Abstract
Serotonin (5-HT) is a neurotransmitter that regulates fundamental aspects of brain development, physiology and behaviour. The serotonin transporter (5-HTT) is deputized to the reuptake of 5-HT from the intersynaptic space in the presynaptic neurons. 5-HTT governs duration and magnitude of 5-HT biological actions, acting as a master regulator of the fine-tuning of 5-HT signalling. Genetic variation at SLC6A4 gene locus, encoding 5-HTT, contributes to alteration in 5-HT reuptake. The 5-HTTLPR/rs25531/rs25532 polymorphisms located in the promoter region of SLC6A4 gene have been associated with stress-related psychopathology and functional brain phenotypes. Besides, further DNA variations in functional regulative elements located at 5′ and 3′ termini of the SLC6A4 gene influence transcriptional and post-transcriptional steps. Recently, epigenetic processes including SLC6A4 promoter methylation and transcript silencing by microRNA were shown to be involved in the aetiology of affective disorders. Furthermore, gene-environment interactions such as early life stress often encompass epigenetic changes, which can stably mark the genome in response to environmental stimuli potentially altering gene expression across lifespan. Therefore, it seems well established that functional variations in the SLC6A4 gene expression can no longer be ascribed to the modulating 5-HTTLPR promoter polymorphism but need to be integrated with the contribution arising from other interactive elements and epigenetic mechanisms. In this review, we discuss genetic and epigenetic layers of regulation affecting SLC6A4 gene expression. An overview of human and cellular studies investigating the impact of these regulatory processes on SLC6A4 gene expression is provided.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Serotonin (5-hydroxytryptamine, 5-HT) is a neurotransmitter synthesized in the serotonergic neurons in the raphe nuclei of the brainstem and hypothalamus of the central nervous system (CNS) [1, 2], in the enterochromaffin cells (ECs) of the gut [3], in serotonergic neurons of the myenteric plexus and in lymphocytes [4]. Through portal circulation, gut-derived 5-HT (approximately 90% of the total 5-HT produced in the body) enters liver and lungs and a fraction gets actively transported to blood platelets, the major site of storage of 5-HT [5].
5-HT is involved in the regulation of fundamental aspects of brain development, physiology and behaviour including mood, emotions, sleep, food intake, platelet coagulation and gastrointestinal function. Serotonergic neurons descending down the spinal cord control muscle activity, and the effects of serotonin in the periphery are perceived in the cardiovascular system, with additional effects in the respiratory system [6]. The functions of serotonin in energy homeostasis range from anoretic effect in the brain to regulation of adipose tissue activity in the periphery [7]. In the CNS, 5-HT is among the many neurotransmitters that contribute to regulate the hypothalamic-pituitary-adrenal axis stress response [8] and has a deep effect on psychological well-being and mood disorders such as depression, schizophrenia and anxiety [9–11].
In the neuroimmune network, through which information is relayed between CNS and immune system by various routes, 5-HT functions as an immunoregulator, modulating immune responses such as T- and B-cell proliferation, inflammatory responses and autoimmune responses [12].
After release from serotonergic neurons, 5-HT binds to post-synaptic 5-HT receptor subtypes to mediate signal transduction. Furthermore, release of 5-HT into synaptic cleft is controlled by presynaptic autoreceptors. The 5-HT transporter (5-HTT) on presynaptic neurons regulates 5-HT neurotransmission by selective reuptake of 5-HT from the synapse, ensuring its recycling into new vesicles [13, 14]. Thus, the 5-HTT in brain and in many peripheral tissues is responsible for the active transport of serotonin into neurons, EC cells, platelets and other cells. The 5-HTT regulates 5-HT level in lymphoid tissues to ensure its proper functioning in innate and adaptive response.
It plays a crucial role in the 5-HT homeostasis through regulation of duration, magnitude and spatial distribution of signals reaching 5-HT receptors, thus acting as a master regulator of the fine-tuning of 5-HT signalling. Dysfunction in this signal pathway has been implicated in a host of neurological diseases [6, 15] and in irritable bowel syndrome (IBS) [16]. Accordingly, 5-HTT is the main target of the class of antidepressant drugs known as selective serotonin reuptake inhibitors (SSRIs) [17].
A large number of studies have been conducted to determine whether genetic variation at solute carrier family 6 (neurotransmitter transporter), member 4 (SLC6A4) gene locus, encoding 5-HTT, contributes to variation in 5-HT reuptake and, thus, to mood and behavioural disease traits. To a large extent, a crucial role for non-coding variants in altering 5-HTT messenger RNA (mRNA) levels has been demonstrated. In particular, a common polymorphic variant located in the promoter region of the SLC6A4 gene, the 5-HTT gene-linked polymorphic region (5-HTTLPR) (rs4795541), has been widely studied and associated to complex neuropsychiatric conditions and traits [15]. An overview of common and uncommon 5-HTTLPR allelic variants that details the genetic architecture and arrangement of repeat elements for the known 5-HTTLPR alleles was recently published [18].
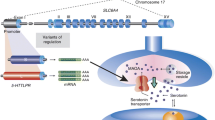
The SLC6A4 gene expression shows greater variation than that expected by the mere influence of the 5-HTTLPR. Notably, at the SLC6A4 locus, DNA variations in functional regulative elements located at the 5′ end of the gene, such as the variable number of tandem repeats (VNTR) in intron 2 and polymorphisms in the 3′-untranslated regions (UTRs), influence transcriptional and post-transcriptional steps. Furthermore, additional layers of regulation might play an even more important regulatory role in SLC6A4 gene expression (Fig. 1). There is actually growing knowledge that variability in the functional features of the human genome at the level above the DNA sequence likely contributes to individual differences in brain function. Among non-sequence-based sources of variability in the genome, epigenetic modifications may play a major role through regulation of molecular machinery involved in the spatio-temporal modulation of gene expression. Epigenetic programming represents a long- and short-term dynamic process counterposed to the static model of DNA variations.
Schematic representation of multiple levels of control acting on transcriptional and post-transcriptional machinery involved in the regulation of the human SLC6A4 gene expression. The 5-HTTLPR depicted here belongs to the L allele (16 repeats) of the SLC6A4 gene promoter. 5-HTTLPR and two SNPs, rs25531 and rs25532, are functional untranslated elements of the promoter repetitive region giving rise to several genetic variants. Individual elements of the 5-HTTLPR are designated by Greek letters according to Nakamura et al.’s nomenclature [19]. Other genetic variants include an intron 2 polymorphism denoted as STin2 VNTR as well as multiple polyadenylation site signals in combination with SNPs in the 3′-UTR. Alternative splicing involving exons 1A, 1B and 1C in specific tissues [20, 21] may also contribute to the regulation of SLC6A4 expression. Overall, hierarchical epigenetic control occurs through the action of miRNAs, methylation and acetylation/deacetylation mechanisms. HDAC histone deacetylase, DNMT DNA methyltransferase, HMT histone methyltransferase
The SLC6A4 locus is of particular interest in the context of epigenetic modifications. Recent research progress on the epigenetic regulation of SLC6A4 has revealed an important role of DNA methylation, long non-coding RNA (lncRNA) and small-non coding RNA such as microRNAs (miRNAs) in various brain disorders [22–24]. Several studies also suggest that both gene × gene (G × G) and gene × environment (G × E) interactions (e.g. the interaction of genes involved in the metabolism and catabolism of 5-HT with neuroreceptor genes or with environmental factors) need to be considered for explaining the effects of SLC6A4 genetic variants on brain circuitry in health and disease [25–27]. Accordingly, G × E interactions, such as those occurring when exposed to early life stress (ELS), encompass epigenetic changes that collectively can stably mark the genome in response to environment, potentially altering gene expression across lifespan and across generations [28–34].
The aim of this study is to take into account genetic and epigenetic layers of regulation affecting the SLC6A4 gene expression. We will provide an overview of human and cellular studies investigating the impact of these regulatory processes on SLC6A4 gene expression that may contribute to the interindividual variability in brain function and that may confer individual differences in psychopathologies susceptibility or resilience. In particular, the review will focus on the serotonin transporter intron 2 (STin2) polymorphism, the 3′-untranslated region (3′-UTR) and the epigenetic processes. While SLC6A4 DNA methylation is the best-understood epigenetic modification, researches devoted to miRNAs and serotonergic transmission are all very recent, so the present article will focus on findings of the past few years.
Method
Identification of Relevant Studies for Inclusion
We searched the PubMed, the National Library of Medicine journal literature search system, for biomedical articles from MEDLINE and life science journals using MeSH and PubMed search tools up to May 2016.
The following keywords were used for the search: “serotonin transporter OR 5HTT OR SLC6A4 OR serotonin promoter transporter AND epigenetic AND miRNA OR microRNA OR miR AND methylation AND non coding RNA OR long non coding RNA OR lnc RNA AND histone AND 3’UTR AND epigenetic process AND STin2 polymorphism”.
