Abstract
TCERG1 is a highly conserved human protein implicated in interactions with the transcriptional and splicing machinery that is associated with neurodegenerative disorders. Biochemical, neuropathological, and genetic evidence suggests an important role for TCERG1 in Huntington’s disease (HD) pathogenesis. At present, the molecular mechanism underlying TCERG1-mediated neuronal effects is unknown. Here, we show that TCERG1 depletion led to widespread alterations in mRNA processing that affected different types of alternative transcriptional or splicing events, indicating that TCERG1 plays a broad role in the regulation of alternative splicing. We observed considerable changes in the transcription and alternative splicing patterns of genes involved in cytoskeleton dynamics and neurite outgrowth. Accordingly, TCERG1 depletion in the neuroblastoma SH-SY5Y cell line and primary mouse neurons affected morphogenesis and resulted in reduced dendritic outgrowth, with a major effect on dendrite ramification and branching complexity. These defects could be rescued by ectopic expression of TCERG1. Our results indicate that TCERG1 affects expression of multiple mRNAs involved in neuron projection development, whose misregulation may be involved in TCERG1-linked neurological disorders.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Global transcriptome analyses suggest that >90% of human multiexon genes undergo alternative splicing [1, 2]; this process contributes greatly to the diversity of cell- and tissue-specific protein expression patterns. A large fraction of these alternatively spliced events occur in the nervous system, highlighting the exquisite and intricate regulation that occurs among neurons, which must respond quickly to many stimuli and perform adequate timely responses [3–5]. Alterations in the regulation of this mRNA biogenesis process cause missplicing of genes that alter gene functions and lead to disease pathology, including diverse neurological disorders [6, 7]. Splicing defects can occur due to mutations in the primary gene sequence that lead to alterations in pre-mRNA splicing, changing splice sites or weakening or strengthening splicing enhancer and silencer elements, or to mutations in core spliceosomal factors or other trans-regulatory factors. Many disorders caused by these mutations have been identified [6]; however, the molecular links between alternative splicing and the disease phenotype and the molecular mechanisms at work remain to be elucidated.
Several neurodegenerative disorders are associated with alterations in dendrite morphology or defects in neuronal development, including changes in dendrite branching patterns, the retraction or loss of dendrite branching, and changes in spine morphology and number. These phenotypes lead to an impairment in neuronal connectivity and are the cause of many neurological and cognitive disorders [8]. One of the mechanisms that has been proposed to explain the effect of these alterations on diseases is the disturbance of the function of transcription and RNA processing factors, which may result in the deregulation of pre-mRNA processing, such as splicing, of important neuronal genes and the subsequent disruption of proper neuronal gene expression [9, 10].
TCERG1, previously named CA150 [11], is a nuclear protein involved in transcription and splicing regulation. TCERG1 binds to transcription and splicing components [12–16], localizes at the interface of nuclear speckles and what are presumably nearby transcription/splicing sites [14, 17], and affects the transcriptional activity of CCAAT/enhancer-binding protein alpha (CEBPA) [18] and the splicing of several splicing reporters [13, 19–22]. Based on these data, TCERG1 has been suggested to couple the transcribing RNA polymerase II (RNAPII) with spliceosome complexes to regulate co-transcriptional splicing events [23, 24]. We recently found support for this hypothesis by showing that TCERG1 regulated the alternative splicing of the BCL2L1 gene by modulating the rate of RNAPII transcription [22]. Interestingly, TCERG1 has been implicated in the pathogenesis of the neurodegenerative disorder Huntington’s disease (HD). TCERG1 interacts with the huntingtin (HTT) protein, is expressed in cells harboring the mutated HTT (striatal and cortical neurons), and has been associated with the morphological deposits related to the disease [25]. TCERG1 may play a neuroprotective role in HD because its overexpression rescues neuronal cell death due to mutant HTT neurotoxicity [26]. These data suggest a pathway that is likely to involve an abnormal interaction of HTT with TCERG1, perhaps by altering ribonucleoprotein functions with deleterious consequences. Moreover, these results suggest an important role for TCERG1 in neuronal development and function. However, little is known about the function of TCERG1 in neurons, where its expression level is high [27].
