Abstract
The guanine deaminase cypin (cytosolic PSD-95 interactor) binds to PSD-95 (postsynaptic density protein 95) and regulates dendrite branching by promoting microtubule polymerization. Here, we identify a novel short isoform of cypin, termed cypinS, which is expressed in mouse and human, but not rat, tissues. Cypin and cypinS mRNA and protein levels peak at P7 and P14 in the mouse brain, suggesting a role for these isoforms during development. Interestingly, although cypinS lacks guanine deaminase activity, overexpression of cypinS increases dendrite branching. This increase occurs further away from soma than do increases resulting from overexpression of cypin. In contrast, overexpression of cypin, but not cypinS, decreases dendritic spine density and maturity. This suggests that changes to spines, but not to dendrites, may be dependent on guanine deaminase activity. Furthermore, overexpression of either cypin or cypinS increases miniature excitatory postsynaptic current (mEPSC) frequency, pointing to a presynaptic role for both isoforms. Interestingly, overexpression of cypinS results in a significantly greater increase in frequency than does overexpression of cypin. Thus, cypin and cypinS play distinct roles in neuronal development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Cypin (cytosolic PSD-95 interactor), also known as nedasin [1], is guanine deaminase (GDA; guanase) [2] and the major interactor of the scaffolding protein postsynaptic density 95 (PSD-95) in the brain. The carboxy terminus of cypin binds to the first two PDZ domains of PSD-95 and decreases PSD-95 localization at postsynaptic sites [2]. Cypin binds to tubulin heterodimers via its collapsin response mediator protein (CRMP) homology domain and promotes microtubule assembly. Overexpression of cypin in hippocampal neurons increases dendrite numbers, and knockdown of cypin decreases dendrites [3]. This action of cypin correlates with GDA activity [3] and zinc binding [4]. Our group has identified signaling molecules that act in concert with cypin to regulate dendritic morphology. For example, we reported that the SNARE-associated protein snapin binds to cypin and disrupts the cypin-tubulin interaction, acting as a negative regulator of cypin-mediated microtubule assembly [5]. Similarly, overexpression of PSD-95 blocks cypin-mediated increases in dendritic branching; however, the binding of cypin to PSD-95 is necessary for stabilization of dendrite growth promoted by cypin [6]. Upstream regulators of cypin include RhoA, a small GTPase, and brain-derived neurotrophic factor (BDNF). Activated RhoA acts as a negative regulator of dendrite branching by decreasing cypin expression in a translation-dependent manner [7]. In contrast, BDNF increases dendritic arborization via CREB-dependent transcriptional activation of cypin [8]. Thus, multiple regulators of dendrite morphology act via cypin, which then regulates microtubule assembly, shaping the dendritic arbor.
Recent evidence suggests a required role for cypin in normal brain function. Genome-wide transcriptome profiling revealed increased expression of cypin in the CA1 region of the hippocampus after oxidative stress [9]. Additionally, nigro-striatal degeneration in a rat model of Parkinson’s disease results in increased expression of cypin in the striatum [10]. Furthermore, overexpression of cypin protects cultured somatic motor neurons from glutamate-induced toxicity [11] and cultured hippocampal neurons from N-methyl-d-aspartate (NMDA) glutamate receptor-induced toxicity by increasing the number of small varicosities on dendrites [12]. Thus, cypin may act in a neuroprotective manner in the injured or diseased brain. Intriguingly, it was recently reported that 23% of mothers of children with maternal autoantibody-related autism (MAR) have autoantibodies with increased reactivity for cypin and/or the related protein collapsin response mediator protein 1 [13]. Taken together, these data suggest that further investigation of cypin function in the brain is warranted to increase our understanding of its role in neuronal development and disease.
In the current study, we identify a short isoform of cypin (cypinS) that lacks guanine deaminase (GDA) activity. Surprisingly, this isoform is expressed in tissue from mouse and human, but not rat. Cypin and cypinS mRNA and protein levels peak at P7 and P14 in the mouse brain, suggesting a role for these isoforms during development when maximal changes to neuronal morphology occur. Our data suggest that the two isoforms play similar but distinct roles in neuronal development. Overexpression of cypinS increases dendrite branching further away from the soma than does overexpression of cypin, and overexpression of cypin significantly decreases spine density and maturity while overexpression of cypinS does not affect spines. Overexpression of cypin or cypinS results in increased mEPSC frequency but not amplitude. Thus, our current data suggest distinct roles for cypin and cypinS in neuronal development.
Results
Analysis of the GDA Gene Predicts Expression of a Short Isoform of Cypin
The GDA gene consists of 14 exons, resulting in a protein product of the 454 amino acid (aa) cypin. An alternative exon 1a exists in GDA, yielding an additional isoform of cypin, which we term cypinS for cypin short. CypinS is encoded by 14 exons with only part of the last exon present, resulting in a shorter 3’ UTR in cypinS mRNA compared to the 3’ UTR in cypin mRNA (Fig. 1a). This isoform is predicted to be expressed only in mouse and human, but not rat, tissues. In fact, we have not observed the presence of this isoform in rat tissue in any of our past studies [2, 3, 7, 8]. As we previously reported [2, 3], cypin protein contains a zinc-binding domain (76–84 amino acids (aa)), a collapsin response mediator protein (CRMP) homology domain (350–403 aa), and carboxy terminal PDZ-binding motif (451–454 aa). CypinS consists of the carboxy terminal 380 aa (75–454 aa) of cypin, thus lacking the amino terminal 74 aa of cypin. Both cypin and cypinS contain His240, His279, and Asp330, three aa that we have reported to be involved in zinc binding ([4]; Fig. 1b).
Schematic of the GDA gene and resulting cypin proteins. a Alternative splicing of GDA, resulting in two isoforms of cypin. Alternative exon 1 for short isoform of cypin (cypinS—Ensembl: ENSMUST00000121725; cypin—Ensembl: ENSMUST00000087600) is indicated in green. This alternative exon is present in mouse and human, but not rat. b Schematic of cypin proteins. The zinc-binding domain (brown) binds zinc [4], the collapsin response mediator protein (CRMP) homology domain (peach) binds tubulin heterodimers [3], and the PDZ-binding motif (red) binds to PSD-95 family members [2]. Additional residues involved in zinc binding, His240, His279, and Asp330, are highlighted (black lines; [4])
Cypin and cypinS mRNA and Protein Are Expressed in Multiple Mouse Brain Regions and Tissues
Cypin and cypinS mRNA differ only in their exon 1 sequences (Fig. 1a). Thus, we constructed primers targeted to the junction of exons 1 and 2 to specifically amplify cypin or cypinS mRNA. Using these primers, we identified unique RT-PCR products for cypin and cypinS mRNA, suggesting the presence of both transcripts in mouse brain (Fig. 2a). To assay for protein expression, we validated the detection of cypinS protein by our polyclonal rabbit cypin antibody (BF6; [8, 12]) by overexpressing cypin and cypinS in HEK293 cells and subjecting lysates from these cells to Western blot analysis (Fig. 2b). Results of this analysis reveal that both isoforms are easily resolved by SDS-polyacrylamide gel electrophoresis and detected by Western blotting. Our data show that cypinS is present in multiple mouse brain regions, including cerebellum, which has low expression of cypin [2, 14], and organs, such as lung, heart, liver, spleen, kidney, and skeletal muscle, which also show expression of cypin (Fig. 2c, d). Similar to what is observed in mouse tissue, cypinS is expressed in human cortical tissue (Fig. 2e).