Articles were also identified using the “related articles” function in PubMed. Furthermore, we found additional papers by performing a manual search of the reference lists of relevant retrieved articles.
Genetic Mechanisms
VNTR Intron STin2
The STin2 polymorphism is a VNTR containing nine (STin2.9), ten (STin2.10) or twelve (STin2.12) copies of a 16- or 17-bp repeat [35]. Sequences of the repeats are showed in Table 1, and structures of the VNTR are reported in Table 2. STin2 alleles affecting SLC6A4 gene expression are summarized in Table 3. In transgenic mice, alleles STin2.10 or STin2.12 function as transcriptional regulatory elements in specific areas of the developing CNS, particularly in the midbrain, hindbrain and neural tube floor plate [38]. These alleles were also found to differ in the strength of their enhancer-like abilities within the developing rostral hindbrain, an area associated with the native SLC6A4 mRNA expression and the maturation of rostral serotoninergic cell clusters at the stage of embryonic development [38]. These findings are corroborated by in vitro data showing that STin2.10 and STin2.12 supported differential levels of reporter gene expression when transformed into embryonic stem (ES) cells [39]. Therefore, the VNTR polymorphism within the intron 2 of SLC6A4 acts as a transcriptional regulator, a characteristic potentially correlated with a susceptibility to affective disorders. Individual repeat elements within the intronic VNTR domain were later shown to differ in their enhancer activity in the ES cell model (c/d and f/d elements supported increased enhancer activity), indicating that not only the number of repeats (STin2.9 showed extremely high level of enhancer activity relative to STin2. 10 and STin2.12 alleles in the JAR cell line) but also the primary DNA sequence of repeat units could affect the transcription of the gene [37].
Several works have demonstrated differential effects of the STin2 and 5-HTTLPR on the expression of reporter genes in clonal cell lines both under normal and addictive (i.e. exposure to cocaine) growth conditions [44–46]. These authors also reported that the transcription CCCTC-binding factor (CTCF) binds and differentially regulates STin2 in vitro [44, 45]. In rat primary cortical cultures, the 5-HTTLPR and STin2 VNTRs could support differential gene expression based on copy number of both VNTRs, when analysed in concert using constructs designed to mimic their endogenous positions in the gene. Hence, the 5-HTTLPR and STin2 VNTRs are likely to be on the same signalling pathway regulated by CTCF [47]. These data on combinatorial potential of VNTRs are consistent with analysis of SLC6A4 expression in lymphoblastoid cell lines, which found evidence of concerted action of the STin2 VNTRs and 5-HTTLPR [40]. Other cellular and human studies investigated functional impact of the STin2 polymorphism in unaffected German and Swedish sample population [41, 48]. No significant effect of the 17-bp VNTR genotype on maximum rate (Vmax) of 5-HT uptake in platelets of 50 male subjects was observed [41]. The result was replicated in cerebrospinal fluid in healthy volunteers where the level of 5-hydroxyindoleacetic acid, the major 5-HT metabolite, was found to be invariant [48]. However, individual STin2.12/ STin2.12 homozygous appeared to have lower uptake affinity than individual STin2.10/STin2.9 heterozygous [41]. These authors reported the same result for the combined analysis of STin2.12 and 5-HTTLPR where no association between the different genotypes of the 5-HTTLPR polymorphism and STin2.12 and the maximum rate of 5-HT uptake into platelets was observed [41].
Hranilovic et al. analysed the effect of STin2 VNTR polymorphism on SLC6A4 mRNA levels in native-expressing cells from schizophrenic patients [40]. Allelic influence of SLC6A4 intron 2 polymorphism was very similar to that of the promoter polymorphism reported by Lesch et al. [49], with low-expressing allele 10 apparently acting as dominant. Test for linear trend showed statistically significant dose effect of the “low-expressing” genotypes on the SLC6A4 mRNA levels, suggesting potential combined influence of the two polymorphic regions on SLC6A4 gene expression [40]. By contrast, Lim and coworkers observed only a weak association for STin2 VNTR in combination with promoter VNTR without statistical significance [50].
In some reports, STin2 polymorphisms and its combined effect with 5-HTTLPR variants has been associated with bipolar affective disorder [51] and lithium response [52] or cognitive dysfunction in major depressive disorder (MDD) [42]. A meta-analysis based on all association studies between schizophrenia and the 5-HTTLPR and STin2 polymorphisms published before April 2004 suggested that the STin2.12 allele is likely a risk factor for schizophrenia susceptibility [53]. Thus, a comprehensive set of markers that fully characterize the linkage disequilibrium relationships at the SLC6A4 gene locus will need to be tested in large well-characterized clinical samples in order to understand the relevance of the SLC6A4 gene polymorphisms in schizophrenia susceptibility [53]. Any changes in the linkage disequilibrium (LD) at this gene locus (Fig. 2) may be associated to different pathologies linked to serotonin transporter. Recent studies have also explored the association between STin2 polymorphism and tobacco use disorder (TUD) or nicotine dependence susceptibility [36, 43, 54]. High expression of SLC6A4, probably due to the presence of the allele STin2.9, led to low 5-HT concentration in the central nervous system and may confer more susceptibility to nicotine dependence [36]. Pizzo de Castro et al. found a remarkable association between the STin2 genetic polymorphism, mood disorders and TUD. The significant comorbidity between mood disorders and TUD, which may be related to both genetic and environmental factors, suggests that biological endophenotypes, i.e. disorders in 5-HT metabolism, may in part underpin the pathophysiology of mood disorders and STin2-related TUD [43]. A recent study showed that comorbid mood disorders and TUD were both associated with specific biomarkers related to oxidative stress (i.e. glutathione S-transferases and paraoxonase 1) and 5-HT pathways [55].
r 2 linkage disequilibrium (LD) coefficient at the SLC6A4 gene locus in healthy Caucasians of the 1000 Genomes Project available at http://www.ensembl.org/
3′-Untranslated Region
SLC6A4 3′-UTR variants play an important role in messenger RNA (mRNA) translation and stability. Mutations in the 3′-UTR of the SLC6A4 mRNA can thus alter the termination site, the polyadenylation (polyA) site signals, the ratio of multiple polyA sites usage and the secondary structure of the 3′ terminus of the mRNA, underlining the multiple ways by which 3′-UTR polymorphisms may cause a deregulated translational control and thereby a disease [56, 57]. The 3′-UTR of the SLC6A4 mRNA contains multiple functional polyadenylation site signals, located at 567 and 690 bp downstream of the stop codon, actually resulting in two mRNA forms that differ by the presence or absence of a 123-bp element [58, 59]. 3′-UTR variants regulating SLC6A4 gene expression are summarized in Table 4. A common SNP T/G (rs3813034) is located in the distal polyA site signal [58] as well as other polymorphic variations in the 3′-UTR (rs34500314, rs13306796, rs1042173 and rs11080121) were reported [61, 62]. Allelic variation at the rs3813034 site did not influence polyadenylation site usage [58]. Recently, Gyawali et al. determined that the rs3813034 alleles lead to different usage of the distal polyadenylation site signal [56]. This is consistent with in vitro polyadenylation studies showing that a T in the position of rs3813034 within the canonical polyA signal (AAUAAA) leads to more efficient polyadenylation than a G [60]. The functional polymorphism rs3813034 hence affects the balance of SLC6A4 polyadenylation forms, suggesting that the distal sequence stabilizes SLC6A4 mRNA either directly through effects on the secondary structure of the messenger or through interactions with RNA-binding proteins or miRNAs [56]. On the other hand, functional studies of the rs1042173 found conflicting effects of this polymorphism on SLC6A4 gene expression [50, 61, 63].
Epigenetic Mechanisms
Epigenetic processes take place through DNA methylation, modifications in the methylation or acetylation status of chromatin-associated histones and gene regulation by non-coding RNAs. DNA methylation and histone modifications encompass conformational changes in DNA and/or chromatin that do not alter the underlying nucleotide sequence, but regulate the molecular machinery involved in the spatio-temporal modulation of gene expression. Non-coding regulatory RNA is emerging as the major architect of neural cell differentiation and nervous system development as well in fine-tuning neuronal plasticity [23]. Unlike the basic DNA sequence, epigenetic marks are often part of the mechanism that drives cell and tissue differentiation. The SLC6A4 gene locus is of particular interest in the context of epigenetic modifications. Epigenetic SLC6A4 gene regulation occurs mainly through direct DNA methylation at CpG islands or presupposes modifications in the acetylation or methylation status of chromatin-associated histones. A post-transcriptional regulation by miRNAs was also recently reported in the literature that has emerged to play important roles in the control of serotonergic transmission.
DNA Methylation
In most cases, methylation at gene promoters leads to silencing of the gene itself. Indeed, methylation occurring within CpG-rich regions near the transcription start-site of a gene tends to have a repressive effect on transcription initiation, and thus correlates with reduced gene expression [64]. Studies investigating effects of methylation on the SLC6A4 gene expression are summarized in Table 5.
