 regulated mammalian genes and analyzed toward databases of mitochondria and stress granule-associated proteins. a MitoMiner annotation of regulated proteins associated to mitochondria. b Stress granule-associated proteins
Discussion
Parkinson’s disease pathogenesis is complex and involves an array of different aspects, including mitochondrial impairment, neuroinflammation, oxidative/nitrosative stress, and dysregulation in protein turnover, events believed to eventually lead to nigral cell loss [48]. DJ-1 has a key function in neuronal protection as a regulator of all previously mentioned aspects of cellular stress. With its broad influence on different cellular pathways, DJ-1 is therefore suggested to be a highly interesting therapeutic target in several neurodegenerative diseases, not exclusively PD. Even though there is a broad understanding of DJ-1 functions, the mechanisms underlying its roles remain largely elusive.
We have established a novel DJ-1 knockout zebrafish line which shows late-stage decrease in overall level of the dopaminergic rate-limiting enzyme, tyrosine hydroxylase, reduction in mitochondrial complex I activity in skeletal muscle, and phenotypic body mass loss associated with PD (Fig. 2) [6, 49, 50]. Zebrafish have become successful vertebrate models for neurodegenerative diseases as most of the molecular mechanisms related to neurological diseases are highly conserved, in addition, their optical transparency, short life cycle, and large production of eggs allow for unique opportunities for live imaging and large-scale drug screening [21].
To identify cellular changes depending on DJ-1 expression, which in its functional absence may lead to later PD-related pathology, we selected brains from early stage adult animals for label-free proteomics analysis. The proteomics analysis showed that DJ-1 deficiency only affected less than 5% of the total identified brain proteins.
Mitochondrial respiratory chain impairment has long been defined as a key feature of PD [6, 49]. Several studies have shown that loss of DJ-1 dysregulates the mitochondrial respiratory chain and in particular targets complex I [51], thus linking DJ-1 to mitochondrial PD pathology. DJ-1 deficiency had no effect on mitochondrial complex I activity at the larval stage, but a gradual decrease in complex I activity in skeletal muscle toward late adulthood was evident (Fig. 2c, d). Interestingly, our proteomic profile of the early adult brain showed that DJ-1 knockout increases the level of complex I components (Fig. 5; Table 2; Online resource 1). At first glance, this seems to oppose the observation of reduced complex I activity which we observed analyzing mitochondria of skeletal muscle from late adults (Fig. 2c, d). However, Miwa et al. have shown that increased amount of complex I components is correlated with less-efficient complex I assembly and increased ROS production [52]. This was observed in old mice, while the opposite was seen in young and diet-restricted animals. Their results agree with the observation of an inverse relationship between complex I activity and ROS production in fibroblasts from patients with inherited complex I disorders [53]. Our data suggest that loss of DJ-1 causes an instability in subunit components of the electron transport chain which affects assembly and activity of the mitochondrial complex I. Less-efficient assembly might further lead to lowered substrate utilization and increased mitochondrial superoxide production [52] .
In the absence of the major redox regulator DJ-1, one would expect an increased oxidative stress level in the DJ-1 null brain and possibly also compensatory redox-regulating pathways to be activated. One way to cope with increased oxidative stress is to increase the availability of the nonenzymatic antioxidant GSH through controlling the expression of glutathione peroxidases (GPXs) and glutathione-S-transferases (GSTs) [54]. In the DJ-1 null brain, both the GSTA1 and GPX1 levels increased (Table 1; Figs. 4 and 5; Online resource 1). This is further supported by the up-regulation of the redox-sensitive protein BCAT (Table 1) which catabolize branched amino acid to glutamate from which GSH is synthetized [35]. We did not however observe any increase in GSTMs (Online resource 1). We have previously shown that increased glial DJ-1 up-regulates GSTm1 [15], a dominant GST isoform in the zebrafish brain [55], most possibly through a Nrf2-dependent pathway [15]. Nrf2 can control the expression levels of both GSTMs in addition to antioxidants such as SOD1 and peroxiredoxins. In our DJ-1 knockout brains, neither of these Nrf2-controlled proteins were regulated (Online resource 1). Thus, a compensatory Nrf2 activation is not supported, but on the other hand, GSH-dependent actions seem to be activated. The absence of a Nrf2 response in DJ-1 null brains also supports our earlier suggestion that DJ-1 may act upstream of Nrf2 in response to neuronal oxidative stress [15].