Expression of cypinS mRNA and protein. a Detection of cypinS mRNA transcripts in mouse brain by qRT-PCR. b Detection of cypin protein isoforms with polyclonal rabbit cypin antibody (BF6, raised against full length cypin; [8, 12]). Overexpression of indicated proteins in HEK293 cells demonstrates that our polyclonal antibody recognizes both cypin and cypinS protein and that the two isoforms are easily resolved by Western blotting. GAPDH is shown as a control for equal protein loading. c Expression of cypin and cypinS proteins in different brain regions. d Expression of cypin and cypinS proteins in different mouse organs. e CypinS is expressed in human brain.
Cypin and cypinS mRNA and Protein Levels Peak at Postnatal Day 7 in Mouse Brain
To identify how cypinS may play a developmental role in brain function, relative mRNA and protein levels were determined in brain from mice at postnatal day (P) 0, 7, 14, 30, and 100 (adult). qRT-PCR results show that cypinS transcript is present at significantly higher levels at P0, P7, and P14 compared to that expressed in adult brain (Fig. 3a). In addition, cypin transcript is highest at P7 (Fig. 3a). Consistent with these results, brain cypin and cypinS protein levels peak at P7 and P14 (Fig. 3b). We determined brain region-specific expression and enrichment in the adult and found that expression of cypinS occurs at similar levels across all regions while cypin is expressed at 20% of these levels in cerebellum (Fig. 3c), consistent with previous reports and by our group and others [2, 14].
Expression profile of cypin and cypinS mRNA and proteins in mouse brain. a Relative mRNA levels (normalized to GAPDH) of cypin and cypinS at developmental time points P0, P7, P14, P30, and P100. mRNA was isolated from four different mouse brains and subjected to qRT-PCR to determine the fold change in mRNA levels compared to adult brain (P100). *p < 0.05; **p < 0.01 compared to P100 for cypinS; #p < 0.05, P100 vs P7 for cypin. p values were determined by two-tailed Student’s t test comparing the 2(−ΔCt) values of the two groups. The p value is adjusted using Benjamini-Hochberg False Discovery Rate. n = 4 biological replicates (male mice), each with three technical replicates. Internal control = GAPDH; calibrator or reference group = P100 (adult mouse brain). DataAssist Software automatically excluded outliers of technical replicates. b Quantification and representative images of blots for relative protein levels of cypin and cypinS at various developmental time points. Cypin and cypinS levels peak at developmental time points P7 and P14. **p < 0.01, P100 vs P7; *p < 0.05, P100 vs P14 (cypin). p = 0.07, P100 vs P7; #p < 0.05, P0 vs P14 (cypinS). c Relative protein levels of cypin and cypinS in different mouse brain regions, demonstrating significantly low levels of cypin in cerebellum compared to whole brain. ****p < 0.0001, whole brain vs cerebellum (cypin). All statistics determined by one-way ANOVA followed by Dunnett’s multiple comparisons test. n = 3–4. Error bars = ± SEM
CypinS Does Not Function as a Guanine Deaminase
Cypin is guanine deaminase (GDA; guanase), an enzyme that converts guanine to xanthine [3, 14]. As we previously reported using colorimetric assay, cypin has GDA activity [3, 14]. Since cypinS lacks the amino terminal 74 aa of cypin but has all aa responsible for zinc binding, a GDA assay was performed to determine whether cypinS has this activity. We transfected COS-7 cells with plasmids encoding GFP, GFP-Cypin, or GFP-CypinS and subjected the cells to the colorimetric GDA assay. Interestingly, unlike cells expressing GFP-Cypin, cells expressing GFP-CypinS do not show GDA activity (Fig. 4a). In addition, we performed Amplex Red GDA assay, which demonstrates kinetics of the GDA reaction rather than activity at a fixed time point (Fig. 4b, c). The Amplex Red assay shows a gradual increase in absorbance with time in extracts from samples cells expressing cypin, while extracts from cells expressing GFP and cypinS do not show increases in absorbance. Taken together, these data suggest that cypin acts as guanine deaminase while cypinS does not. Thus, cypinS may not play a role in guanine metabolism.
CypinS does not function as a guanine deaminase. a COS-7 cells were transfected with pEGFP-C1, pEFGP-C1-Cypin or pEGFP-C1-CypinS, fixed, and subjected to guanine deaminase assay. −guanine is a negative control lacking guanine substrate in the reaction. Cypin has guanine deaminase activity while cypinS lacks guanine deaminase activity. Scale bar = 100 μm. b Western blot analysis for detection of protein expression of GFP, cypin-GFP, and cypinS-GFP in transfected HEK293 cells. c Amplex Red colorimetric assay to analyze guanine deaminase activity [4, 44]. HEK293 cell lysate (50 μg) was subjected to the assay. Lysate from HEK293 cells expressing GFP and cypin were used as negative and positive controls, respectively. Lysate from cells expressing cypinS does not demonstrate guanine deaminase activity (n = 3). Error bars = ± SEM
Overexpression of CypinS Increases Dendrite Branching in Hippocampal Neurons
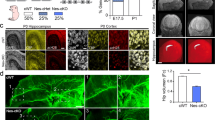
Overexpression of cypin increases primary and secondary dendrite numbers in cultured hippocampal neurons, and GDA activity correlates with this increase [3]. Since cypinS lacks GDA activity (Fig. 4), we asked whether cypinS regulates dendrite branching. Cypin and cypinS were overexpressed on DIV7 in cultured rat hippocampal neurons, and dendrite branching was analyzed at DIV12 using Sholl analysis [15,16,17]. It is important to note that cypinS is not expressed in rat neurons [2, 3, 7, 8], and thus, we used these neurons to determine the role for cypinS in a null background. Overexpression of cypinS results in increased dendrite branching more distal from the cell body than the resulting increase in dendrites resulting from overexpression of cypin (Fig. 5a–c). Overexpression of either cypin or cypinS results in increased tertiary and higher order dendrites, total dendrites, branch points, and terminal points (Fig. 5d–i). Thus, cypinS overexpression results in increases in overall dendrite arborization that is similar to the increase promoted by cypin. Importantly, our data suggest that the GDA activity of cypin is not required for increases in dendrites.