Effects of Environment on SLC6A4 Gene DNA Methylation
Epigenetic marks are partially heritable [83] even though they can change in response to environmental stimuli. G × E studies have begun to reveal relationship between SLC6A4 gene DNA methylation, 5-HTTLPR, stressful life events and influence on gene expression as well on susceptibility for psychopathology [26, 27, 74, 84–89]. The impact of early adversity on the susceptibility to psychiatric disorders in later life is influenced by environmental factors (nature of stressors, time of exposure in development and severity and cumulative exposure effects) as well as by biological factors (gender, age, predisposing genetic polymorphisms in genes associated with mood regulation and stress response). Accordingly, several studies in human and non-human primates showed that methylation patterns within and near the SLC6A4 gene differ as a function of early or recent life stress or trauma. The SLC6A4 promoter region susceptible of methylation is illustrated in Fig. 3.
Localization of CpG island and AluJB element in the SLC6A4 transcriptional control region on chromosome 17q11. a Schematic picture of the SLC6A4 transcriptional control region with the functional 5-HTTLPR and the embedded rs25531/rs25532 polymorphisms, the AluJb element and the CpG island overlapping the first exon (exon 1A). The exon lA and the alternative exon 1B placed approximately 14-kb downstream [20, 90] are also shown. b The sequence of AluJb and position of eight CpG sites (boxed and in bold) according to Dannlowski et al. [78] are shown. The sequence of CpG island is displayed with CpG sites (boxed), the putative TATA box (in bold) and exon 1A (shadowed in grey). The CpG 1–9 sites are numbered according to the study of Domschke et al. [76] and comprise the 1–5 CpGs analysed in the study by Alasaari et al. [71], the 1–9 CpGs analysed by Devlin et al. [91] and all CpGs (1–5 and 7–8) analysed by Kang et al. and Kim et al., respectively [75, 92]. Modified from Fig. 1 Domschke et al. [76] with permission of Oxford University Press
In a non-human primate model, Kinnally et al. reported an association between animal’s stress response and SLC6A4 methylation in LL individuals who had experienced an ELS. Therefore, greater DNA methylation conferred a genomic background of “risk” in the context of early stress [65]. Wang and coworkers clearly showed a tight correlation between peripheral SLC6A4 promoter DNA methylation states and brain 5-HT synthesis in healthy adult males with different levels of ELS. Significantly higher level of methylation was measured at specific CpG sites in both T cells and monocytes isolated from higher-level childhood physical aggression [66].
Effects of 5-HTTLPR on SLC6A4 DNA Promoter Methylation
In vitro studies using 49 human lymphoblasts cell lines reported that methylation status of a 799-bp CpG island surrounding untranslated exon 1A (Fig. 3) negatively affects mRNA transcription. The magnitude of this effect was dependent on the 5-HTTLPR genotype (increased methylation in S carriers), so that the S allele was associated with lower amount of mRNA transcription [67]. In a further study, which used a much more precise method of quantitating methylation and four times the number of cells, the genotypic effect was not found, suggesting that effect is small or that the previous findings were a false positive [68]. Nevertheless, this study nicely demonstrated that greater amount of the CpG island methylation was associated with decreased mRNA transcription. Into human placental choriocarcinoma JAR cells, methylation of SLC6A4 promoter suppressed reporter transcriptional activity, supporting a functional role of DNA methylation in SLC6A4 promoter regulation [66]. Both complete and partial (as little as 10%) methylation of SLC6A4 promoter resulted in reduced reporter gene expression in the same recipient cell line [69]. An experimental Rhesus macaques model was used by Kinnally and coworkers to investigate epigenetic regulation of SLC6A4 gene. Carriers of the Rhesus macaques low-expressing 5-HTTLPR alleles (S allele) exhibited higher mean SLC6A4 CpG methylation, which was associated with lower peripheral blood mononuclear cells (PBMCs) SLC6A4 expression [70].
Interaction Between Environment and 5-HTTLPR: Effects on SLC6A4 DNA Promoter Methylation
Alasaari and coworkers reported that DNA methylation levels in the promoter region of SLC6A4 varied between high and low work stress environments among female nurses. Blood leucocytes from individuals working in a high work stress environment showed 21–65% lower levels of methylation compared to individuals in a low work stress environment. Decreased methylation may lead to increased transcriptional activity of the gene, increased reuptake of 5-HT from synaptic clefts and termination of the activity of 5-HT. Authors hypothesized that hypomethylation could represent an adaptation mechanism for stress [71]. A pilot investigation of a peripheral cell marker of epigenetic risk for depression also showed elevated buccal cell SLC6A4 methylation in individuals who carried 5-HTTLPR S allele [69]. Even though no association between depressive symptoms and SLC6A4 methylation was revealed, depressive symptoms were more common among individuals with S allele. These results are in agreement with a study showing higher SLC6A4 methylation in blood cells of S allele carriers, but only in the case of less unresolved trauma [72]. In summary, 5-HTTLPR genotype appears to further differentiate SLC6A4 methylation as some studies reported increased methylation in S carriers [67, 68, 70–72, 74]. This suggests that methylation of S allele may exacerbate the impact of ELS [93], although some reported the reverse pattern in relation to unresolved trauma [72]. On the other hand, recent findings showing the S allele and prenatal/early adversity associated with decreased peripheral SLC6A4 mRNA levels in an additive manner further indicated that these effects appeared to be largely independent of methylation profiles within the SLC6A4 promoter-associated CpG island [73]. A very recent study combined all of the putative elements of a molecular G × E interaction, i.e. SLC6A4 promoter methylation, 5-HTTLPR genotype, stressful life events and cortisol response [74]. In vivo blood-based evaluation of SLC6A4 or glucocorticoid receptor NR3C1 mRNA expression at baseline as a function of either 5-HTTLPR genotype or life stress was performed. In contrast to Wankerl et al.’s study, [73], neither significant differential SLC6A4 gene expression nor additive effect between these two variables were demonstrated [74]. On the one hand, this study showed increased mRNA level in the LL allele carriers whereas the S group individual’s expression level remain unchanged indicating a dynamic “5-HTTLPR-based” regulation of SLC6A4 expression following exposure to stressor [74]. Overall, both early and recent life stress correlated positively with site-specific SLC6A4 methylation as a function of 5-HTTLPR genotype, as higher methylation level was measured in S group but not in LL participants. Considering that the 5-HTTLPR S allele negatively moderates the effect of life events on depression [85], these findings are in line with previous studies that associated higher SLC6A4 methylation with depression [68, 75, 94]. However, studies by Wankerl et al. [73] and by Duman and Canli [74] have one limitation since they enrolled homogeneous sample of healthy Caucasian males and may not generalize to other ethnic group or women or individuals with psychopathology.
Pharmacoepigenetic Studies
Two pharmacoepigenetic studies investigated the impact of DNA methylation patterns in the SLC6A4 promoter region in mediating antidepressant treatment response in an Asian population sample [75] and Caucasian patients [76] with MDD. Higher SLC6A4 promoter methylation status significantly associated with a range of childhood adversity and correlated with family history of depression and higher perceived stress, but it did not predict antidepressant treatment outcome [75]. In this respect, the analysis of Domschke et al. [76] was somewhat in contrast to Kang et al.’s study [75] showing that DNA hypomethylation of the SLC6A4 transcriptional control region might impair antidepressant treatment response possibly via increased 5-HTT expression and consequently decreased 5-HT availability [76]. In a very recent study, Iga et al. examined the association of SLC6A4 gene promoter methylation and 5-HTTLPR genotype before antidepressant treatment and expression before and after treatment in a sample of Japanese patients with MDD [77]. SLC6A4 mRNA expression was significantly higher in unmedicated MDD patients compared with healthy controls and was significantly decreased after 8 weeks of antidepressant treatment. The mean methylation level was significantly higher in patients compared with controls, consistent with previous studies examining the same methylation sites [68]. They also found no association between the mean SLC6A4 expression level and the 5-HTTLPR genotype in patients or controls. Notably, decreased methylation levels at two CpG sites (CpG3 and CpG5, see Fig. 3) were related to increased depressive symptoms.