Almost half of the regulated proteins in DJ-1 knockout brains were either known as mitochondrial or predicted to be mitochondrial (Fig. 6). Apart from those associated to mitochondrial bioenergetic function, some of the down-regulated proteins (ABCE1, TBC1d8b, and KIF1b) could be linked to mitophagy and mitochondrial dynamics (Tables 1 and 3) [37, 45, 47]. Mitochondrial dysregulation, and specifically mitophagy, is closely linked with both spontaneous and the hereditary forms of PD [56], and PD-associated proteins PINK1, Parkin, and DJ-1 have been shown to be implicated in removal of dysfunctional mitochondria [14, 57]. The down-regulated ATP-binding cassette, subfamily E (OABP), member 1 (ABCE1) protein (Table 1; verified in Fig. 4) was recently shown to be down-regulated at the level of transcription in PD patients [37]. ABCE1 is a novel actor in the early stage of mitophagy, and through ubiquitination by NOT4, it recruits the autophagy machinery [37]. Mitophagy seems to be controlled through simultaneously occurring PINK1-regulated quality control of mitochondrial localized translation and ABCE1 ubiquitination. DJ-1 has been shown to complex with PINK1 and Parkin [58]. We also observed CNOT1, a component of the CCR4-NOT complex [59], to be down-regulated (Online resource 1; Acc. No. F1QFA6) in DJ-1-deficient brains. Thus, DJ-1 might have a double directed influence on mitophagy.
DJ-1 has been shown to regulate basal autophagy and facilitate neuronal protection through the extracellular signal-regulated kinase (ERK1/2) pathway [14]. Our results show that map2k2b, which belongs to the ERK1/2 pathway, is dysregulated in the DJ-1 null brain (Table 3). Also, picalma, a regulator of endocytosis and autophagy [46], was shown to be down-regulated in the knockout brains (Table 3). Taken together, our DJ-1 knockout model shows a general impairment in proteins controlling basal mitophagy and autophagy needed to maintain cellular homeostasis and protecting cells from dysfunctional mitochondria.
Apart from being associated to mitochondria, almost half of the regulated proteins in the DJ-1-deficient brain could be linked to translational control and stress granules (Figs. 5 and 6; Tables 1, 2, and 3). DJ-1 has long been known to be a part of a RNA-binding complex and to have RNA-binding activity in dopaminergic cells [60]. Recently, DJ-1 was reported to localize to stress granules and interact with several stress granule components upon induction of oxidative stress [61]. Stress granules are membrane-less pro-survival protective structures that assemble in response to a variety of environmental stresses and disassemble after the stress has resolved. A number of stress granule proteins are implicated in age-related diseases, especially neurodegenerative diseases [62]. Thus, the variety of roles assigned to DJ-1 may therefore include also a regulatory role in stress granules.
Neurodegeneration-triggered astrogliosis has been linked to neuroinflammation observed in PD [63], but the role of this astrogliosis is largely debated, as to whether this process is harmful or neuroprotective [64]. Astrogliosis is generally associated with up-regulation of astrocytic marker proteins such as gfap and vimentin [65]. In our DJ-1 knockout brains, there was no significant difference between wild-type and DJ-1 null brain levels of gfap (Fig. 4; Online resource 1). Both in the early and late sampled brains, there was great individual variation in gfap levels in the DJ-1 animals. Neither did the levels of vimentin differ between DJ-1 null and wild type (Online resource 1). These results support the observation of Tong et al. [65] who observed a large sample variation in glial markers among PD patient samples. Also in DJ-1 knockout mice, Choi et al. found no distinction in gfap levels between wild type and knockout [66]. In fact, they showed that DJ-1 neuroprotection was mediated through astrogliosis and suggested that DJ-1 deficiency caused defects in astrocyte function to repair of neuronal damage.
Pro-inflammatory nitrosative damage is a pathological hallmark of PD [67, 68], and we have previously shown that increased astrocytic DJ-1 expression can inhibit the generation of protein nitrosylation [15]. Recently, Sun et al. showed that nitric oxide levels were higher in PINK1-deficient glia compared with control after inflammatory induction by lipopolysaccharide [69]. Argininosuccinate lyase (asl) is the only enzyme capable to produce arginine, the substrate for nitric oxide generation by nitric oxide synthase isoforms [39]. We had detectable levels of asl only in DJ-1-deficient brains (Table 2). This indicates that DJ-1 deficiency may lay the basis for accelerated NO production and as PINK1 also influence inflammatory-induced neuronal death. On the other hand, we also found complement complex 3 (C3) and possibly its downstream activation fragments (C3a and C3b) (our results do not distinguish between C3 and its fragments), components involved in inflammatory response [70], to be down-regulated in DJ-1 null brains (Fig. 5; Table 1). C3 deficiency has been shown to be neuroprotective and reduce neuroinflammation [71]. Thus, the observed down-regulation in complement C3 might be an early response to cellular stress induced by the lack of DJ-1.