Overexpression of cypin or cypinS results in increased dendrite branching in rat hippocampal neurons. Hippocampal neurons were cultured from rat embryos at 18 days gestation. Neurons were transfected on DIV7 with pEGFP-C1, pEGFP-C1-Cypin, or pEGFP-C1-CypinS, co-transfected with cDNA encoding mRFP (to fill the dendritic arbor), and fixed at DIV12. a Representative inverted mRFP images. b Sholl analysis of neurons expressing indicated proteins. c Overexpression of cypin or cypinS results in increased proximal dendritic arborization compared to control neurons expressing GFP. Blue solid line: p < 0.05, control vs cypin; red solid line: p < 0.05, control vs cypinS, determined by two-way ANOVA followed by Tukey’s multiple comparisons test. d–i Overexpression of cypin or cypinS increases tertiary and higher order dendrites, total number of dendrites, branch points, and terminal points. *p < 0.05 (terminal points); **p < 0.01 (tertiary and higher order dendrites, total number of dendrites); ***p < 0.001 (branch points), control vs cypin; *p < 0.05 (tertiary and higher order dendrites, total dendrites, and branch points); **p < 0.01 (terminal points), control vs cypinS. Determined by one-way ANOVA followed by Tukey’s multiple comparisons test. Error bar = ±SEM. n = 38 (control), n = 39 (cypin), and n = 38 (cypinS). Scale bar = 50 μm
Overexpression of Cypin, but Not cypinS, Alters Spine Density and Morphology
To determine whether the subcellular localization of cypinS differs from that of cypin, we performed synaptosomal fractionation. Cypin is present in all fractions, including cellular cytosol, crude synaptosomes, synaptic cytosol, and synaptic membranes. In contrast, although cypinS is also present in crude synaptosomes and synaptic cytosol, it is present in synaptic membranes where cypin is present at a lower level than in other fractions (Fig. 6a). Furthermore, since PSD-95 is enriched in the synaptic membrane fraction [2] and cypinS contains a carboxy terminal PDZ-binding motif, we performed co-immunoprecipitation in mouse brain lysate. As expected, cypin and cypinS co-immunoprecipitate with PSD-95 (Fig. 6b). Thus, based on these results, we hypothesize that cypinS plays a role in spine development due to its interaction with both PSD-95 and its localization at synaptic membranes. To test our hypothesis, we overexpressed cypin or cypinS in rat hippocampal neurons at DIV14, and spine number and morphology were assessed at DIV17. Overexpression of cypin results in a significant decrease in spine density and the number of mature spines. In contrast, cypinS had a marginal but non-significant effect on spine density (p = 0.077; Fig. 6c–f). Overexpression of either cypin or cypinS had no effect on spine length or width (Fig. 6g, h). Our results suggest that cypinS may not play a major role in spinogenesis, especially since rat neurons do not express cypinS.
Overexpression of cypin, but not cypinS, alters spine density and morphology. a Western blot analysis demonstrating the presence of cypin and cypinS in synaptosomal fractions from brains of young (P14) and adult (P120) mice. b Co-immunoprecipitation of cypin and cypinS with PSD-95 from whole brain lysate. PSD-95 is detected as multiple bands, consistent with previous reports. Cypin and cypinS are detected in PSD-95 immunoprecipitates with a smaller molecular weight band below where cypinS is detected, representing a modified cypinS or possible degradation product. c Representative images of dendritic spines at DIV17 from neurons transfected with pEGFP-C1 (control), pEGFP-C1-Cypin, or pEGFP-C1-CypinS. d Total spines/10 μm within a 60 μm segment away from the soma in neurons expressing indicated proteins. **p < 0.01, control vs cypin; p = 0.077, control vs cypinS (total spines) determined by one-way ANOVA followed by Tukey’s multiple comparisons test. e Mature spines/10 μm within a 60-μm segment away from the soma in neurons expressing indicated proteins. *p < 0.05, control vs cypin; p = 0.096, control vs cypinS (number of mature spines) determined by one-way ANOVA followed by Tukey’s multiple comparisons test. f–h Overexpression of cypin or cypinS has no effect on immature spine density, spine width, or spine length. n = 14 (control), n = 16 (cypin), n = 16 (cypinS) from three independent rat cultures. Error bar = ± SEM. Scale bar = 5 μm
Overexpression of Cypin and cypinS Increases mEPSC Frequency
Since both cypin and cypinS are present in synaptic cytosol fractions where synaptic vesicles are present, we asked whether overexpression of either isoform affects neuronal electrophysiology. Rat hippocampal neurons were transduced at DIV 14 with lentivirus encoding GFP, GFP-Cypin, or GFP-CypinS, and patch clamp analysis was performed at DIV 21. Interestingly, overexpression of cypin or cypinS resulted in a significant increase in the frequency of mEPSCs with no effect on amplitude (Fig. 7a–c). In addition, the effect of overexpression of cypinS on frequency is significantly higher than that of overexpression of cypin. Taken together with dendrite branching and spine morphology data, our results suggest that cypin and cypinS regulate both postsynaptic and presynaptic sites in hippocampal neurons in distinct manners.
Overexpression of cypin or cypinS results in increased frequency of miniature excitatory postsynaptic currents (mEPSCs). mEPSCs were recorded on DIV21 from rat hippocampal neurons overexpressing GFP (n = 16), cypin (n = 15), or cypinS (n = 13). a Representative trace recordings. b, c Bar graph analysis of mEPSC frequency and amplitude. *p < 0.05, control vs cypin; ****p < 0.0001, control vs cypinS; #p < 0.05, cypin vs cypinS. p values determined by Kruskal-Wallis test followed by Dunn’s multiple comparisons test. Error bars = ± SEM
Discussion
In the current work, we report the identification of a novel isoform of cypin, which we term cypinS. Interestingly, cypinS is expressed in human and mouse tissues, but not in rat tissues. CypinS is expressed at lower levels than cypin in mouse brain tissues except in the cerebellum, where cypin is expressed at very low levels [2, 14]. Cypin functions as a guanine deaminase [3, 14]; however, cypinS lacks this activity. We found that mRNA and protein levels of cypin and cypinS peak at P7-P14 in mouse brain, suggesting an important role for these proteins during neuronal development when maximum changes in morphology occur. Expression of either cypin or cypinS increases dendrite branching, which we previously reported to be dependent on the CRMP homology region [3] that binds to tubulin heterodimers and promotes microtubule assembly [18, 19]. In contrast, only overexpression of cypin results in changes to dendritic spines. Overexpression of either isoform results in increases in frequency of mEPSCs; however, overexpression of cypinS promotes a significantly higher increase in mEPSC frequency. Taken together, these data suggest different pre- and postsynaptic roles for cypin and cypinS.
Until now, we proposed that the guanine deaminase activity of cypin is important for its effects on dendritogenesis. Previous results from our laboratory suggest that mutation of residues that bind zinc eliminate the GDA activity and the dendrite-promoting activity of cypin [3, 4], leading us to conclude that the two activities are related. However, based on our work presented here, it appears that zinc binding, but not GDA activity, is necessary for cypin-promoted changes to dendrites. Evolutionary analysis performed by our laboratory led to a model by which an ancestral cytidine deaminase-like protein evolved by gene duplication followed by acquisition of PDZ-binding motifs and a zinc-binding domain [4]. This model suggests that a new but distinct function was acquired by the mammalian guanine deaminase cypin.