Imaging Epigenetics
Studies combining neuroimaging and epigenetics (i.e. imaging epigenetics) have provided novel insights into the contributions of DNA sequence variation to individual differences in brain function, behaviour and risk for psychopathology [88]. Regarding SLC6A4 gene, in particular, a brain imaging study indicates that SLC6A4 methylation status in the T cells and monocytes is associated with in vivo measures of 5-HT synthesis in the orbitofrontal cortex [66]. This study strongly suggests that peripheral SLC6A4 DNA methylation could be a marker of central 5-HT function and the feasibility of using methylation at specific CpG sites of SLC6A4 as non-invasive biomarkers of 5-HT synthesis and behaviours associated with altered 5-HT function such as aggression. Higher methylation of particular CpG sites was associated with lower in vivo measures of brain 5-HT in the orbitofrontal cortex [66]. Increased SLC6A4 promoter methylation was associated with greater threat-related amygdala reactivity, possibly reflecting decreased SLC6A4 gene expression and, consequently, reduced regional 5-HT reuptake. In addition, methylation of CpG sites that most strongly associated with amygdala activity was negatively correlated with SLC6A4 mRNA concentrations in post-mortem amygdala tissue [79], consistent with recent works demonstrating a substantial correlation between the blood and brain methylomes [66, 95]. Very recent findings suggest that relative methylation status of the SLC6A4 proximal promoter region is a reliable predictor of amygdala reactivity across time. Using prospective longitudinal epigenetic, neuroimaging and behavioural data from adolescents, Swartzl and coworkers demonstrated that changes in gene methylation associated with lower socioeconomic status (SES) to predict changes in risk-related brain function. Specifically, lower SES predicted change in SLC6A4 proximal promoter methylation, which in turn predicted change in threat-related reactivity of the centromedial amygdala [81]. Frodl and coworkers reported an interactive effect between the SLC6A4 promoter polymorphism and childhood adversity on brain structure specifically in patients with depression, suggesting diagnosis-specific epigenetic regulatory effects [96]. They hypothesized that higher peripheral SLC6A4 DNA methylation was associated with lower brain reactivity. Further investigations explored whether effects of methylation on SLC6A4 gene affect brain function to a larger extent in patients than controls, based on the fact that methylation of SLC6A4 might be more pronounced in patients with MDD [32]. In a study involving 25 patients with MDD, SLC6A4 methylation was associated with differential modulation of activity in the brain insula, operculum, hippocampus and amygdala during emotional attention processing [80]. This study provided further validation for particular SLC6A4 DNA methylation states as peripheral markers of brain functional states [80]. A following study in depressed patients reported that greater DNA methylation in specific CpG sites at the SLC6A4 promoter in peripheral cells was independently associated with childhood trauma, depression, and smaller hippocampal volume. The relatively strong association between peripheral methylation of SLC6A4 regulatory region and hippocampal volumes thus suggested that SLC6A4 methylation might be an underlying physiological mechanism of how gene and environment interact to affect hippocampal development [82]. Recently, an Alu element of subtype AluJb was identified in the promoter region between 5-HTTLPR and the CpG island, which was shown to be relevant for methylation [78]. The AluJb element belongs to the family of short interspersed nuclear elements (SINE) and contains several CpG sites (Fig. 3). Lower AluJb methylation was associated with lower hippocampal grey matter volumes. Such lower Alu methylation might influence SLC6A4 expression via different mechanisms that might involve exonization (alternative exons provisioning) [97] or transcription factor binding such as Paired box protein Pax-6 (PAX6), which is of particular importance in brain and CNS development [98].
Histone Modification
Histone acetylation/deacetylation is an essential epigenetic mechanism that controls chromatin structure, DNA accessibility to transcription factors and modulation of gene expression [99, 100]. Potential of the 5-HTTLPR genotype to modulate epigenetic remodelling, such as regulation of chromatin structure and DNA-binding activity of transcription factors, was assessed in serotonergic JAR cell line [46]. CTCF bound to both L and S alleles under normal growth conditions, whereas after exposure to cocaine, CTCF bound only to the L allele. The lack of CTCF binding to the S allele correlated to methyl-binding protein, MeCP2 binding, which in turn recruited the histone deacetylase (HDAC) suggesting that MeCP2 acts as a negative transcriptional regulator. Association of the MeCP2 and the corepressor HDAC to the S allele may block the regulatory effect of the S allele on the expression of the SLC6A4 gene. Additionally to the binding of CTCF and MeCP2, effects of cocaine were associated with increased association of positive H3K4m2 histone mark and the SLC6A4 proximal promoter region encompassing the 5′-HTTLPR. This suggests that cocaine stimulus may determine an epigenetic remodelling, thus enhancing transcriptional rate, which in turn increase of RNA II polymerase binding to the promoter region. Histone modifications were also detected within the SLC6A4 gene after cocaine exposure. In particular, the H3K36m3 histone marker showed increased association with the SLC6A4 exon2/intron2 region. However, acetylation status of H3 at lysine 9 was increased, reflecting a positive effect of cocaine on histone modifications. The authors hypothesized that H3K36 methylation and subsequent involvement in deacetylation activity may be indicative of repression of spurious intragenic transcription in the SLC6A4 exon2/intron2 region or that the presence of heterozygous STin2 alleles in JAR cells could lead to different histone modifications after cocaine treatment [46]. A model representing the working hypothesis for these epigenetic modifications over the SLC6A4 proximal promoter and intron 2 under basal conditions and following cocaine treatment was presented [46]. In line with these findings, human intestinal Caco-2 cell line showed reduced SLC6A4 expression following treatment with HDAC inhibitors [101]. Caco-2 cells represent an excellent in vitro model system to study SLC6A4 regulation as they display anatomic and functional similarities to ileal enterocytes and also show a serotonergic phenotype. Interestingly, HDAC inhibition by butyrate or trichostatin A (TSA) decreased the SLC6A4 gene promoter 1 activity (hSLC6A4 p1, upstream of exon 1A). The decrease in the 5-HTT expression by HDAC inhibition was also recapitulated in an in vivo model. Similar to the in vitro model, in an in vivo model of mice, pectin feeding (which results in high colonic butyrate levels by anaerobic fermentation) decreased SLC6A4 mRNA expression in the ileum and distal colon. These data indicate a novel mechanism of down-regulation of intestinal SLC6A4 by epigenetic mechanisms involving HDAC2 inhibition and increased association of histone H3 or H4 acetylation with human SLC6A4 promoter 1 [101]. A schematic representation of this mechanism is shown in Fig. 4.
Schematic representation of effects of histone modification on SLC6A4 gene expression. a In human intestinal Caco-2 cell line and in a mouse model, HDAC inhibitors butyrate and TSA decrease SLC6A4 expression by increasing association of acetylated histone H3 or H4 with hSLC6A4 p1 (upstream of exon 1A), resulting in decrease in promoter activity. Specific inhibition of HDAC2 mimicked the effects of butyrate or TSA in decreasing SLC6A4 expression [101]. b Effects of early stress and age on methylated histone 3 (H3K4me3) binding levels to DNA extracted from hippocampi of Rhesus macaques. Peer-reared monkeys exhibited lower levels of binding when compared to their mother-reared counterparts. Among 5-HTTLPR short allele carriers, cerebrospinal fluid 5-hydroxyindoleacetic acid (5-HIAA) levels were lower in stress-exposed subjects [102]
One commonly studied epigenetic mark is the histone 3 protein that is trimethylated at lysine 4 (modified histone, H3K4me3). This is a histone that marks “active” promoters and would therefore be expected to facilitate or promote gene expression. Lindell et al. examined the effects of early stress on H3K4me3 binding at the SLC6A4 promoter and the interactions of stress with 5-HTTLPR genotype in tissue derived from stress-sensitive brain regions (hippocampus) of Rhesus macaques that had been reared in the presence or absence of stress [102]. They found a decline in H3K4me3 from preadolescence to post-adolescence and lower levels in peer-reared monkeys when compared to their mother-reared counterparts, as well effects of genotype on H3K4me3 binding. These findings are depicted in Fig. 4.