The observed up-regulation of C-terminal domain small phosphatase 1 (CTDSP1) might also be a stress response to DJ-1 deficiency (Table 2). CTDSP1 has a function in stabilizing the repressor element 1 silencing transcription factor (REST) complex which in turn promotes neuroprotection by suppressing genes involved in oxidative stress [72]. This supports the observations of CTDSP1 transcript being up-regulated in substantia nigra of PD patients [41] and REST-deficient mice to be more vulnerable to MPTP-induced nigral loss [73].
DJ-1 has been shown to modulate α-synuclein accumulation and toxicity [74]. Alpha-synuclein is not found in zebrafish, but zebrafish γ1-synuclein is proposed to be closest to human α-synuclein [75]. We did not observe any change in the levels of γ1-synuclein or other synuclein isoforms in the DJ-1-deficient brains (Online resource 1). However, an uncharacterized protein with encoded by cc2d1a was identified in the DJ-1 null and not wild type (Table 2). CC2D1A is involved in the oligomerization of α-synuclein and in its absence of there a decrease in α-synuclein oligomerization [76].
Conclusion
We have established a novel DJ-1/park7 knockout zebrafish line, which at late adult stage shows a reduction in tyrosine hydroxylase levels, reduced mitochondrial complex I activity in skeletal muscle, and weight loss. Using label-free mass spectrometry, we identified 4091 proteins from young adult brains, of which less than 5% were altered in expression level due to DJ-1 loss. Among these were novel DJ-1-regulated proteins that could be linked to mitochondrial function and mitophagy as well as stress response and inflammation, and therefore be key actors in the induction of later pathogenesis. Early adult knockout animals exhibited an increased abundance of subunits of the mitochondrial respiratory chain and an age-dependent down-regulation of complex I activity, a cellular condition that can increase the amount of mitochondrial superoxide production over time. As zebrafish are ideal vertebrate models for live imaging, drug screening, and genetic manipulation, our model can be used as a tool to further understand the complex neuroprotective role of DJ-1 and how it can be manipulated.
Availability of Data and Materials
The authors declare that the data supporting the findings of this study are available within the article and its online resource files.
Abbreviations
- CRISPR-Cas9:
-
Clustered regularly spaced palindromic repeat-associated protein 9
- PAM:
-
Protospacer adjacent motif
- gRNA:
-
Guide RNA
- dpf:
-
Days post-fertilization
- KO:
-
Knockout (park7−/−)
- bp:
-
Base pair
- WT:
-
Tübingen AB wild type
- DA:
-
Dopamine
- gst:
-
Glutathione S-transferase
- abce1:
-
ATP-binding cassette, subfamily E, member 1
- th:
-
Tyrosine hydroxylase
- gfap:
-
Glial fibrillary acidic protein
- GPX:
-
Glutathione peroxidases
- Nrf2:
-
Nuclear factor (erythroid-derived 2)-like 2
- ERK1/2:
-
Extracellular signal-regulated kinase
- asl:
-
Argininosuccinate lysate
- C3:
-
Complement complex 3
- CTDSP1:
-
C-terminal domain small phosphatase 1
References
Ariga H, Iguchi-Ariga SMM (2017) DJ-1/PARK7 protein—Parkinson’s disease, cancer and oxidative stress-induced diseases. Adv Exp Med Biol 1037. https://doi.org/10.1007/978-981-10-6583-5
Bonifati V, Rizzu P, van Baren MJ, Schaap O, Breedveld GJ, Krieger E, Dekker MC, Squitieri F et al (2003) Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science 299(5604):256–259. https://doi.org/10.1126/science.1077209
Choi J, Sullards CM, Olzmann JA, Rees HD, Weintraub ST, Bostwick DE, Gearing M, Levey AI et al (2006) Oxidative damage of DJ-1 is linked to sporadic Parkinson and Alzheimer diseases. J Biol Chem 281(16):10816–10824. https://doi.org/10.1074/jbc.m509079200
Takahashi-Niki K, Niki T, Iguchi-Ariga SMM, Ariga H (2017) Transcriptional regulation of DJ-1. Adv Exp Med Biol 1037:89–95. https://doi.org/10.1007/978-981-10-6583-5_7