CypinS is present in mouse and human but not rat. Although the human genome shares greater than 97% genes with the mouse and rat genomes [20, 21], maps of conserved synteny between human, mouse, and rat genomes suggest that there are differences in the genome when comparing human to mouse and human to rat [21]. In this same study, 278 orthologous segments were identified between human and rat, with 280 between human and mouse [21]. Thus, while cypin may function as a guanine deaminase and regulator of dendritic morphology in all three species, cypinS may act to further shape neuronal function in the mouse and human. Specifically, cypinS regulates distal dendrite branching away from soma while cypin shapes the proximal arbor, suggesting a GDA-independent role for cypinS in mouse and human.
CypinS has a shorter 3’UTR compared to cypin (Fig. 1a), and we have shown that protein expression of cypinS is only 6- to 10-fold lower than that of cypin while mRNA expression is 1000- to 40,000-fold lower than that of cypin (Fig. 3a). Although a number of studies have reported high correlation between reference mRNA and protein levels [22, 23], other studies have shown poor correlation between mRNA and protein levels, where magnitudes of difference in mRNA exist for the same levels of protein expression and vice versa [23, 24]. There are multiple factors, including ribosomal occupancy, half-lives of proteins, and codon adaptation index, that explain the absence of a correlation between mRNA and protein expression [23]. The 3’UTR of mRNA is involved in post-transcriptional regulation of gene expression [25]. Adenosine (A) and uridine (U) rich elements, also called AU rich elements (AREs), are present in 3’UTRs of mRNA [26]. AREs consist of one or more AUUUA pentamers and are involved in the regulation of mRNA stability and changes to translation [26, 27]. The 3’UTRs of cypin and cypinS differ, and thus, we used the AREsite database to identify AREs in the 3’UTRs of cypin and cypinS transcripts [28]. There are ten AUUUA pentamer motifs in the 3’UTR of cypin and five AUUUA pentamer motifs in the 3’UTR of cypinS (Supplementary Fig. 1). This difference in the number of AUUUA pentamers in the AREs of the 3’UTRs of cypin and cypinS mRNAs may be responsible for different stabilities of both mRNAs, and thus may explain the lack of correlation between mRNA and protein expression. Since this is one of many mechanisms that may be responsible for our observation, why cypin and cypinS protein levels do not correlate with transcript levels will be a focus of future studies.
Zinc is involved in the regulation of dendrite branching by promoting stability of microtubules [29]. In fact, in a rat model of zinc deficiency, there is impaired dendritic differentiation of basket and stellar cells [30]. Zinc also regulates multiple zinc finger proteins and transcriptional factors that shape the dendritic arbor [31,32,33,34,35]. CypinS may bind to zinc, as does cypin [4], since the two isoforms share all residues that coordinate for zinc binding. We posit that cypin and cypinS may act as zinc sensors to increase dendrites. Zinc not only plays an important role in normal synaptic function, but when altered, it plays a role in a host of brain disorders, such as Alzheimer’s disease, depression, Parkinson’s disease, Huntington’s disease, amyotrophic lateral sclerosis, and prion disease (reviewed in [36]). Furthermore, excess zinc mediates toxicity due to brain injury or stroke (reviewed in [36]). In line with these data, we previously reported that overexpression of cypin in cultured hippocampal neurons protects them from N-methyl-d-aspartate-mediated death [12]. It is possible that in this scenario, cypin acts to sense zinc and mediate protective changes to dendrites [12].
Cypin also binds to PSD-95 through its PDZ-binding motif and regulates PSD-95 synaptic targeting [2]. Moreover, overexpression of cypin decreases spine density (Fig. 6d and [12]). We have now demonstrated that cypinS is present in crude synaptosomes, synaptic cytosol, and synaptic membranes (Fig. 6a). Like cypin, cypinS contains a PDZ-binding motif on its carboxy terminus, and our data suggest that cypinS binds to PSD-95 (Fig. 6b). Surprisingly, we found that the overexpression of cypinS has no significant effect on spine density (Fig. 6d), suggesting that cypinS functions distinctly from cypin in the regulation of spine density and morphology.
All previous work in our laboratory focused on cypin function in dendrites; however, cypin is also present in the axon [2], and histochemical assays for GDA activity in rat brain reveals the presence of cypin in the mossy fibers in the CA3 region of hippocampus [14], consistent with our data in mouse brain (Patel and Firestein, unpublished data). For the first time, we report the effects of cypin and cypinS overexpression on neuronal electrophysiology. Overexpression of either cypin or cypinS increases the frequency of mEPSCs with no effect on amplitude. This lack of effect on amplitude despite decreased spine density mediated by cypin overexpression may reflect homeostatic compensatory activities. Although cypinS alters mEPSC frequency without having an effect on spine density, this discrepancy may reflect a cell autonomous function for cypinS. However, since we used lentivirus to transduce neurons to overexpress cypinS, our transduction efficiency was 100%. Thus, we cannot rule out whether changes in mEPSC frequency include a cell non-autonomous component. Moreover, overexpression of cypinS results in a significantly greater increase in frequency of mEPSCs than does overexpression of cypin. The higher frequency of mEPSCs is caused by increased release of neurotransmitters [37]. We previously reported that cypin binds to snapin, a SNARE-associated protein involved in modulating neurotransmitter release by regulating vesicle docking and fusion [38], via the CRMP homology domain of cypin [5]. Thus, the increases in mEPSC frequency observed when cypin or cypinS is overexpressed may be due, in part, to binding of snapin. Furthermore, release of vesicular pools of neurotransmitter is zinc-dependent (i.e., [39,40,41,42,43]). As cypin binds to zinc and cypinS retains all zinc binding sites, cypin and cypinS may act as zinc sensors to regulate the vesicular pool and neuronal electrophysiology. Thus, cypin and cypinS may play roles in zinc-dependent pathways.
Taken together, we not only have identified a novel cypin isoform but we have also uncovered new presynaptic roles for both isoforms. As summarized in Fig. 8, our data suggest that the two isoforms of cypin play similar but distinct roles in neuronal development. Cypin is a guanine deaminase, but cypinS does not have this enzymatic activity. Both isoforms regulate dendrite branching; however, cypin regulates proximal dendrite branching while cypinS regulates distal dendrite branching. Cypin regulates spine number while overexpression of cypinS has no effect on spines. Both isoforms regulate presynaptic release of neurotransmitters, but the overexpression of cypinS promotes a greater release than does overexpression of cypin. Although both isoforms of cypin are predicted to bind snapin, the lack of the 74 amino terminal amino acids in cypinS may contribute to different effects on snapin-mediated synaptic vesicle release. For example, cypin may have additional binding partners and regulators at its amino terminus than does cypinS, resulting in distinct regulation of function or three-dimensional folding, conferring unique functions to each cypin isoform.