In the current study, we used whole-transcriptome analysis to identify endogenous TCERG1 gene targets. We found that TCERG1 affects the transcription and alternative splicing of numerous genes. Cells derived from the nervous lineage relative to other tissues possess a larger proportion of TCERG1-dependent differentially expressed genes, especially those involved in cytoskeleton dynamics and neurite outgrowth. This prompted us to examine the role of TCERG1 in the nervous system. We characterized the function of TCERG1 in neurons using shRNAs to specifically silence TCERG1 expression. Our results suggest that TCERG1 is required for normal neurite development in cultured cells. These findings provide novel insight into the molecular function of TCERG1 in neurons and suggest how abnormal regulation of the transcription and/or alternative splicing of TCERG1-specific targets may play a role in the pathogenesis of TCERG1-associated neurological disorders.
Materials and Methods
Cell Culture, siRNA and Plasmid Transfection, and shRNA Transduction
The HEK293T, SH-SY5Y, and C2C12 cell lines were grown under standard conditions. SH-SY5Y and C2C12 cells, which were kindly provided by J. Ortín (CNB, Madrid) and F. Ventura (University of Barcelona), respectively, were plated in collagen-coated plates. The T-REx-HEK293 cell lines have been described previously [28]. For RNAi-mediated depletion of TCERG1, HEK293T cells were grown to approximately 50% confluence and transfected using the Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s protocols with 60 nM (final concentration) of either of the following siRNA duplexes: siEGFP (5′-CUACAACAGCCACAACGE-3′) or siTCERG1 (5′-GGAGUUGCACAAGAUAGUU-3′) [20]. The cells were harvested 72 h after transfection. For plasmid transfection, SH-SY5Y cells were transfected with the appropriate amounts of the indicated constructs using JetPRIME reagent.
For lentiviral infection, HEK293T cells growing in a 9.6-cm dish were transfected with the packaging, envelope, and shRNA vector DNA (1.5 μg each) using LipoD293 reagent (1:3 ratio of DNA/reagent) according to the manufacturer’s protocol. After 72 h, the medium was collected, and the virus was concentrated by ultracentrifugation at 50.000×g for 90 min at 4 °C. Viral particles were added to the SH-SY5Y cells. Transduced cells were selected with 1 μg/ml puromycin for a first round of selection and routinely cultured in the presence of 1.5 μg/ml puromycin.
Hippocampal neurons were dissected from E17.5 CD1 mice (undetermined sex) in HBSS containing 0.6% glucose and 10 mM HEPES. The culture protocols employed have been described previously [29]. Primary cultures were transfected at 3 or 7 days in vitro (DIV) using a CalPhos Mammalian Transfection Kit (Clontech) and analyzed at 7 or 14 DIV, respectively.
Western Blot Analysis
A fraction of total cells was lysed in cold T7 buffer (20 mM HEPES [pH 7.9], 150 mM EDTA, 1% NP-40, 1 mM dithiothreitol, protease inhibitor mixture (Complete, Roche), and 1 mM PMSF), as previously described [22]. The proteins were separated by SDS-PAGE, transferred to a nitrocellulose membrane (Amersham Biosciences), and incubated with specific antibodies against TCERG1 [14] or CDK9 (catalog no. sc-484, Santa Cruz Biotechnology) as a control at a 1:2000 dilution. Peroxidase-conjugated secondary antibodies (Perkin Elmer Life Science) were used, and the bound antibodies were detected by enhanced chemiluminescence (Perkin Elmer Life Science).
Plasmids
The following viral vectors were purchased from Sigma: pLKO.1, pLKO.1-TCERG1-sh9 (5′-GCCAAGAATTTAGACTCAGAA-3′), and pLKO.1-TCERG1-sh4 (5′-TCCTAAAGAAGAGCCTATAAA-3′). To generate the TCERG1 mutant vector resistant to sh9 (mut-TCERG1), wild-type TCERG1 cDNA was subcloned from the pEFBOST7-TCERG1 vector [30] into the pcDNA3-EGFP vector (Addgene) using PCR. A QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene) was then used to change nucleotides 2590 (T to C), 2591 (T to C), 2593 (A to G), 2599 (A to C), and 2602 (A to G) using the following primer: 5′- GTACATTGAAAAAATAGCCAAGAACCTGGACTCCGAGAAAGAAAAGGAGCTTG-3′. The construct was verified by DNA sequencing. Plasmids were transformed into DH5α cells for selection.
RNA Sample Preparation and Exon Array Processing
Total RNA was isolated from HEK293T cells from four independent experiments in triplicate with TRIzol reagent (Invitrogen) and RNeasy columns (Qiagen). Samples were subjected to quality tests, labeled, and hybridized to Affymetrix GeneChIP Human Exon 1.0ST Array (Affymetrix) according to the manufacturer’s recommendations in the Genomic Unit of CABIMER (Sevilla, Spain). Dataset analysis and visualization were performed using EASANA (GenoSplice, Paris, France), which is based on FAST DB annotations [31, 32]. Array data are available at the Gene Expression Omnibus (accession number GSE73815).