Xu C-Y, Kang W-Y, Chen Y-M, Jiang T-F, Zhang J, Zhang L-N, Ding J-Q, Liu J et al (2017) DJ-1 inhibits α-synuclein aggregation by regulating chaperone-mediated autophagy. Front Aging Neurosci 9:308. https://doi.org/10.3389/fnagi.2017.00308
Franco-Iborra S, Vila M, Perier C (2018) Mitochondrial quality control in neurodegenerative diseases: focus on Parkinson’s disease and Huntington’s disease. Front Neurosci 12:342–342. https://doi.org/10.3389/fnins.2018.00342
Moscovitz O, Ben-Nissan G, Fainer I, Pollack D, Mizrachi L, Sharon M (2015) The Parkinson’s-associated protein DJ-1 regulates the 20S proteasome. Nat Commun 6:6609. https://doi.org/10.1038/ncomms7609
Kyung JW, Kim JM, Lee W, Ha TY, Cha SH, Chung KH, Choi DJ, Jou I et al (2018) DJ-1 deficiency impairs synaptic vesicle endocytosis and reavailability at nerve terminals. Proc Natl Acad Sci U S A 115(7):1629–1634. https://doi.org/10.1073/pnas.1708754115
Raninga PV, Di Trapani G, Tonissen KF (2017) The multifaceted roles of DJ-1 as an antioxidant. Adv Exp Med Biol 1037:67–87. https://doi.org/10.1007/978-981-10-6583-5_6
Canet-Aviles RM, Wilson MA, Miller DW, Ahmad R, McLendon C, Bandyopadhyay S, Baptista MJ, Ringe D et al (2004) The Parkinson’s disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc Natl Acad Sci U S A 101(24):9103–9108. https://doi.org/10.1073/pnas.0402959101
Kim RH, Smith PD, Aleyasin H, Hayley S, Mount MP, Pownall S, Wakeham A, You-Ten AJ et al (2005) Hypersensitivity of DJ-1-deficient mice to 1-methyl-4-phenyl-1,2,3,6-tetrahydropyrindine (MPTP) and oxidative stress. Proc Natl Acad Sci U S A 102(14):5215–5220. https://doi.org/10.1073/pnas.0501282102
Dave KD, De Silva S, Sheth NP, Ramboz S, Beck MJ, Quang C, Switzer RC, Ahmad SO et al (2014) Phenotypic characterization of recessive gene knockout rat models of Parkinson’s disease. Neurobiol Dis 70:190–203. https://doi.org/10.1016/j.nbd.2014.06.009
Chandran JS, Lin X, Zapata A, Hoke A, Shimoji M, Moore SO, Galloway MP, Laird FM et al (2008) Progressive behavioral deficits in DJ-1-deficient mice are associated with normal nigrostriatal function. Neurobiol Dis 29(3):505–514. https://doi.org/10.1016/j.nbd.2007.11.011
Krebiehl G, Ruckerbauer S, Burbulla LF, Kieper N, Maurer B, Waak J, Wolburg H, Gizatullina Z et al (2010) Reduced basal autophagy and impaired mitochondrial dynamics due to loss of Parkinson’s disease-associated protein DJ-1. PLoS One 5(2):e9367. https://doi.org/10.1371/journal.pone.0009367
Frøyset AK, Edson AJ, Gharbi N, Khan EA, Dondorp D, Bai Q, Tiraboschi E, Suster ML et al (2018) Astroglial DJ-1 over-expression up-regulates proteins involved in redox regulation and is neuroprotective in vivo. Redox Biol 16:237–247. https://doi.org/10.1016/j.redox.2018.02.010
De Miranda BR, Rocha EM, Bai Q, Ayadi A, Hinkle D, Burton EA, Greenamyre TJ (2018) Astrocyte-specific DJ-1 overexpression protects against rotenone-induced neurotoxicity in a rat model of Parkinson’s disease. Neurobiol Dis 115:101–114. https://doi.org/10.1016/j.nbd.2018.04.008
van Horssen J, Drexhage JA, Flor T, Gerritsen W, van der Valk P, de Vries HE (2010) Nrf2 and DJ1 are consistently upregulated in inflammatory multiple sclerosis lesions. Free Radic Biol Med 49(8):1283–1289. https://doi.org/10.1016/j.freeradbiomed.2010.07.013
Hanagasi HA, Giri A, Kartal E, Guven G, Bilgic B, Hauser AK, Emre M, Heutink P et al (2016) A novel homozygous DJ1 mutation causes parkinsonism and ALS in a Turkish family. Parkinsonism Relat Disord 29:117–120. https://doi.org/10.1016/j.parkreldis.2016.03.001
Lev N, Barhum Y, Ben-Zur T, Aharony I, Trifonov L, Regev N, Melamed E, Gruzman A et al (2015) A DJ-1 based peptide attenuates dopaminergic degeneration in mice models of Parkinson’s disease via enhancing Nrf2. PLoS One 10(5):e0127549. https://doi.org/10.1371/journal.pone.0127549
Molcho L, Ben-Zur T, Barhum Y, Offen D (2018) DJ-1 based peptide, ND-13, promote functional recovery in mouse model of focal ischemic injury. PLoS One 13(2):e0192954–e0192954. https://doi.org/10.1371/journal.pone.0192954