RNA-Level Regulation: miRNAs
miRNAs are small non-coding RNAs, particularly abundant in the nervous system, that play pivotal roles as post-transcriptional regulators in neurogenesis, synapse development and plasticity in the brain. They also operate a spatio-temporal translational control of target mRNA during dendritic morphogenesis, regulating the expression of hundreds of genes that govern critical aspects of neuroplasticity and synapse function [24]. Recent works demonstrated that miRNAs participate to post-transcriptional mechanisms that regulate SLC6A4 mRNA translation, as illustrated in Fig. 5. Studies investigating effects of miRNAs regulation on SLC6A4 gene expression are described below and are summarized in Table 6. In silico computational target prediction and in vitro luciferase reporter assay identified miR-16 as miRNA with complementarity to the 3′-UTR of the SLC6A4 mRNA [104]. Consistent with a putative role of miR-16 as a negative regulator of SLC6A4 translation, Baudry et al. demonstrated that the miR-16 regulated the expression of SLC6A4 gene in the murine 1C11 neuroectodermal cell line, which can differentiate into either serotonergic or noradrenergic neuronal cells. 1C11 cells expressed a low level of miR-16, which increased along the noradrenergic pathway, whereas the level did not change along the serotonergic program. Similarly, lower basal levels of miR-16 were found in mouse serotonergic raphe nuclei vs noradrenergic locus coeruleus at physiological conditions. In mice, chronic SSRI fluoxetine treatment augments the level of miR-16 in serotonergic raphe nuclei by antagonizing Wnt signalling, as inferred from increase in glycogen synthase kinase-3β (GSK3β) and thereby negatively regulates SLC6A4 gene expression. This increase in miR-16 levels was accompanied by a decrease in levels of pri/pre-miRNA16, suggesting an influence on maturation rather than transcription. When infused into the locus coeruleus, fluoxetine failed to induce any change in miR-16 expression, which is in agreement with the lack of SLC6A4 expression in noradrenergic neurons under basal conditions. In contrast, raphe responds to fluoxetine treatment by releasing the neurotrophic protein S100β, which in turn acts on the noradrenergic neurons of the locus coeruleus, lowering miR-16 levels accompanied by the induction of 5-HTT protein expressions. Hence, the locus coeruleus responds to fluoxetine injection in raphe by switching on serotonergic functions. Thus, up-regulation of miR-16 plays a role in silencing SLC6A4 transcripts during noradrenergic differentiation and miR-16 may contribute to the therapeutic action of SSRI antidepressants in monoaminergic neurons [104]. The observation that chronic administration of the SSRI fluoxetine induces miR-16, which translationally represses maturation of mRNA encoding 5-HTT, is an effect probably related to the delayed onset of its antidepressant properties. Recently, Moya and coworkers showed that miR-15 as well as miR-16 regulated SLC6A4 expression both in human and rat tissues [103]. This post-transcriptional control was characterized in reporter assays in human JAR and rat RN46A cell lines, which endogenously express these two miRNA species. In addition, using lentiviral particles to obtain sustained miRNA overexpression, endogenous 5-HTT protein levels were effectively reduced in JAR cells 4 days post-transduction [103]. Interestingly, reporter constructs made with two SLC6A4 3′-UTR molecular haplotype (i.e. the SNPs rs1042173 and rs3813034) resulted in no significant differences with regard to miR-15a or miR-16 translational repression. This result confirmed a previous report showing that the reduced expression of the reporter mRNA carrying SLC6A4 3′-UTR was not affected by polymorphism rs1042173, which lies in the putative binding site of miR-545, another negative regulator of SLC6A4 translation [107]. The SNP rs11080121, a minor allele of rs1042173, in the 3′ untranslated region of SLC6A4 seems to disrupt a conserved binding site for miR-590-3p. When Montasser and colleagues tested the association between hot flashes (HFs) in both European-American and African-American premenopausal and perimenopausal women and genetic variants in SLC6A4 gene, they found that disruption of the miR-590-3p binding site leads to higher expression of SLC6A4. Consequent depletion of 5-HT in synaptic clefts triggers the presynaptic autoreceptor feedback mechanism to produce more 5-HT, which is protective against HFs [62]. Other emerging evidence on this layer of SLC6A4 gene post-transcriptional control arise from a study that elucidated the role of a specific miRNA in regulating the central 5-HT system activity, under “baseline” and challenged conditions [106]. In vitro luciferase assays and mutation studies revealed a strong repressive effect for miR-135 on both SLC6A4 and 5-hydroxytryptamine receptor-1A (HTR1A) gene transcripts. A series of experiments in which the miR-135 levels were functionally manipulated in vivo to assess the effects on animal behaviour was performed. In humans, a significant decrease in miR-135a levels in the blood and brain of depressed patients, compared to match controls, was observed. Authors then proposed miR-135 as an essential regulatory element responsible for maintaining intact serotonergic tone under normal conditions and essential for the brain response to antidepressants. According to this theory, increased levels of miR-135 would repress an array of 5-HT system-related transcripts, including SLC6A4 and presynaptic HTR1A levels, causing an increase in 5-HT in the synaptic cleft, which is associated with decreases in depressive symptoms [106].
Very recently, Xiu-Jun Liao and colleagues demonstrated that microRNA-24 inhibits serotonin reuptake transporter expression and aggravates irritable bowel syndrome, indicating that miR-24 plays a role in the pathogenesis of IBS via regulation of 5-HTT expression [105].
Conclusions and Perspectives
It seems currently well established that variations in the levels of 5-HTT may not strictly represent different transcriptional activity of the SLC6A4 gene arising from the 5-HTTLPR L/S genotype. Cellular and tissue contexts may result in differences in SLC6A4 gene expression and consequently in distinct availability of 5-HTT protein. Careful consideration of epigenetic mechanisms will be required to account for previously unexplained variability in brain function than sequence-based variation alone. DNA methylation patterns within the SLC6A4 gene promoter as well as the SLC6A4 mRNA translational repression by miRNAs highlight different levels of regulation. Furthermore, the newly emerged family of the lncRNAs may provide an additional layer of regulation for fine-tuning of miRNA function through affecting epigenetic processes, particularly in the brain [23].
Epigenetic modifications can change in response to environmental stimuli too. Epigenetic modifications can moderate the impact of the individual’s genotype on biological processes such as brain function. Therefore, environmentally induced methylation appears to affect sensitivity to stress and reactivity to life stressors [26], with effect on the individual differences in psychopathology susceptibility or resilience [74, 82].
Recent works have revealed considerable correlation across peripheral and neural cells for both gene expression and DNA methylation patterns, as well for miRNA levels [66, 71, 80, 95, 106]. Consequently, results obtained from peripheral blood cells can be translated to neural tissue districts.
Genome-wide association studies currently evidence that changes in gene expression, rather than protein-coding sequence variations, largely contribute to the risk for complex genetic traits. In particular, with regard to psychiatric disorders, genetic variations in non-coding regulatory regions may result in differential transcriptional responses to developmental signals and environmental/psychosocial stressors [108]. A deeper appreciation of the hierarchical interaction among the multiple actors of gene expression, such as regulatory elements, transcription factors and epigenetic machinery will be beneficial to better understand SLC6A4 gene regulation levels that support neurological processes.
References
Hornung JP (2003) The human raphe nuclei and the serotonergic system. J Chem Neuroanat 26(4):331–343
Hensler JG (2012) Basic neurochemistry: principles of molecular, cellular and medical neurobiology, eighth edition