Computational Analysis
The 419 hits detected at the alternative splicing level were manually inspected using the EASANA Visualization Module and aligned using the UCSC Genome Browser Blat alignment tool. The number of endogenous targets was reduced to 356. The DAVID Functional Annotation tool [33, 34] was used to examine TCERG1 transcript- and exon-level target genes for over-representation in GO categories and biological pathways. Cassette exon sequences were analyzed using the MEME program found in the MEME suite v.4.11.1 (http://meme-suite.org/tools/meme). In total, 37 and 29 up- and down-regulated sequences, respectively, and the same number of control sequences were analyzed separately. The analysis settings were adjusted to a “site distribution” of “any number of repetitions” per sequence to search up to 10 motifs and to search in the given strand (appropriate for RNA). We compared the similarity of the significant motifs found in cassette exons up- or down-regulated with the motif found within random exons using the Tomtom tool and discarded the sequences with significant similarity (P < 0.05). We also used the Tomtom tool to find the predicted RNA binding proteins that would preferentially bind to these conserved motifs. The GC content was calculated for each exon as the percentage of G and C nucleotides divided by the total number of nucleotides within the exon. Unpaired Student’s t test was used for statistical analysis.
Experimental Validation by RT-qPCR Analysis
Total RNA was isolated using TRIzol. RNA quantity and purity were assessed with a NanoDrop spectrophotometer (Thermo Scientific). The RNA was digested with 10 U of RNase-free DNase I (Roche) for 30 min at 37 °C. After DNase inactivation at 70 °C for 5 min, 1 μg of RNA was reverse transcribed by the Moloney murine leukemia virus RT (Invitrogen) for 1 h at 37 °C using random hexamer primers in a 20-μl reaction according to the manufacturer’s instructions. Negative control reactions (without reverse transcriptase) were also performed. One-tenth of the resulting cDNA was amplified by PCR using primers designed to amplify a 100–160 bp region. The primers were designed using the Primer3Plus web interface [35]. The specificity of the primers was confirmed using the in silico tools available at the UCSC Genome Bioinformatics site (http://genome.ucsc.edu/cgi-bin/hgPcr) and the Friendly Alternative Splicing and Transcripts Database (http://www.fast-db.com/fastdb2/frame_2.html). The quantification of the transcripts was performed by real-time PCR using iQ™ SYBR® Green Supermix (Bio-Rad) and an iCycler iQ™ thermal cycler station (Bio-Rad). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal gene control for transcript level validation because its mRNA level did not change upon TCERG1 knockdown by exon array or RT-qPCR analysis. The following thermal cycler program was used: 95 °C for 3′, 40 amplification cycles of 95 °C for 30″, (annealing temperature appropriate for each primer pair) for 30″, and 72 °C for 45″, followed by melting curve analysis. Melting curves and agarose gel analysis confirmed the presence of one specific product of the correct size and the absence of primer dimers. For transcript-level targets, target mRNA was quantified relative to GAPDH using the ΔΔCt method [36]:
where E is the PCR efficiency and ΔCt = (the cycle threshold [Ct] for EGFP treatment) – (Ct for TCERG1 knockdown). Duplicate reactions were performed with each primer pair for each cDNA from three independent silencing experiments. For exon-level targets, the transcript variant of interest was quantified relative to GAPDH and to an exon common to all transcript variants, which was quantified relative to GAPDH, using the same formula.
Immunofluorescence
Cells were grown on coverslips and fixed with 4% paraformaldehyde and 4% sucrose in PBS (pH 7.4) for 20 min at 37 °C. The cells were washed and permeabilized three times with PBS containing 0.5% Triton X-100 for 5 min at room temperature and blocked in PBS containing 10% BSA and 0.5% Triton X-100 for 1 h at room temperature. The cells were incubated with 50 μg/ml Phalloidin-Tetramethylrhodamine B isothiocyanate (TRITC) (Sigma) in PBS containing 10% BSA for 1 h at room temperature (humidity chamber). The cells were subsequently washed three times with 0.1% BSA in PBS, three times with PBS, and once with distilled water; lastly, the cells were rinsed with 70% ethanol. Coverslips were mounted onto glass slides using the ProLong Gold Antifade reagent with DAPI (Life Technologies). Images were acquired with an inverted Leica SP2 confocal microscope using an HCX PL APO CS 63.0 × 1.40 OIL UV objective. All images were digitally processed for presentation using Image J and Adobe Photoshop CS5 software.