Vaz RL, Outeiro TF, Ferreira JJ (2018) Zebrafish as an animal model for drug discovery in Parkinson’s disease and other movement disorders: a systematic review. Front Neurol 9:347. https://doi.org/10.3389/fneur.2018.00347
Gagnon JA, Valen E, Thyme SB, Huang P, Akhmetova L, Ahkmetova L, Pauli A, Montague TG et al (2014) Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One 9(5):e98186. https://doi.org/10.1371/journal.pone.0098186
Varshney GK, Carrington B, Pei W, Bishop K, Chen Z, Fan C, Xu L, Jones M et al (2016) A high-throughput functional genomics workflow based on CRISPR/Cas9-mediated targeted mutagenesis in zebrafish. Nat Protoc 11(12):2357–2375. https://doi.org/10.1038/nprot.2016.141
Meeker ND, Hutchinson SA, Ho L, Trede NS (2007) Method for isolation of PCR-ready genomic DNA from zebrafish tissues. BioTechniques 43(5):610, 612, 614. https://doi.org/10.2144/000112619
Jha P, Wang X, Auwerx J (2016) Analysis of mitochondrial respiratory chain supercomplexes using blue native polyacrylamide gel electrophoresis (BN-PAGE). Curr Protoc Mouse Biol 6(1):1–14. https://doi.org/10.1002/9780470942390.mo150182
Nasuti C, Carloni M, Fedeli D, Gabbianelli R, Stefano A, Serafina C, Silva I, Domingues V et al (2013) Effects of early life permethrin exposure on spatial working memory and on monoamine levels in different brain areas of pre-senescent rats. Toxicology 303:162–168. https://doi.org/10.1016/j.tox.2012.09.016
Cannazza G, Stefano DA, Mosciatti B, Braghiroli D, Baraldi M, Pinnen F, Sozio P, Benatti C et al (2005) Detection of levodopa, dopamine and its metabolites in rat striatum dialysates following peripheral administration of l-DOPA prodrugs by mean of HPLC-EC. J Pharm Biomed Anal 36(5):1079–1084. https://doi.org/10.1016/j.jpba.2004.09.029
Chatterjee D, Gerlai R (2009) High precision liquid chromatography analysis of dopaminergic and serotoninergic responses to acute alcohol exposure in zebrafish. Behav Brain Res 200(1):208–213. https://doi.org/10.1016/j.bbr.2009.01.016
Frøyset A, Khan E, Fladmark K (2016) Quantitative proteomics analysis of zebrafish exposed to sub-lethal dosages of β-methyl-amino-L-alanine (BMAA). Sci Rep 6(1). https://doi.org/10.1038/srep29631
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A et al (2015) STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43(D1):D447–D452. https://doi.org/10.1093/nar/gku1003
Kasprzyk A (2011) BioMart: driving a paradigm change in biological data management. Database 2011(0):bar049. https://doi.org/10.1093/database/bar049
Smith AC, Robinson AJ (2016) MitoMiner v3.1, an update on the mitochondrial proteomics database. Nucleic Acids Res 44(D1):D1258–D1261. https://doi.org/10.1093/nar/gkv1001
Youn J-Y, Dunham WH, Hong S, Knight J, Bashkurov M, Chen GI, Bagci H, Rathod B et al (2018) High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol Cell 69(3):517–532.e11. https://doi.org/10.1016/j.molcel.2017.12.020
Markmiller S, Soltanieh S, Server KL, Mak R, Jin W, Fang MY, Luo E-C, Krach F et al (2018) Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell 172(3):590–376537088. https://doi.org/10.1016/j.cell.2017.12.032
McBrayer SK, Mayers JR, DiNatale GJ, Shi DD, Khanal J, Chakraborty AA, Sarosiek KA, Briggs KJ et al (2018) Transaminase inhibition by 2-hydroxyglutarate impairs glutamate biosynthesis and redox homeostasis in glioma. Cell 175(1):101–116.e25. https://doi.org/10.1016/j.cell.2018.08.038
Shao J, Gao F, Zhang B, Zhao M, Zhou Y, He J, Ren L, Yao Z et al (2017) Aggregation of SND1 in stress granules is associated with the microtubule cytoskeleton during heat shock stimulus. Anat Rec 300(12):2192–2199. https://doi.org/10.1002/ar.23642
Wu Z, Wang Y, Lim J, Liu B, Li Y, Vartak R, Stankiewicz T, Montgomery S et al (2018) Ubiquitination of ABCE1 by NOT4 in response to mitochondrial damage links co-translational quality control to PINK1-directed mitophagy. Cell Metab 28(1):130–1440000000. https://doi.org/10.1016/j.cmet.2018.05.007