Gill RK, Pant N, Saksena S, Singla A, Nazir TM, Vohwinkel L, Turner JR, Goldstein J et al (2008) Function, expression, and characterization of the serotonin transporter in the native human intestine. Am J Phys Gastrointest Liver Physiol 294(1):G254–G262. doi:10.1152/ajpgi.00354.2007
Faraj BA, Olkowski ZL, Jackson RT (1994) Expression of a high-affinity serotonin transporter in human lymphocytes. Int J Immunopharmacol 16(7):561–567
Anderson GM, Feibel FC, Cohen DJ (1987) Determination of serotonin in whole blood, platelet-rich plasma, platelet-poor plasma and plasma ultrafiltrate. Life Sci 40(11):1063–1070
Murphy DL, Lerner A, Rudnick G, Lesch KP (2004) Serotonin transporter: gene, genetic disorders, and pharmacogenetics. Mol Interv 4(2):109–123. doi:10.1124/mi.4.2.8
Namkung J, Kim H, Park S (2015) Peripheral serotonin: a new player in systemic energy homeostasis. Mol Cells 38(12):1023–1028. doi:10.14348/molcells.2015.0258
Herman JP, Prewitt CM, Cullinan WE (1996) Neuronal circuit regulation of the hypothalamo-pituitary-adrenocortical stress axis. Crit Rev Neurobiol 10(3–4):371–394
Lesch K-P (2010) The role of serotonin transporter in modeling psychiatric disorders: focus on depression, emotion regulation, and the social brain. Experimental Models in Serotonin Transporter Research. Cambridge University Press. doi:10.1017/CBO9780511729935.012
Lin SH, Lee LT, Yang YK (2014) Serotonin and mental disorders: a concise review on molecular neuroimaging evidence. Clin Psychopharmacol Neurosci 12(3):196–202. doi:10.9758/cpn.2014.12.3.196
Murphy DL, Moya PR, Fox MA, Rubenstein LM, Wendland JR, Timpano KR (2013) Anxiety and affective disorder comorbidity related to serotonin and other neurotransmitter systems: obsessive-compulsive disorder as an example of overlapping clinical and genetic heterogeneity. Philos Trans Royal Soc London Series B, Biol Sci 368(1615):20120435. doi:10.1098/rstb.2012.0435
Jaiswal P, Mohanakumar KP, Rajamma U (2015) Serotonin mediated immunoregulation and neural functions: complicity in the aetiology of autism spectrum disorders. Neurosci Biobehav Rev 55:413–431. doi:10.1016/j.neubiorev.2015.05.013
Rudnick G, Clark J (1993) From synapse to vesicle: the reuptake and storage of biogenic amine neurotransmitters. Biochim Biophys Acta 1144(3):249–263
Daws LC, Toney GM (2007) High-speed chronoamperometry to study kinetics and mechanisms for serotonin clearance in vivo. In: Michael AC, Borland LM (eds) Electrochemical methods for neuroscience. CRC Press, Boca Raton (FL)
Gelernter J (2014) SLC6A4 polymorphism, population genetics, and psychiatric traits. Hum Genet 133(4):459–461. doi:10.1007/s00439-013-1412-2
Colucci R, Blandizzi C, Bellini M, Ghisu N, Tonini M, Del Tacca M (2008) The genetics of the serotonin transporter and irritable bowel syndrome. Trends Mol Med 14(7):295–304. doi:10.1016/j.molmed.2008.05.001
Owens MJ, Morgan WN, Plott SJ, Nemeroff CB (1997) Neurotransmitter receptor and transporter binding profile of antidepressants and their metabolites. J Pharmacol Exp Ther 283(3):1305–1322
Iurescia S, Seripa D, Rinaldi M (2016) Role of the 5-HTTLPR and SNP promoter polymorphisms on serotonin transporter gene expression: a closer look at genetic architecture and in vitro functional studies of common and uncommon allelic variants. Mol Neurobiol. doi:10.1007/s12035-015-9409-6
Nakamura M, Ueno S, Sano A, Tanabe H (2000) The human serotonin transporter gene linked polymorphism (5-HTTLPR) shows ten novel allelic variants. Mol Psychiatry 5(1):32–38
Bradley CC, Blakely RD (1997) Alternative splicing of the human serotonin transporter gene. J Neurochem 69(4):1356–1367
Ozsarac N, Santha E, Hoffman BJ (2002) Alternative non-coding exons support serotonin transporter mRNA expression in the brain and gut. J Neurochem 82(2):336–344
Sugawara H, Bundo M, Ishigooka J, Iwamoto K, Kato T (2013) Epigenetic regulation of serotonin transporter in psychiatric disorders. J Genet Genomics 40(7):325–329. doi:10.1016/j.jgg.2012.10.002
Barry G (2014) Integrating the roles of long and small non-coding RNA in brain function and disease. Mol Psychiatry 19(4):410–416. doi:10.1038/mp.2013.196
Dreyer JL (2010) New insights into the roles of microRNAs in drug addiction and neuroplasticity. Genome Med 2(12):92. doi:10.1186/gm213
Canli T, Lesch KP (2007) Long story short: the serotonin transporter in emotion regulation and social cognition. Nat Neurosci 10(9):1103–1109. doi:10.1038/nn1964
Caspi A, Hariri AR, Holmes A, Uher R, Moffitt TE (2010) Genetic sensitivity to the environment: the case of the serotonin transporter gene and its implications for studying complex diseases and traits. Am J Psychiatry 167(5):509–527. doi:10.1176/appi.ajp.2010.09101452
Munafo MR, Durrant C, Lewis G, Flint J (2009) Gene X environment interactions at the serotonin transporter locus. Biol Psychiatry 65(3):211–219. doi:10.1016/j.biopsych.2008.06.009
Franklin TB, Russig H, Weiss IC, Graff J, Linder N, Michalon A, Vizi S, Mansuy IM (2010) Epigenetic transmission of the impact of early stress across generations. Biol Psychiatry 68(5):408–415. doi:10.1016/j.biopsych.2010.05.036
McGowan PO, Sasaki A, D’Alessio AC, Dymov S, Labonte B, Szyf M, Turecki G, Meaney MJ (2009) Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci 12(3):342–348. doi:10.1038/nn.2270
Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M et al (2004) Epigenetic programming by maternal behavior. Nat Neurosci 7(8):847–854. doi:10.1038/nn1276
Booij L, Tremblay RE, Szyf M, Benkelfat C (2015) Genetic and early environmental influences on the serotonin system: consequences for brain development and risk for psychopathology. J Psychiatr Neurosci: JPN 40(1):5–18
Booij L, Wang D, Levesque ML, Tremblay RE, Szyf M (2013) Looking beyond the DNA sequence: the relevance of DNA methylation processes for the stress-diathesis model of depression. Philos Trans R Soc Lond Ser B Biol Sci 368(1615):20120251. doi:10.1098/rstb.2012.0251
Lupien SJ, McEwen BS, Gunnar MR, Heim C (2009) Effects of stress throughout the lifespan on the brain, behaviour and cognition. Nat Rev Neurosci 10(6):434–445. doi:10.1038/nrn2639
Sun H, Kennedy PJ, Nestler EJ (2013) Epigenetics of the depressed brain: role of histone acetylation and methylation. Neuropsychopharmacol: Off Publ Am Coll Neuropsychopharmacol 38(1):124–137. doi:10.1038/npp.2012.73
Battersby S, Ogilvie AD, Smith CA, Blackwood DH, Muir WJ, Quinn JP, Fink G, Goodwin GM et al (1996) Structure of a variable number tandem repeat of the serotonin transporter gene and association with affective disorder. Psychiatr Genet 6(4):177–181
de Lima KW, Guembarovski RL, Oda JM, Ramos G, Oliveira BV, Cavalli IJ, de Souza Fonseca Ribeiro EM, Goncalves MS et al (2012) Association between the STin2 VNTR polymorphism and smoking behavior in oral cancer patients and healthy individuals. Clin Exp Med 12(1):13–19. doi:10.1007/s10238-011-0140-y
Lovejoy EA, Scott AC, Fiskerstrand CE, Bubb VJ, Quinn JP (2003) The serotonin transporter intronic VNTR enhancer correlated with a predisposition to affective disorders has distinct regulatory elements within the domain based on the primary DNA sequence of the repeat unit. Eur J Neurosci 17(2):417–420
MacKenzie A, Quinn J (1999) A serotonin transporter gene intron 2 polymorphic region, correlated with affective disorders, has allele-dependent differential enhancer-like properties in the mouse embryo. Proc Natl Acad Sci U S A 96(26):15251–15255
Fiskerstrand CE, Lovejoy EA, Quinn JP (1999) An intronic polymorphic domain often associated with susceptibility to affective disorders has allele dependent differential enhancer activity in embryonic stem cells. FEBS Lett 458(2):171–174
Hranilovic D, Stefulj J, Schwab S, Borrmann-Hassenbach M, Albus M, Jernej B, Wildenauer D (2004) Serotonin transporter promoter and intron 2 polymorphisms: relationship between allelic variants and gene expression. Biol Psychiatry 55(11):1090–1094. doi:10.1016/j.biopsych.2004.01.029
Kaiser R, Muller-Oerlinghausen B, Filler D, Tremblay PB, Berghofer A, Roots I, Brockmoller J (2002) Correlation between serotonin uptake in human blood platelets with the 44-bp polymorphism and the 17-bp variable number of tandem repeat of the serotonin transporter. Am J Med Genet 114(3):323–328
Sarosi A, Gonda X, Balogh G, Domotor E, Szekely A, Hejjas K, Sasvari-Szekely M, Faludi G (2008) Association of the STin2 polymorphism of the serotonin transporter gene with a neurocognitive endophenotype in major depressive disorder. Prog Neuro-Psychopharmacol Biol Psychiatry 32(7):1667–1672. doi:10.1016/j.pnpbp.2008.06.014