Neurite Length Quantification
Neurite extension and soma area were assessed in the acquired images using gvSIG software (http://www.gvsig.org). For these experiments, the SH-SY-5Y cells were seeded at the same confluence (5.2 × 105 and 2.6 × 105 cells in 100 mm culture dishes without or with retinoic acid, respectively) and allowed to grow for 48 h prior to image collection. Approximately 13 images were captured via light microscopy using a ×20 objective. At least 30 cells were measured for each replicate using gvSIG software. To create a pixel/μm relationship, several images were acquired with the Neubauer chamber and the lengths of two lines of different known lengths were measured in pixels.
We used NeuronStudio software for semi-automatic measurement and analysis of the primary culture images [37], followed by manual inspection of automatic tracings and, if necessary, manual editing of the tracings. All images were treated with the same image filters. The NeuronJ plug-in (ImageJ) was used for the rescue experiments, and only green channel images were measured to analyze neurites belonging to single cells.
Statistical Analysis
Statistical analysis was performed using Prism 5.0 software (GraphPad). Two-tailed paired Student’s t tests were used to compare the samples and their respective controls in RT-qPCR, overexpression experiments, and primary cultures. One-way ANOVA was used to quantify neurite length and soma area in SH-SY5Y cells. The P values are represented by asterisks (*P = 0.01–0.05; **P = 0.001–0.01; and ***P < 0.001). The absence of an asterisk indicates that the change relative to the control is not statistically significant.
Results
TCERG1 Depletion Results in Significant Changes in Both Transcript and Exon Levels
To investigate the global role of TCERG1 in the regulation of gene expression, we performed a genome-wide analysis of transcript- and exon-level changes in HEK293T cells treated with an siRNA targeting TCERG1 (siTCERG1) compared to mock siRNA (siEGFP)-treated control cells. Treatment of cells with siTCERG1 (60 nM) reduced TCERG1 expression (Fig. 1a). Quantitative RT-PCR (RT-qPCR) confirmed that knockdown with siTCERG1 led to a decrease in mRNA levels (see below). Total RNA was prepared from siEGFP- and siTCERG1-treated cells from four independent experiments. RNA samples were used to measure global transcriptome and alternative splicing changes using the Affymetrix Human Exon 1.0 Array, which features probesets corresponding to exonic, intronic, and untranslated (UTR) regions of all well-annotated human genes [38]. Therefore, variations in expression levels and changes in pre-mRNA splicing can be measured to analyze the effect of TCERG1 knockdown on gene expression. Transcript- and exon-level changes were identified with the EASANA algorithm (see the “Materials and Methods”) [27]. Changes in transcript levels in response to TCERG1 depletion were detected for 128 protein-coding genes (fold-change ≥1.5; P ≤ 0.05) of 16,762 genes tested (Table S1). Of the 128 regulated genes, 108 (84.5%) were down-regulated and 20 (15.5%) were up-regulated upon TCERG1 knockdown compared to control cells (Fig. 1b). Differential exon levels were detected in 314 genes (fold-change ≥1.5; P ≤ 0.05) of the 16,762 genes tested (Fig. 1b), including 356 alternative transcriptional or splicing events (Table S1). Additionally, 34 genes showed changes in more than one exon, and six (C19orf48, GPR125, PCLO, PHF6, TTC39A, and UBAP2) exhibited up- and down-regulation. After manually inspecting the 356 TCERG1 depletion-mediated exon-level changes, we assessed individual types of alternative transcriptional or splicing events. We found 14 alternative 3′ splice sites (4%) and 16 alternative 5′ splice sites (4%), 50 intron retentions (14%), 60 alternative first exons (17%), 68 alternative last exons (19%), 71 cassette exons (skipped exons, 20%), and 77 patterns in which the change could not be explained by a clear event (22%) (Fig. 1c). The data show that TCERG1 regulates different types of alternative transcriptional or splicing events, indicating its broad role in the regulation of pre-mRNA processing. Notably, the largest number of exon-level targets corresponded to cassette exons followed by alternative last/first exons and intron retention events. The majority of events (236 of 356) were up-regulated upon TCERG1 knockdown, which may indicate a role for TCERG1 as a repressor (Fig. 1d). We also observed changes in both transcript and exon levels for 28 genes (Fig. 1e). This result suggests that TCERG1 may regulate these genes by coupling transcription and alternative splicing, a mechanism to which TCERG1 has been previously linked [22]. However, the number of genes that showed changes in both transcript and exon levels was not significantly higher than would be expected by chance alone (23 genes, P = 0.05).