Jinghui X, Xiaodi F, Mengqiu P, Xiao Z, Zhaoyu C, Dongmei W, Xiaomei Z, Qiong C et al (2018) Mitochondrial creatine kinase is decreased in the serum of idiopathic Parkinson’s disease patients. Aging Dis. https://doi.org/10.14336/ad.2018.0615
Nagamani S, Erez A, Lee B (2012) Argininosuccinate lyase deficiency. Genet Med 14(5):501–507. https://doi.org/10.1038/gim.2011.1
Rak M, Rustin P (2014) Supernumerary subunits NDUFA3, NDUFA5 and NDUFA12 are required for the formation of the extramembrane arm of human mitochondrial complex I. FEBS Lett 588(9):1832–1838. https://doi.org/10.1016/j.febslet.2014.03.046
Bossers K, Meerhoff G, Balesar R, Dongen JW, Kruse CG, Swaab DF, Verhaagen J (2009) Analysis of gene expression in Parkinson’s disease: possible involvement of neurotrophic support and axon guidance in dopaminergic cell death. Brain Pathol 19(1):91–107. https://doi.org/10.1111/j.1750-3639.2008.00171.x
Hayes SD, Liu H, MacDonald E, Sanderson CM, Coulson JM, Clague MJ, Urbé S (2012) Direct and indirect control of mitogen-activated protein kinase pathway-associated components, BRAP/IMP E3 ubiquitin ligase and CRAF/RAF1 kinase, by the deubiquitylating enzyme USP15. J Biol Chem 287(51):43007–43018. https://doi.org/10.1074/jbc.m112.386938
Simon M, Richard EM, Wang X, Shahzad M, Huang VH, Qaiser TA, Potluri P, Mahl SE et al (2015) Mutations of human NARS2, encoding the mitochondrial asparaginyl-tRNA synthetase, cause nonsyndromic deafness and Leigh syndrome. PLoS Genet 11(3):e1005097. https://doi.org/10.1371/journal.pgen.1005097
Ayuso P, Martínez C, Lorenzo-Betancor O, Pastor P, Luengo A, Jiménez-Jiménez FJ, Alonso-Navarro H, Villalba MT et al (2011) A polymorphism located at an ATG transcription start site of the heme oxygenase-2 gene is associated with classical Parkinson’s disease. Pharmacogenet Genomics 21(9):565–571. https://doi.org/10.1097/fpc.0b013e328348f729
Potting C, Crochemore C, Moretti F, Nigsch F, Schmidt I, Manneville C, Carbone W, Knehr J et al (2018) Genome-wide CRISPR screen for PARKIN regulators reveals transcriptional repression as a determinant of mitophagy. Proc Natl Acad Sci U S A 115(2):E180–E189. https://doi.org/10.1073/pnas.1711023115
Ando K, Tomimura K, Sazdovitch V, Suain V, Yilmaz Z, Authelet M, Ndjim M, Vergara C et al (2016) Level of PICALM, a key component of clathrin-mediated endocytosis, is correlated with levels of phosphotau and autophagy-related proteins and is associated with tau inclusions in AD, PSP and Pick disease. Neurobiol Dis 94:32–43. https://doi.org/10.1016/j.nbd.2016.05.017
Nangaku M, Sato-Yoshitake R, Okada Y, Noda Y, Takemura R, Yamazaki H, Hirokawa N (1994) KIF1B, a novel microtubule plus end-directed monomeric motor protein for transport of mitochondria. Cell 79(7):1209–1220. https://doi.org/10.1016/0092-8674(94)90012-4
Cuenca L, Gil-Martinez AL, Cano-Fernandez L, Sanchez-Rodrigo C, Estrada C, Fernandez-Villalba E, Herrero MT (2018) Parkinson’s disease: a short story of 200 years. Histol Histopathol:18073. https://doi.org/10.14670/hh-18-073
Schapira AHV, Cooper JM, Dexter D, Clark JB, Jenner P, Marsden CD (1990) Mitochondrial complex I deficiency in Parkinson's disease. J Neurochem 54(3):823–827. https://doi.org/10.1111/j.1471-4159.1990.tb02325.x
Chen H, Zhang SM, Hernán MA, Willett WC, Ascherio A (2003) Weight loss in Parkinson’s disease. Ann Neurol 53(5):676–679. https://doi.org/10.1002/ana.10577
Lopez-Fabuel I, Martin-Martin L, Resch-Beusher M, Azkona G, Sanchez-Pernaute R, Bolaños JP (2017) Mitochondrial respiratory chain disorganization in Parkinson’s disease-relevant PINK1 and DJ1 mutants. Neurochem Int 109:101–105. https://doi.org/10.1016/j.neuint.2017.03.023
Miwa S, Jow H, Baty K, Johnson A, Czapiewski R, Saretzki G, Treumann A, von Zglinicki T (2014) Low abundance of the matrix arm of complex I in mitochondria predicts longevity in mice. Nat Commun 5(1):3837. https://doi.org/10.1038/ncomms4837
Verkaart S, Koopman WJ, van Emst-de Vries SE, Nijtmans LG, van den Heuvel LW, Smeitink JA, Willems PH (2007) Superoxide production is inversely related to complex I activity in inherited complex I deficiency. Biochim Biophys Acta 1772(3):373–381. https://doi.org/10.1016/j.bbadis.2006.12.009