Pizzo de Castro MR, Vargas Nunes SO, Guembarovski RL, Ariza CB, Oda JM, Vargas HO, Piccoli de Melo LG, Watanabe MA et al (2015) STin2 VNTR polymorphism is associated with comorbid tobacco use and mood disorders. J Affect Disord 172C:347–354. doi:10.1016/j.jad.2014.10.023
Klenova E, Scott AC, Roberts J, Shamsuddin S, Lovejoy EA, Bergmann S, Bubb VJ, Royer HD et al (2004) YB-1 and CTCF differentially regulate the 5-HTT polymorphic intron 2 enhancer which predisposes to a variety of neurological disorders. J Neurosci: Off J Soc Neurosci 24(26):5966–5973. doi:10.1523/JNEUROSCI.1150-04.2004
Roberts J, Scott AC, Howard MR, Breen G, Bubb VJ, Klenova E, Quinn JP (2007) Differential regulation of the serotonin transporter gene by lithium is mediated by transcription factors, CCCTC binding protein and Y-box binding protein 1, through the polymorphic intron 2 variable number tandem repeat. J Neurosci: Off J Soc Neurosci 27(11):2793–2801. doi:10.1523/JNEUROSCI.0892-06.2007
Vasiliou SA, Ali FR, Haddley K, Cardoso MC, Bubb VJ, Quinn JP (2012) The SLC6A4 VNTR genotype determines transcription factor binding and epigenetic variation of this gene in response to cocaine in vitro. Addict Biol 17(1):156–170. doi:10.1111/j.1369-1600.2010.00288.x
Ali FR, Vasiliou SA, Haddley K, Paredes UM, Roberts JC, Miyajima F, Klenova E, Bubb VJ et al (2010) Combinatorial interaction between two human serotonin transporter gene variable number tandem repeats and their regulation by CTCF. J Neurochem 112(1):296–306. doi:10.1111/j.1471-4159.2009.06453.x
Jonsson EG, Nothen MM, Gustavsson JP, Neidt H, Bunzel R, Propping P, Sedvall GC (1998) Polymorphisms in the dopamine, serotonin, and norepinephrine transporter genes and their relationships to monoamine metabolite concentrations in CSF of healthy volunteers. Psychiatry Res 79(1):1–9
Lesch KP, Bengel D, Heils A, Sabol SZ, Greenberg BD, Petri S, Benjamin J, Muller CR et al (1996) Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science 274(5292):1527–1531
Lim JE, Papp A, Pinsonneault J, Sadee W, Saffen D (2006) Allelic expression of serotonin transporter (SERT) mRNA in human pons: lack of correlation with the polymorphism SERTLPR. Mol Psychiatry 11(7):649–662. doi:10.1038/sj.mp.4001797
Kunugi H, Hattori M, Kato T, Tatsumi M, Sakai T, Sasaki T, Hirose T, Nanko S (1997) Serotonin transporter gene polymorphisms: ethnic difference and possible association with bipolar affective disorder. Mol Psychiatry 2(6):457–462
Tharoor H, Kotambail A, Jain S, Sharma PS, Satyamoorthy K (2013) Study of the association of serotonin transporter triallelic 5-HTTLPR and STin2 VNTR polymorphisms with lithium prophylaxis response in bipolar disorder. Psychiatr Genet 23(2):77–81. doi:10.1097/YPG.0b013e32835d6fad
Fan JB, Sklar P (2005) Meta-analysis reveals association between serotonin transporter gene STin2 VNTR polymorphism and schizophrenia. Mol Psychiatry 10(10):928–938 . doi:10.1038/sj.mp.4001690891
Pizzo de Castro MR, Maes M, Guembarovski RL, Ariza CB, Reiche EM, Vargas HO, Vargas MM, de Melo LG et al (2014) SLC6A4 STin2 VNTR genetic polymorphism is associated with tobacco use disorder, but not with successful smoking cessation or smoking characteristics: a case control study. BMC Genet 15:78. doi:10.1186/1471-2156-15-78
Vargas Nunes SO, Pizzo de Castro MR, Moreira EG, Guembarovski RL, Barbosa DS, Vargas HO, Piccoli de Melo LG, Bortolasci CC et al (2015) Association of paraoxonase (PON)1 activity, glutathione S-transferase GST T1/M1 and STin.2 polymorphisms with comorbidity of tobacco use disorder and mood disorders. Neurosci Lett 585:132–137. doi:10.1016/j.neulet.2014.11.002
Gyawali S, Subaran R, Weissman MM, Hershkowitz D, McKenna MC, Talati A, Fyer AJ, Wickramaratne P et al (2010) Association of a polyadenylation polymorphism in the serotonin transporter and panic disorder. Biol Psychiatry 67(4):331–338. doi:10.1016/j.biopsych.2009.10.015
Murphy DL, Moya PR (2011) Human serotonin transporter gene (SLC6A4) variants: their contributions to understanding pharmacogenomic and other functional GxG and GxE differences in health and disease. Curr Opin Pharmacol 11(1):3–10. doi:10.1016/j.coph.2011.02.008
Battersby S, Ogilvie AD, Blackwood DH, Shen S, Muqit MM, Muir WJ, Teague P, Goodwin GM et al (1999) Presence of multiple functional polyadenylation signals and a single nucleotide polymorphism in the 3′ untranslated region of the human serotonin transporter gene. J Neurochem 72(4):1384–1388
Heils A, Teufel A, Petri S, Seemann M, Bengel D, Balling U, Riederer P, Lesch KP (1995) Functional promoter and polyadenylation site mapping of the human serotonin (5-HT) transporter gene. J Neural Transm Gen Sect 102(3):247–254
Sheets MD, Ogg SC, Wickens MP (1990) Point mutations in AAUAAA and the poly (A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res 18(19):5799–5805
Vallender EJ, Priddy CM, Hakim S, Yang H, Chen GL, Miller GM (2008) Functional variation in the 3′ untranslated region of the serotonin transporter in human and rhesus macaque. Genes Brain Behav 7(6):690–697. doi:10.1111/j.1601-183X.2008.00407.x
Montasser ME, Ziv-Gal A, Brown JP, Flaws JA, Merchenthaler I (2015) A potentially functional variant in the serotonin transporter gene is associated with premenopausal and perimenopausal hot flashes. Menopause 22(1):108–113. doi:10.1097/GME.0000000000000291
Seneviratne C, Huang W, Ait-Daoud N, Li MD, Johnson BA (2009) Characterization of a functional polymorphism in the 3′ UTR of SLC6A4 and its association with drinking intensity. Alcohol Clin Exp Res 33(2):332–339. doi:10.1111/j.1530-0277.2008.00837.x
Jones PA (2012) Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13(7):484–492. doi:10.1038/nrg3230
Kinnally EL, Feinberg C, Kim D, Ferguson K, Leibel R, Coplan JD, John Mann J (2011) DNA methylation as a risk factor in the effects of early life stress. Brain Behav Immun 25(8):1548–1553. doi:10.1016/j.bbi.2011.05.001
Wang D, Szyf M, Benkelfat C, Provencal N, Turecki G, Caramaschi D, Cote SM, Vitaro F et al (2012) Peripheral SLC6A4 DNA methylation is associated with in vivo measures of human brain serotonin synthesis and childhood physical aggression. PLoS One 7(6):e39501. doi:10.1371/journal.pone.0039501
Philibert R, Madan A, Andersen A, Cadoret R, Packer H, Sandhu H (2007) Serotonin transporter mRNA levels are associated with the methylation of an upstream CpG island. Ame J Med Genet Part B, Neuropsychiatr Genet: Off Publ Int Soc Psychiatr Genet 144B(1):101–105. doi:10.1002/ajmg.b.30414
Philibert RA, Sandhu H, Hollenbeck N, Gunter T, Adams W, Madan A (2008) The relationship of 5HTT (SLC6A4) methylation and genotype on mRNA expression and liability to major depression and alcohol dependence in subjects from the Iowa Adoption Studies. Am J Med Genet Part B, Neuropsychiatr Genet: Off Publ Int Soc Psychiatr Genet 147B(5):543–549. doi:10.1002/ajmg.b.30657
Olsson CA, Foley DL, Parkinson-Bates M, Byrnes G, McKenzie M, Patton GC, Morley R, Anney RJ et al (2010) Prospects for epigenetic research within cohort studies of psychological disorder: a pilot investigation of a peripheral cell marker of epigenetic risk for depression. Biol Psychol 83(2):159–165. doi:10.1016/j.biopsycho.2009.12.003
Kinnally EL, Capitanio JP, Leibel R, Deng L, LeDuc C, Haghighi F, Mann JJ (2010) Epigenetic regulation of serotonin transporter expression and behavior in infant rhesus macaques. Genes Brain Behav 9(6):575–582. doi:10.1111/j.1601-183X.2010.00588.x
Alasaari JS, Lagus M, Ollila HM, Toivola A, Kivimaki M, Vahtera J, Kronholm E, Harma M et al (2012) Environmental stress affects DNA methylation of a CpG rich promoter region of serotonin transporter gene in a nurse cohort. PLoS One 7(9):e45813. doi:10.1371/journal.pone.0045813
van Ijzendoorn MH, Caspers K, Bakermans-Kranenburg MJ, Beach SR, Philibert R (2010) Methylation matters: interaction between methylation density and serotonin transporter genotype predicts unresolved loss or trauma. Biol Psychiatry 68(5):405–407. doi:10.1016/j.biopsych.2010.05.008
Wankerl M, Miller R, Kirschbaum C, Hennig J, Stalder T, Alexander N (2014) Effects of genetic and early environmental risk factors for depression on serotonin transporter expression and methylation profiles. Transl Psychiatry 4:e402. doi:10.1038/tp.2014.37
Duman EA, Canli T (2015) Influence of life stress, 5-HTTLPR genotype, and SLC6A4 methylation on gene expression and stress response in healthy Caucasian males. Biol Mood Anxiety Disord 5:2. doi:10.1186/s13587-015-0017-x
Kang HJ, Kim JM, Stewart R, Kim SY, Bae KY, Kim SW, Shin IS, Shin MG et al (2013) Association of SLC6A4 methylation with early adversity, characteristics and outcomes in depression. Prog Neuro-Psychopharmacol Biol Psychiatry 44:23–28. doi:10.1016/j.pnpbp.2013.01.006