Summary of the specific effects of overexpression of cypin or cypinS on dendrites, spines, and electrophysiology. Cypin is guanine deaminase while cypinS does not have this enzymatic activity. Cypin overexpression increases proximal dendrite branching, and overexpression of cypinS increases distal dendrite branching. Cypin overexpression decreases spine density, while overexpression of cypinS has no effect on spine density. Overexpression of cypin or cypinS increases mEPSC frequency, with cypinS overexpression showing significantly greater mEPSC frequency than that observed with cypin overexpression
Experimental Procedures
Ethical Approval
All studies using animals were approved by the Rutgers Institutional Animal Care and Use Committee.
Western Blot and Quantification
Mouse (C57BL/6J; no. 000664; Jackson Laboratory) brain tissue was homogenized in 0.5 ml TEE (20 mM Tris-HCl, pH 7.4, 1 mM EDTA, 1 mM EGTA, pH 7.4) for each 100 mg wet weight of tissue and incubated with an equal volume of 2× RIPA buffer (100 mM Tris-HCl, pH 7.4, 300 mM NaCl, 1% deoxycholate, 2% NP-40, 0.2% SDS, 2 mM EDTA, pH 7.4) containing 1 mM phenylmethylsulfonyl fluoride (PMSF) and protease inhibitor cocktail and rocked at 4 °C for 1 h on a Nutator. Insoluble material was pelleted at 12,000×g at 4 °C for 15 min. All protein extracts were evaluated for protein concentration using the Pierce BCA protein assay kit (Thermo Scientific) following the manufacturer’s protocol. Proteins (12–15 μg) were resolved on a 10% SDS-polyacrylamide gel and transferred to polyvinylidene difluoride (PVDF) membrane. The membrane was blocked with 5% bovine serum albumin in TBST (20 mM Tris pH 7.5, 150 mM NaCl, 0.1% Tween-20) for 1 h. The blots were probed with the indicated antibodies in TBST containing 3% BSA: polyclonal rabbit anti-cypin (1:500; BF6) [8, 12], monoclonal mouse anti-PSD-95 (1:1000; K28/43, UC Davis/NIH NeuroMab Facility), polyclonal chicken anti-GFP (1:1000, A10262, Invitrogen (Thermo Fisher Sci.)), and monoclonal mouse anti-GAPDH (1:1000; MAB374, Millipore). Experiments were repeated at least three times. Blots were scanned, and intensities of bands were quantified using the ImageJ software from NIH. The number of pixels of absolute intensity of bands was normalized to the intensity of those for GAPDH (internal controls). Human cortical tissue was provided by the Human Brain and Spinal Fluid Resource Center (Los Angeles, CA).
Analysis of Cypin and cypinS mRNA
mRNA was isolated from different mouse brain tissues using the Trizol reagent (Invitrogen) following the manufacturer’s protocol. A reaction of 2 μg of RNA in a total volume of 20 μl was converted to cDNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) following the manufacturer’s protocol. cDNA (10 μl) was used to perform real-time PCR using SYBR Green Real-Time master mix following the manufacturer’s protocol. The following primer pairs (F for forward, R for reverse) were used for real-time PCR reactions: mouse GAPDH: (F) AGGTCGGTGTGAACGGATTTG, (R) TGTAGACCATGTAGTTGAGGTCA; mouse cypin (Ensembl: ENSMUST00000087600): (F) GTCTTCCGAGGGACTTTCGTC, (R) ATGGTTTGAAGCACCACTCCT; mouse cypinS (Ensembl: ENSMUST00000121725): (F) AGAGCAAGCCCATTGCTGTG (R) ATGGTTTGAAGCACCACTCCT. ABI 7900 HT sequence detection system (Applied Biosystems) was used to record the Ct values. SDS 2.4, RQ Manager 1.2.1 and DataAssist V3.01 (Applied Biosystems) software packages were used to generate RQ (relative quantitation or fold change) and p values using two-tailed Student’s t test comparing the 2(−ΔCt) values of the two groups. The p value was adjusted using Benjamini-Hochberg False Discovery Rate. Four biological replicates were performed, with each of them having three technical replicates. GAPDH served as the control for levels of total mRNA analyzed, and mRNAs levels from brain from mice at postnatal day 100 served as the reference group.
Guanine Deaminase Activity Assay
COS-7 cells were grown at a density of 15,000 cells/cm2 on glass coverslips. Cells were transfected with cDNA encoding GFP, GFP-Cypin, or GFP-CypinS using Lipofectamine 2000 (Invitrogen) following the manufacturer’s protocol. Forty-eight hours after transfection, cells were washed with an ice-cold phosphate-buffered saline (PBS), and images were taken using a fluorescence microscope. The protocol for the guanine deaminase assay was modified from Paletzki [14], as we have previously reported [3]. Cells were fixed for 1 h in 3% glutaraldehyde in 0.05 M sodium cacodylate buffer, pH 7.8. Endogenous xanthine was removed by incubating the coverslips in 2.5 U/ml xanthine oxidase (Sigma) in bicine buffer for 15–30 min at 37 °C followed by two 5 min rinses in bicine buffer. Cells were incubated in substrate solution (8 mM guanine, 0.625 U/ml xanthine oxidase, 0.9% 4-nitro blue tetrazolium chloride (NBT; Sigma) in 0.1 M bicine pH 7.8) for 1–3 h at 37 °C. The reaction was stopped by washing the cells in distilled water twice for 2 min, and coverslips were mounted onto slides. Control reactions were performed without guanine.
Amplex Red Assay
HEK293 cells were plated at one million cells/well of six well plate on DIV0 and transfected with indicated plasmids using Lipofectamine 2000 (Invitrogen) on DIV1. Cells were lysed in GDA lysis buffer (150 mM NaCl, 25 mM Tris-HCl, pH 7.4, and 1 mM PMSF) on DIV3 to prepare lysate. Fifty micrograms of lysate were subjected to guanine deaminase assay using GDA lysis buffer containing 0.025 U/ml xanthine oxidase (Sigma), 0.002 U/ml peroxidase (Sigma), 50 μM Amplex Red reagent (Molecular Probes), and 16 mM guanine (Sigma). Samples were centrifuged at 9500×g for 1 min at to remove insoluble guanine. A negative control without guanine were also used. All samples were incubated at 37 °C, and absorbance was recorded at 571 nm using a single beam Genesys 10 UV/Vis Spectrophotometer (Spectronic, UK) at indicated time intervals [4, 44].
Hippocampal Neuronal Culture and Transfection
Hippocampal neuronal cultures were prepared from rat embryos at 18 days gestation as previously described [45]. Neurons were cultured in Neurobasal medium (Life Technologies) supplemented with B27, l-glutamine, penicillin, and streptomycin. Neurons were plated on to glass coverslips coated with poly-d-lysine (0.1 mg/ml, Sigma) in a 37 °C incubator with 5% CO2. Neurons were plated at a density of 105,000 cells/cm2 for dendrite branching experiments and at a density of 50,000 cells/cm2 for spine analysis. Neurons were transfected with pEGFP-C1, pEGFP-C1-Cypin, or pEGFP-C1-CypinS using Lipofectamine LTX+PLUS (Invitrogen) following the manufacturer’s protocol for dendrite branching and using calcium phosphate transfection for spine analysis [45]. For all conditions, neurons were co-transfected with pGW1-mRFP to visualize dendrites and axon. For dendrite branching experiments, neurons were transfected at 7 days in vitro (DIV7) and fixed with 4% paraformaldehyde (PFA) in PBS at DIV12. For spine analysis, neurons were transfected on DIV14 and fixed on DIV17. Cells were immunostained with chicken anti-GFP (1:250; Thermo Fisher Sci.), rabbit anti-RFP (1:250; Rockland), and mouse anti-MAP2 (1:250; BD Pharmingen). Immunostaining was visualized with Cy2-, Cy3-, or Cy5-conjugated secondary antibodies (1:500; Thermo Fisher Sci.).