Gorrini C, Harris IS, Mak TW (2013) Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov 12(12):931–947. https://doi.org/10.1038/nrd4002
Glisic B, Mihaljevic I, Popovic M, Zaja R, Loncar J, Fent K, Kovacevic R, Smital T (2015) Characterization of glutathione-S-transferases in zebrafish (Danio rerio). Aquat Toxicol 158:50–62. https://doi.org/10.1016/j.aquatox.2014.10.013
Hsieh C-H, Shaltouki A, Gonzalez AE, Bettencourt da Cruz A, Burbulla LF, St. Lawrence E, Schüle B, Krainc D et al (2016) Functional impairment in Miro degradation and mitophagy is a shared feature in familial and sporadic Parkinson’s disease. Cell Stem Cell 19(6):709–724. https://doi.org/10.1016/j.stem.2016.08.002
Nguyen TN, Padman BS, Lazarou M (2016) Deciphering the molecular signals of PINK1/Parkin mitophagy. Trends Cell Biol 26(10):733–744. https://doi.org/10.1016/j.tcb.2016.05.008
Xiong H, Wang D, Chen L, Choo Y, Ma H, Tang C, Xia K, Jiang W et al (2009) Parkin, PINK1, and DJ-1 form a ubiquitin E3 ligase complex promoting unfolded protein degradation. J Clin Invest 119(3):650–660. https://doi.org/10.1172/jci37617
Ito K, Takahashi A, Morita M, Suzuki T, Yamamoto T (2011) The role of the CNOT1 subunit of the CCR4-NOT complex in mRNA deadenylation and cell viability. Protein Cell 2(9):755–763. https://doi.org/10.1007/s13238-011-1092-4
Hod Y, Pentyala SN, Whyard TC, El-Maghrabi RM (1999) Identification and characterization of a novel protein that regulates RNA–protein interaction. J Cell Biochem 72(3):435–444. https://doi.org/10.1002/(sici)1097-4644(19990301)72:3<435::aid-jcb12>3.0.co;2-h
Repici M, Hassanjani M, Maddison DC, Garção P, Cimini S, Patel B, Szegö ÉM, Straatman KR et al (2018) The Parkinson’s disease-linked protein DJ-1 associates with cytoplasmic mRNP granules during stress and neurodegeneration. Mol Neurobiol:1–17. https://doi.org/10.1007/s12035-018-1084-y
Gan L, Cookson MR, Petrucelli L, Spada AR (2018) Converging pathways in neurodegeneration, from genetics to mechanisms. Nat Neurosci 21(10):1300–1309. https://doi.org/10.1038/s41593-018-0237-7
Booth HDE, Hirst WD, Wade-Martins R (2017) The role of astrocyte dysfunction in Parkinson’s disease pathogenesis. Trends Neurosci 40(6):358–370. https://doi.org/10.1016/j.tins.2017.04.001
Verkhratsky A, Rodriguez JJ, Parpura V (2013) Astroglia in neurological diseases. Future Neurol 8(2):149–158. https://doi.org/10.2217/fnl.12.90
Tong J, Ang L-C, Williams B, Furukawa Y, Fitzmaurice P, Guttman M, Boileau I, Hornykiewicz O et al (2015) Low levels of astroglial markers in Parkinson’s disease: relationship to α-synuclein accumulation. Neurobiol Dis 82:243–253. https://doi.org/10.1016/j.nbd.2015.06.010
Choi D-JJ, Eun J-HH, Kim BG, Jou I, Park SM, Joe E-HH (2017) A Parkinson’s disease gene, DJ-1, repairs brain injury through Sox9 stabilization and astrogliosis. Glia 66(2):445–458. https://doi.org/10.1002/glia.23258
Tsang A, Chung K (2009) Oxidative and nitrosative stress in Parkinson’s disease. Biochim Biophys Acta Mol basis Dis 1792(7):643–650. https://doi.org/10.1016/j.bbadis.2008.12.006
Chung K, David KK (2010) Emerging roles of nitric oxide in neurodegeneration. Nitric Oxide 22(4):290–295. https://doi.org/10.1016/j.niox.2010.02.002
Sun D-Z, Song C-Q, Xu Y-M, Wang R, Liu W, Liu Z, Dong X-S (2018) Involvement of PINK1/Parkin-mediated mitophagy in paraquat-induced apoptosis in human lung epithelial-like A549 cells. Toxicol in Vitro 53:148–159. https://doi.org/10.1016/j.tiv.2018.08.009
Orsini F, Blasio D, Zangari R, Zanier ER, Simoni M-G (2014) Versatility of the complement system in neuroinflammation, neurodegeneration and brain homeostasis. Front Cell Neurosci 8:380. https://doi.org/10.3389/fncel.2014.00380
Shi Q, Chowdhury S, Ma R, Le KX, Hong S, Caldarone BJ, Stevens B, Lemere CA (2017) Complement C3 deficiency protects against neurodegeneration in aged plaque-rich APP/PS1 mice. Sci Transl Med 9(392):eaaf6295. https://doi.org/10.1126/scitranslmed.aaf6295