Domschke K, Tidow N, Schwarte K, Deckert J, Lesch KP, Arolt V, Zwanzger P, Baune BT (2014) Serotonin transporter gene hypomethylation predicts impaired antidepressant treatment response. Int J Neuropsychopharmacol / Off Sci J Coll Int Neuropsychopharmacologicum 17(8):1167–1176. doi:10.1017/S146114571400039X
Iga JI, Watanabe SY, Numata S, Umehara H, Nishi A, Kinoshita M, Inoshita M, Shimodera S et al (2016) Association study of polymorphism in the serotonin transporter gene promoter, methylation profiles, and expression in patients with major depressive disorder. Hum Psychopharmacol 31(3):193–199. doi:10.1002/hup.2527
Dannlowski U, Kugel H, Redlich R, Halik A, Schneider I, Opel N, Grotegerd D, Schwarte K et al (2014) Serotonin transporter gene methylation is associated with hippocampal gray matter volume. Hum Brain Mapp 35(11):5356–5367. doi:10.1002/hbm.22555
Nikolova YS, Koenen KC, Galea S, Wang CM, Seney ML, Sibille E, Williamson DE, Hariri AR (2014) Beyond genotype: serotonin transporter epigenetic modification predicts human brain function. Nat Neurosci 17(9):1153–1155. doi:10.1038/nn.3778
Frodl T, Szyf M, Carballedo A, Ly V, Dymov S, Vaisheva F, Morris D, Fahey C et al (2015) DNA methylation of the serotonin transporter gene (SLC6A4) is associated with brain function involved in processing emotional stimuli. J Psychiatry Neurosci: JPN 40(5):296–305
Swartz JR, Hariri AR, Williamson DE (2016) An epigenetic mechanism links socioeconomic status to changes in depression-related brain function in high-risk adolescents. Mol Psychiatry. doi:10.1038/mp.2016.82
Booij L, Szyf M, Carballedo A, Frey EM, Morris D, Dymov S, Vaisheva F, Ly V et al (2015) DNA methylation of the serotonin transporter gene in peripheral cells and stress-related changes in hippocampal volume: a study in depressed patients and healthy controls. PLoS One 10(3):e0119061. doi:10.1371/journal.pone.0119061
Bernstein BE, Meissner A, Lander ES (2007) The mammalian epigenome. Cell 128(4):669–681. doi:10.1016/j.cell.2007.01.033
Bennett AJ, Lesch KP, Heils A, Long JC, Lorenz JG, Shoaf SE, Champoux M, Suomi SJ et al (2002) Early experience and serotonin transporter gene variation interact to influence primate CNS function. Mol Psychiatry 7(1):118–122. doi:10.1038/sj/mp/4000949
Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, McClay J, Mill J et al (2003) Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science 301(5631):386–389. doi:10.1126/science.1083968
Green JG, McLaughlin KA, Berglund PA, Gruber MJ, Sampson NA, Zaslavsky AM, Kessler RC (2010) Childhood adversities and adult psychiatric disorders in the national comorbidity survey replication I: associations with first onset of DSM-IV disorders. Arch Gen Psychiatry 67(2):113–123. doi:10.1001/archgenpsychiatry.2009.186
Varese F, Smeets F, Drukker M, Lieverse R, Lataster T, Viechtbauer W, Read J, van Os J et al (2012) Childhood adversities increase the risk of psychosis: a meta-analysis of patient-control, prospective- and cross-sectional cohort studies. Schizophr Bull 38(4):661–671. doi:10.1093/schbul/sbs050
Nikolova YS, Hariri AR (2015) Can we observe epigenetic effects on human brain function? Trends Cogn Sci 19(7):366–373. doi:10.1016/j.tics.2015.05.003
Jawahar MC, Murgatroyd C, Harrison EL, Baune BT (2015) Epigenetic alterations following early postnatal stress: a review on novel aetiological mechanisms of common psychiatric disorders. Clin Epigenetics 7:122. doi:10.1186/s13148-015-0156-3
Mortensen OV, Thomassen M, Larsen MB, Whittemore SR, Wiborg O (1999) Functional analysis of a novel human serotonin transporter gene promoter in immortalized raphe cells. Brain Res Mol Brain Res 68(1–2):141–148
Devlin AM, Brain U, Austin J, Oberlander TF (2010) Prenatal exposure to maternal depressed mood and the MTHFR C677T variant affect SLC6A4 methylation in infants at birth. PLoS One 5(8):e12201. doi:10.1371/journal.pone.0012201
Kim JM, Stewart R, Kang HJ, Kim SW, Shin IS, Kim HR, Shin MG, Kim JT et al (2013) A longitudinal study of SLC6A4 DNA promoter methylation and poststroke depression. J Psychiatr Res 47(9):1222–1227. doi:10.1016/j.jpsychires.2013.04.010
Beach SR, Brody GH, Todorov AA, Gunter TD, Philibert RA (2011) Methylation at 5HTT mediates the impact of child sex abuse on women’s antisocial behavior: an examination of the Iowa adoptee sample. Psychosom Med 73(1):83–87. doi:10.1097/PSY.0b013e3181fdd074
Zhao J, Goldberg J, Bremner JD, Vaccarino V (2013) Association between promoter methylation of serotonin transporter gene and depressive symptoms: a monozygotic twin study. Psychosom Med 75(6):523–529. doi:10.1097/PSY.0b013e3182924cf4
Tylee DS, Kawaguchi DM, Glatt SJ (2013) On the outside, looking in: a review and evaluation of the comparability of blood and brain “-omes”. Am J Med Genet Part B, Neuropsychiatr Genet: Off Publ Int Soc Psychiatr Genet 162B(7):595–603. doi:10.1002/ajmg.b.32150
Frodl T, Reinhold E, Koutsouleris N, Donohoe G, Bondy B, Reiser M, Moller HJ, Meisenzahl EM (2010) Childhood stress, serotonin transporter gene and brain structures in major depression. Neuropsychopharmacol: Off Publ Am Coll Neuropsychopharmacol 35(6):1383–1390. doi:10.1038/npp.2010.8
Sela N, Mersch B, Hotz-Wagenblatt A, Ast G (2010) Characteristics of transposable element exonization within human and mouse. PLoS One 5(6):e10907. doi:10.1371/journal.pone.0010907
Polak P, Domany E (2006) Alu elements contain many binding sites for transcription factors and may play a role in regulation of developmental processes. BMC Genomics 7:133. doi:10.1186/1471-2164-7-133
Verdone L, Caserta M, Di Mauro E (2005) Role of histone acetylation in the control of gene expression. Biochem Cell Biol = Biochimie et biologie cellulaire 83(3):344–353. doi:10.1139/o05-041
Fuks F (2005) DNA methylation and histone modifications: teaming up to silence genes. Curr Opin Genet Dev 15(5):490–495. doi:10.1016/j.gde.2005.08.002
Gill RK, Kumar A, Malhotra P, Maher D, Singh V, Dudeja PK, Alrefai W, Saksena S (2013) Regulation of intestinal serotonin transporter expression via epigenetic mechanisms: role of HDAC2. Am J Physiol Cell Physiol 304(4):C334–C341. doi:10.1152/ajpcell.00361.2012
Lindell SG, Yuan Q, Zhou Z, Goldman D, Thompson RC, Lopez JF, Suomi SJ, Higley JD et al (2012) The serotonin transporter gene is a substrate for age and stress dependent epigenetic regulation in rhesus macaque brain: potential roles in genetic selection and gene x environment interactions. Dev Psychopathol 24(4):1391–1400. doi:10.1017/s0954579412000788
Moya PR, Wendland JR, Salemme J, Fried RL, Murphy DL (2013) miR-15a and miR-16 regulate serotonin transporter expression in human placental and rat brain raphe cells. Int J Neuropsychopharmacol/ Off Sci J Coll Int Neuropsychopharmacologicum 16(3):621–629. doi:10.1017/S1461145712000454
Baudry A, Mouillet-Richard S, Schneider B, Launay JM, Kellermann O (2010) miR-16 targets the serotonin transporter: a new facet for adaptive responses to antidepressants. Science 329(5998):1537–1541. doi:10.1126/science.1193692
Liao XJ, Mao WM, Wang Q, Yang GG, Wu WJ, Shao SX (2016) MicroRNA-24 inhibits serotonin reuptake transporter expression and aggravates irritable bowel syndrome. Biochem Biophys Res Commun 469(2):288–293. doi:10.1016/j.bbrc.2015.11.102
Issler O, Haramati S, Paul ED, Maeno H, Navon I, Zwang R, Gil S, Mayberg HS et al (2014) MicroRNA 135 is essential for chronic stress resiliency, antidepressant efficacy, and intact serotonergic activity. Neuron 83(2):344–360. doi:10.1016/j.neuron.2014.05.042
Jensen KP, Covault J, Conner TS, Tennen H, Kranzler HR, Furneaux HM (2009) A common polymorphism in serotonin receptor 1B mRNA moderates regulation by miR-96 and associates with aggressive human behaviors. Mol Psychiatry 14(4):381–389. doi:10.1038/mp.2008.15
Barr CL, Misener VL (2016) Decoding the non-coding genome: elucidating genetic risk outside the coding genome. Genes Brain Behav 15(1):187–204. doi:10.1111/gbb.12269
Acknowledgments
This study was supported by “Progetto CNR–DSB.AD007.107” to MR and by “Progetto CNR-DSB.AD007.090” to SI.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Iurescia, S., Seripa, D. & Rinaldi, M. Looking Beyond the 5-HTTLPR Polymorphism: Genetic and Epigenetic Layers of Regulation Affecting the Serotonin Transporter Gene Expression. Mol Neurobiol 54, 8386–8403 (2017). https://doi.org/10.1007/s12035-016-0304-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-016-0304-6