Dendrite Branching and Sholl Analysis
Imaging was performed using an EVOS-FL fluorescence imaging system (Thermo Fisher Sci.). All images were analyzed using the ImageJ software (NIH) and MATLAB (MathWorks). Sholl analysis was performed using the Bonfire program developed by the Firestein laboratory [15,16,17]. In brief, concentric circles were drawn around the soma every 6 μm and intersections with dendrite branches were counted. All data were exported from MATLAB to Excel for quantification. Statistical differences (p < 0.5) were determined by two-way or one-way ANOVA followed by Tukey’s multiple comparisons test using the GraphPad Prism6 software. The experimenter was blinded to conditions during all Sholl and data analyses.
Synaptosomal Fractionation
One hemisphere from an adult or P14 male mouse brain was lysed in 1.2 ml sucrose buffer (10 mM HEPES, pH 7.5, 1.5 mM MgCl2, 320 mM sucrose, 5 mM EDTA, 5 mM dithiothreitol (DTT), 0.1 mM PMSF, 10 μM MG132, PMSF, and 1× protease inhibitors). The lysate was centrifuged for 10 min at 1000×g to remove nuclei and cellular debris. The supernatant was centrifuged for 15 min at 10,000×g to pellet crude synaptosomes and a supernatant containing soluble proteins. The pellet was resuspended in sucrose buffer containing 1% Triton X-100 for 1 h at 4 °C followed by centrifugation for 30 min at 40,000×g using a TLA 100.3 fixed angle rotor in a Beckman Coulter Optima LTX centrifuge to isolate a pellet containing synaptic membranes and supernatant containing synaptic cytosol [46].
Immunoprecipitation
Mouse whole brain lysate was prepared in 1× RIPA buffer with PMSF and protease inhibitor cocktail at 4 °C as mentioned previously. Protein (2 mg) was used to immunoprecipitate PSD-95 with 5 μg anti-ms-PSD-95 (K28/43, NeuroMab) and 50 μl Protein G magnetic beads (GE Healthcare). Five micrograms of normal mouse IgG were used for control immunoprecipitation. Precipitated protein was eluted from antibody-bead complexes using 2× SDS-PAGE sample buffer (125 mM Tris, pH 6.8, 20% glycerol, 4% sodium dodecyl sulfate, 0.01% bromophenol blue, 5% β-mercaptoethanol) and subjected to Western blot analysis.
Dendritic Spine Analysis
Images of dendritic segments were taken with a 60× Plan Apo oil-immersion objective (NA 1.4) using a Yokogawa CSU-10 spinning disk confocal head attached to an inverted fluorescence microscope (Olympus IX50). X-Y and Z-resolution were set as 0.067–0.067 and 0.1 μm, respectively, to define dendritic spines. All images were analyzed using the ImageJ software (NIH). The number of protrusions was counted from two randomly chosen secondary or tertiary dendrites per neuron. Spines along dendritic segments were counted at distances 20 to 80 μm from the soma. Spines were manually counted from at least 14 neurons for each experimental condition. The following criteria were used in excel to classify spines: mushroom spines have total length < 5 μm and head width/neck width > 1.5; thin spines have total length < 5 μm and head width/neck width > 1 but < 1.5; stubby spines have total length < 1 μm and head width/neck width < 1; filopodia have total length > 1 and head width/neck width < 1. Mushroom and thin/stubby are mature and immature spines, respectively. Analysis was performed with the experimenter blinded to the condition.
Lentivirus Production
cDNA encoding cypin or cypinS was cloned into the pHUG lentiviral vector (gift from Dr. Christoph Proschel) at the BsrG1 site using the Infusion HD Cloning kit (Takara Bio USA, Inc.) by following the manufacturer’s protocol. In brief, on day 1, HEK293 cells were plated at the density of 6.5 × 106 cells/T75 flask. On day 2, cells were transfected by the calcium phosphate method [45] with lentiviral plasmids for expression of GFP, GFP-cypin, or GFP-cypinS and PAX2 and VSV packaging vectors. On day 5, supernatant was collected and centrifuged at 1500×g for 5 min to remove dead cells and debris. The supernatant was transferred into a new tube containing 5× PEG-it virus precipitation solution (System Biosciences), thus diluting 1:4, followed by incubation at 4 °C for 2 days to concentrate the virus. On day 7, the mixture was centrifuged at 1500×g for 30 min at 4 °C to pellet the virus. The supernatant was carefully removed, and the pellet was resuspended in 150–200 μl sterile PBS and aliquoted into 10 μl fractions stored at − 80 °C [47].
Electrophysiology
All electrophysiological recordings were performed as we previously described [48]. In brief, hippocampal neurons were transduced on DIV 14 with lentivirus encoding GFP, GFP-cypin, or GFP-cypinS, and recording was performed on DIV 20-21. Whole cell patch-clamp recordings were made on the soma of hippocampal neurons. For recordings, cells were bathed in artificial cerebrospinal fluid (ACSF) containing (in mM) the following: 140 NaCl, 5 KCl, 2 CaCl2, 2 MgCl2, 10 HEPES, and 10 glucose (pH 7.4 adjusted with NaOH; 290–310 mOsmol). Recording electrodes (3–5 MΩ) contained a K-based internal solution composed of (in mM): 126 K-gluconate, 4 KCl, 10 HEPES, 4 ATP-Mg, 0.3 GTP-Na2, 10 phosphocreatine, and 10 QX-314 bromide (pH 7.2; 280–300 mOsmol). In order to record miniature excitatory postsynaptic currents (mEPSCs), action potentials were blocked with 1 μM tetrodotoxin (Tocris, R&D Systems) and GABA inhibitory currents with 50 μM picrotoxin (Tocris, R&D Systems). The membrane potential was held at − 70 mV throughout all experiments. Data were amplified and filtered at 2 kHz by a patch-clamp amplifier (MultiClamp 700B), digitalized (Digidata1440A), stored, and analyzed by pCLAMP (Molecular Devices, Union City, CA). Data were discarded when the input resistance changed > 20% during recording.