Hwang J-Y, Zukin SR (2018) REST, a master transcriptional regulator in neurodegenerative disease. Curr Opin Neurobiol 48:193–200. https://doi.org/10.1016/j.conb.2017.12.008
Yu M, Suo H, Liu M, Cai L, Liu J, Huang Y, Xu J, Wang Y et al (2013) NRSF/REST neuronal deficient mice are more vulnerable to the neurotoxin MPTP. Neurobiol Aging 34(3):916–927. https://doi.org/10.1016/j.neurobiolaging.2012.06.002
Zondler L, Miller-Fleming L, Repici M, Gonçalves S, Tenreiro S, Rosado-Ramos R, Betzer C, Straatman KR et al (2014) DJ-1 interactions with α-synuclein attenuate aggregation and cellular toxicity in models of Parkinson’s disease. Cell Death Dis 5(7):e1350. https://doi.org/10.1038/cddis.2014.307
Milanese C, Sager JJ, Bai Q, Farrell TC, Cannon JR, Greenamyre TJ, Burton EA (2012) Hypokinesia and reduced dopamine levels in zebrafish lacking β- and γ1-synucleins. J Biol Chem 287(5):2971–2983. https://doi.org/10.1074/jbc.m111.308312
Gonçalves SA, Macedo D, Raquel H, Simões PD, Giorgini F, Ramalho JS, Barral DC, Moita L et al (2016) shRNA-based screen identifies endocytic recycling pathway components that act as genetic modifiers of alpha-synuclein aggregation, secretion and toxicity. PLoS Genet 12(4). https://doi.org/10.1371/journal.pgen.1005995
Acknowledgments
We wish to thank Puja Thiel for performing the zebrafish injections and Mikkel Espolin Birkeland for dissection of zebrafish. Mass spectrometry analysis was carried out at the Proteomics Core Facility at the University of Bergen (PROBE). The CRISPR-Cas9 plasmids were received from the Suster Lab at Uni Research AS. The authors also wish to thank Sushma Grellscheid and Thomas Stevenson for discussions regarding stress granules.
Funding
This research was funded by grants from Advokat Rolf Sandberg Rebergs and Ellen Marie Rebergs legat (KEF), Det alminnelige naturvitenskapelige forskningsfond, UiB (AJE), and COST action 16112 (KEF and ADS).
Author information
Authors and Affiliations
Contributions
AJE and KEF designed the study and wrote the paper. AJE, AKF, HAH, IE, and EAK performed the experiments. ADS performed the dopamine analysis. AJE, KEF, and AKF analyzed the data.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no conflict of interest.
Ethical Approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. All procedures performed in studies involving animals were in accordance with the ethical standards of the University of Bergen where the studies were conducted. Establishment of the park7−/− (KO) line and euthanization of adult fish were approved by the Norwegian National Animal Research Authority at Mattilsynet (FOTS ID8039 and ID14039).
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Online resource 1
Proteins identified by mass spectrometry from park7−/− and wild-type 3-month-old zebrafish brains. Proteins identified by site or reverse were removed in addition to removal of potential contaminants. Proteins with a score less than 4 and those identified by only a single unique peptide were also removed. In order to determine the fold change, a two-sample Student’s t test was run (S0 = 2; p value threshold = 0.05) on proteins with at least 3 valid values in both KO and WT samples. Those proteins that did not meet the criteria for the t test have a fold change and p value not determined (n.d.). LFQ intensities are present after log2 transformation, and invalid values are shown as NaN. KO, park7−/−; WT, wild type. (XLSX 665 kb)
Online resource 2
All peptides identified by mass spectrometry from park7−/− (KO) and wild-type (WT) adult zebrafish brains. LFQ intensities are presented after log2 transformation. (XLSX 7513 kb)
Rights and permissions
About this article
Cite this article
Edson, A.J., Hushagen, H.A., Frøyset, A.K. et al. Dysregulation in the Brain Protein Profile of Zebrafish Lacking the Parkinson’s Disease-Related Protein DJ-1. Mol Neurobiol 56, 8306–8322 (2019). https://doi.org/10.1007/s12035-019-01667-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-019-01667-w