References
Kuwahara H, Araki N, Makino K, Masuko N, Honda S, Kaibuchi K, Fukunaga K, Miyamoto E et al (1999) A novel NE-dlg/SAP102-associated protein, p51-nedasin, related to the amidohydrolase superfamily, interferes with the association between NE-dlg/SAP102 and N-methyl-d-aspartate receptor. J Biol Chem 274(45):32204–32214. https://doi.org/10.1074/jbc.274.45.32204
Firestein BL, Brenman JE, Aoki C, Sanchez-Perez AM, El-Husseini AE, Bredt DS (1999) Cypin: a cytosolic regulator of PSD-95 postsynaptic targeting. Neuron 24(3):659–672. https://doi.org/10.1016/S0896-6273(00)81120-4
Akum BF, Chen M, Gunderson SI, Riefler GM, Scerri-Hansen MM, Firestein BL (2004) Cypin regulates dendrite patterning in hippocampal neurons by promoting microtubule assembly. Nat Neurosci 7(2):145–152. https://doi.org/10.1038/nn1179
Fernandez JR, Welsh WJ, Firestein BL (2008) Structural characterization of the zinc binding domain in cytosolic PSD-95 interactor (cypin): role of zinc binding in guanine deamination and dendrite branching. Proteins 70(3):873–881. https://doi.org/10.1002/prot.21683
Chen M, Lucas KG, Akum BF, Balasingam G, Stawicki TM, Provost JM, Riefler GM, Jornsten RJ et al (2005) A novel role for snapin in dendrite patterning: interaction with cypin. Mol Biol Cell 16(11):5103–5114. https://doi.org/10.1091/mbc.E05-02-0165
Charych EI, Akum BF, Goldberg JS, Jornsten RJ, Rongo C, Zheng JQ, Firestein BL (2006) Activity-independent regulation of dendrite patterning by postsynaptic density protein PSD-95. J Neurosci 26(40):10164–10176. https://doi.org/10.1523/JNEUROSCI.2379-06.2006
Chen H, Firestein BL (2007) RhoA regulates dendrite branching in hippocampal neurons by decreasing cypin protein levels. J Neurosci 27(31):8378–8386. https://doi.org/10.1523/JNEUROSCI.0872-07.2007
Kwon M, Fernandez JR, Zegarek GF, Lo SB, Firestein BL (2011) BDNF-promoted increases in proximal dendrites occur via CREB-dependent transcriptional regulation of cypin. J Neurosci 31(26):9735–9745. https://doi.org/10.1523/JNEUROSCI.6785-10.2011
Wang X, Pal R, Chen XW, Kumar KN, Kim OJ, Michaelis EK (2007) Genome-wide transcriptome profiling of region-specific vulnerability to oxidative stress in the hippocampus. Genomics 90(2):201–212. https://doi.org/10.1016/j.ygeno.2007.03.007
Fuller HR, Hurtado ML, Wishart TM, Gates MA (2014) The rat striatum responds to nigro-striatal degeneration via the increased expression of proteins associated with growth and regeneration of neuronal circuitry. Proteome Sci 12(1):20. https://doi.org/10.1186/1477-5956-12-20
Hedlund E, Karlsson M, Osborn T, Ludwig W, Isacson O (2010) Global gene expression profiling of somatic motor neuron populations with different vulnerability identify molecules and pathways of degeneration and protection. Brain 133(Pt 8):2313–2330. https://doi.org/10.1093/brain/awq167
Tseng CY, Firestein BL (2011) The role of PSD-95 and cypin in morphological changes in dendrites following sublethal NMDA exposure. J Neurosci 31(43):15468–15480. https://doi.org/10.1523/JNEUROSCI.2442-11.2011
Braunschweig D, Krakowiak P, Duncanson P, Boyce R, Hansen RL, Ashwood P, Hertz-Picciotto I, Pessah IN et al (2013) Autism-specific maternal autoantibodies recognize critical proteins in developing brain. Transl Psychiatry 3(7):e277. https://doi.org/10.1038/tp.2013.50
Paletzki RF (2002) Cloning and characterization of guanine deaminase from mouse and rat brain. Neuroscience 109(1):15–26. https://doi.org/10.1016/S0306-4522(01)00352-9
Kutzing MK, Langhammer CG, Luo V, Lakdawala H, Firestein BL (2010) Automated Sholl analysis of digitized neuronal morphology at multiple scales. J Vis Exp 45(45). https://doi.org/10.3791/2354
Langhammer CG, Previtera ML, Sweet ES, Sran SS, Chen M, Firestein BL (2010) Automated Sholl analysis of digitized neuronal morphology at multiple scales: whole cell Sholl analysis versus Sholl analysis of arbor subregions. Cytometry A 77(12):1160–1168. https://doi.org/10.1002/cyto.a.20954
Sweet ES, Langhammer CL, Kutzing MK, Firestein BL (2013) Semiautomated analysis of dendrite morphology in cell culture. Methods Mol Biol 1018:261–268. https://doi.org/10.1007/978-1-62703-444-9_24
Gu Y, Ihara Y (2000) Evidence that collapsin response mediator protein-2 is involved in the dynamics of microtubules. J Biol Chem 275(24):17917–17920. https://doi.org/10.1074/jbc.C000179200
Fukata Y, Itoh TJ, Kimura T, Menager C, Nishimura T, Shiromizu T, Watanabe H, Inagaki N et al (2002) CRMP-2 binds to tubulin heterodimers to promote microtubule assembly. Nat Cell Biol 4(8):583–591. https://doi.org/10.1038/ncb825
Mural RJ, Adams MD, Myers EW, Smith HO, Miklos GLG, Wides R, Halpern A, Li PW et al (2002) A comparison of whole-genome shotgun-derived mouse chromosome 16 and the human genome. Science 296(5573):1661–1671. https://doi.org/10.1126/science.1069193
Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, Scott G, Steffen D et al (2004) Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428(6982):493–521. https://doi.org/10.1038/nature02426
Futcher B, Latter GI, Monardo P, McLaughlin CS, Garrels JI (1999) A sampling of the yeast proteome. Mol Cell Biol 19(11):7357–7368. https://doi.org/10.1128/MCB.19.11.7357
Greenbaum D, Colangelo C, Williams K, Gerstein M (2003) Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol 4(9):117. https://doi.org/10.1186/gb-2003-4-9-117
Gygi SP, Rochon Y, Franza BR, Aebersold R (1999) Correlation between protein and mRNA abundance in yeast. Mol Cell Biol 19(3):1720–1730. https://doi.org/10.1128/MCB.19.3.1720
Barrett LW, Fletcher S, Wilton SD (2012) Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. Cell Mol Life Sci 69(21):3613–3634. https://doi.org/10.1007/s00018-012-0990-9
Barreau C, Paillard L, Osborne HB (2005) AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res 33(22):7138–7150. https://doi.org/10.1093/nar/gki1012
Chen CY, Shyu AB (1995) AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci 20(11):465–470. https://doi.org/10.1016/S0968-0004(00)89102-1
Gruber AR, Fallmann J, Kratochvill F, Kovarik P, Hofacker IL (2011) AREsite: a database for the comprehensive investigation of AU-rich elements. Nucleic Acids Res 39(Database issue):D66–D69. https://doi.org/10.1093/nar/gkq990
Perrin L, Roudeau S, Carmona A, Domart F, Petersen JD, Bohic S, Yang Y, Cloetens P et al (2017) Zinc and copper effects on stability of tubulin and actin networks in dendrites and spines of hippocampal neurons. ACS Chem Neurosci 8(7):1490–1499. https://doi.org/10.1021/acschemneuro.6b00452
Dvergsten CL, Johnson LA, Sandstead HH (1984) Alterations in the postnatal development of the cerebellar cortex due to zinc deficiency. III. Impaired dendritic differentiation of basket and stellate cells. Brain Res 318(1):21–26
Shi L, Lin S, Grinberg Y, Beck Y, Grozinger CM, Robinson GE, Lee T (2007) Roles of Drosophila Kruppel-homolog 1 in neuronal morphogenesis. Dev Neurobiol 67(12):1614–1626. https://doi.org/10.1002/dneu.20537
Clark SG, Chiu C (2003) C. elegans ZAG-1, a Zn-finger-homeodomain protein, regulates axonal development and neuronal differentiation. Development 130(16):3781–3794. https://doi.org/10.1242/dev.00571
Li W, Wang F, Menut L, Gao FB (2004) BTB/POZ-zinc finger protein abrupt suppresses dendritic branching in a neuronal subtype-specific and dosage-dependent manner. Neuron 43(6):823–834. https://doi.org/10.1016/j.neuron.2004.08.040
Kuo TY, Chen CY, Hsueh YP (2010) Bcl11A/CTIP1 mediates the effect of the glutamate receptor on axon branching and dendrite outgrowth. J Neurochem 114(5):1381–1392. https://doi.org/10.1111/j.1471-4159.2010.06852.x
Koyama Y, Hattori T, Nishida T, Hori O, Tohyama M (2015) Alterations in dendrite and spine morphology of cortical pyramidal neurons in DISC1-binding zinc finger protein (DBZ) knockout mice. Front Neuroanat 9:52. https://doi.org/10.3389/fnana.2015.00052
Prakash A, Bharti K, Majeed ABA (2015) Zinc: indications in brain disorders. Fundam Clin Pharmacol 29(2):131–149. https://doi.org/10.1111/fcp.12110
Turrigiano GG, Leslie KR, Desai NS, Rutherford LC, Nelson SB (1998) Activity-dependent scaling of quantal amplitude in neocortical neurons. Nature 391(6670):892–896. https://doi.org/10.1038/36103
Ilardi JM, Mochida S, Sheng ZH (1999) Snapin: a SNARE-associated protein implicated in synaptic transmission. Nat Neurosci 2(2):119–124. https://doi.org/10.1038/5673
Arons MH, Lee K, Thynne CJ, Kim SA, Schob C, Kindler S, Montgomery JM, Garner CC (2016) Shank3 is part of a zinc-sensitive signaling system that regulates excitatory synaptic strength. J Neurosci : Off J Soc Neurosci 36(35):9124–9134. https://doi.org/10.1523/JNEUROSCI.0116-16.2016
Westbrook GL, Mayer ML (1987) Micromolar concentrations of Zn2+ antagonize NMDA and GABA responses of hippocampal neurons. Nature 328(6131):640–643. https://doi.org/10.1038/328640a0
Mayer ML, Vyklicky L Jr (1989) The action of zinc on synaptic transmission and neuronal excitability in cultures of mouse hippocampus. J Physiol 415(1):351–365. https://doi.org/10.1113/jphysiol.1989.sp017725
Lu YM, Taverna FA, Tu R, Ackerley CA, Wang YT, Roder J (2000) Endogenous Zn(2+) is required for the induction of long-term potentiation at rat hippocampal mossy fiber-CA3 synapses. Synapse 38(2):187–197. https://doi.org/10.1002/1098-2396(200011)38:2<187::AID-SYN10>3.0.CO;2-R
Li Y, Hough CJ, Suh SW, Sarvey JM, Frederickson CJ (2001) Rapid translocation of Zn(2+) from presynaptic terminals into postsynaptic hippocampal neurons after physiological stimulation. J Neurophysiol 86(5):2597–2604. https://doi.org/10.1152/jn.2001.86.5.2597
Fernandez JR, Sweet ES, Welsh WJ, Firestein BL (2010) Identification of small molecule compounds with higher binding affinity to guanine deaminase (cypin) than guanine. Bioorg Med Chem 18(18):6748–6755. https://doi.org/10.1016/j.bmc.2010.07.054
Kwon M, Firestein BL (2013) DNA transfection: Calcium phosphate method. Methods Mol Biol 1018:107–110. https://doi.org/10.1007/978-1-62703-444-9_10
Chao HW, Tsai LY, Lu YL, Lin PY, Huang WH, Chou HJ, Lu WH, Lin HC et al (2013) Deletion of CPEB3 enhances hippocampus-dependent memory via increasing expressions of PSD95 and NMDA receptors. J Neurosci 33(43):17008–17022. https://doi.org/10.1523/JNEUROSCI.3043-13.2013
Mendenhall A, Lesnik J, Mukherjee C, Antes T, Sengupta R (2012) Packaging HIV- or FIV-based lentivector expression constructs and transduction of VSV-G pseudotyped viral particles. J Vis Exp 62(62):e3171. https://doi.org/10.3791/3171
Hernandez K, Swiatkowski P, Patel MV, Liang C, Dudzinski NR, Brzustowicz LM, Firestein BL (2016) Overexpression of isoforms of nitric oxide synthase 1 adaptor protein, encoded by a risk gene for schizophrenia, alters actin dynamics and synaptic function. Front Cell Neurosci 10:6. https://doi.org/10.3389/fncel.2016.00006
Funding
This research was supported by National Science Foundation grant IOS-1353724 to BLF. MVP was supported in part by a Predoctoral Fellowship from the New Jersey Commission on Brain Injury Research #CBIR15FEL009. PS was supported in part by National Institutes of Health Biotechnology Training Grant T32 GM008339-20 and a Predoctoral Fellowship from the New Jersey Commission on Brain Injury Research #CBIR16FEL013. ARR was supported by the National Institutes of Health Biotechnology Training Grant T32 GM008339-20. KC was supported in part by a Rutgers University Aresty Research Fellowship.
Author information
Authors and Affiliations
Contributions
MVP and BLF designed experiments and wrote the manuscript with input from PS. MVP performed biochemistry and morphology experiments and analyzed resulting data. MK identified cypinS in mouse and performed preliminary experiments. PS performed electrophysiology experiments and analyzed all resulting data. KC worked with MVP to perform cloning of the plasmids and the biochemistry experiments to validate BF6 antibody against cypinS. ARR provided cultured neurons for morphology experiments and performed imaging of dendrite branching experiments. BLF supervised the project.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflicts of interest.
Electronic Supplementary Material
Supplementary Fig. 1
(DOCX 156kb).
Rights and permissions
About this article
Cite this article
Patel, M.V., Swiatkowski, P., Kwon, M. et al. A Novel Short Isoform of Cytosolic PSD-95 Interactor (Cypin) Regulates Neuronal Development. Mol Neurobiol 55, 6269–6281 (2018). https://doi.org/10.1007/s12035-017-0849-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-017-0849-z