Identification of differentially expressed genes upon TCERG1 knockdown. a TCERG1 knockdown in HEK293T cells. HEK293T cells were transfected with control (siEGFP) or TCERG1 (siTCERG1) siRNA duplexes. At approximately 72 h after transfection, a sample of the cell lysates was resolved on SDS-PAGE gels, and the proteins were transferred to membranes to detect TCERG1 and CDK9 protein expression. b Number of transcripts and exons that were differentially expressed under TCERG1 knockdown conditions. Up- and down-regulated genes are depicted in red and green, respectively. c Classification of the alternative transcriptional or splicing events found in the array after manually inspecting the 356 TCERG1 depletion-mediated exon-level changes (314 genes). d Number of up- (red) or down-regulated (green) alternative transcriptional or splicing events (up and down refers to a greater or lesser inclusion of the exon in the mature transcript, respectively). e Venn diagram illustrating the overlap between the changes identified at the transcript and exon levels. The comparison identified 28 genes with both transcript- and exon-level changes
We used RT-qPCR to verify the differential expression of selected targets identified after the bioinformatics analysis of the microarray data, which showed changes at the transcription or splicing levels or both. These genes were also selected because of their implication in cytoskeleton dynamics and neuron development (see below). We first selected ten genes showing changes in overall transcript levels, some of which also showed changes at the exon level. We validated all ten genes (Fig. 2a). We also analyzed five putative exon-level target genes using primers spanning alternatively spliced exons. We validated four genes tested that displayed the same direction of change as the array prediction. ATM did not show a significant change and thus was not considered as having a validated change (Fig. 2b). Of note, TCERG1 regulates its own message, generating an isoform with a longer first exon upon its silencing (Fig. 2b). The observed changes in transcription and splicing were also tested in the T-REx-HEK293 cell line, in which expression of control shRNA and shRNA targeting TCERG1 can be induced by adding tetracycline. Analysis of cell lysates of these induced cells showed that cells containing shRNAs targeting TCERG1 expressed significantly lower levels of TCERG1 protein compared to cells containing control shRNAs [28] (Fig. S1C). We first validated Vimentin (VIM), in which the retention of two introns (seven and eight) in its 3′ end leads to an alternative longer 3′UTR. Primers were designed to anneal at intron 7 and at the end of exon 8, thus covering the alternative 3′ UTR (Fig. S1A), and RT-PCR results confirmed the presence of decreased levels of this isoform upon TCERG1 knockdown (Fig. S1B). TCERG1 depletion correlated with increased protein levels (Fig. S1C). To our knowledge, this is the first report of a correlation between retention of these VIM introns and protein expression levels. We also analyzed and validated three more target genes with putative changes in gene expression in these cells (Fig. S1D).
RT-qPCR analyses for several transcript- (a) and exon-level (b) changes predicted to be differentially expressed in TCERG1-depleted cells. The transcript isoform (Iso, gray bars) was quantified relative to an mRNA common to all transcript variants (black bars). GAPDH was selected as a reference gene for the validations (see Experimental Procedures). Each gene target is shown as a schematic representation, with exons (black boxes) and introns (gray boxes) and with the predicted alternative processing event; arrows indicate primer locations. Relative expression levels and fold changes were log2 transformed for visual evaluation. The data are from at least three independent experiments (means ± SEM). **P = 0.001–0.01; ***P < 0.001
Most of the alternative transcriptional or splicing events regulated by TCERG1 were identified in the cassette exon category (Fig. 1c). To investigate the common features associated with these alternatively spliced regions, we examined the sequences within or flanking the regulated exon. First, we analyzed sequences in the regulated exon and compared these sequences to a background of TCERG1-non-regulated cassette exon events from the exon array analysis. Additionally, we analyzed a set of randomly selected constitutive exons proximal to the TCERG1-regulated cassette exons. We did not observe substantial differences in the GT-AG rule of the 5′ and 3′ splice sites (Fig. S2A) or in the exon length among the analyzed set of sequences (Fig. S2B). We also examined the position of the regulated exons relative to the 5′ and 3′ ends of the pre-mRNA. We observed a similar distribution along the pre-mRNA between the control and the up-regulated cassette exon events (mean of 51%, P = 0.926) and a different distribution for the down-regulated exons toward the 3′ end of the pre-mRNA (mean of 65%, P < 0.05) (Fig. S2C). Next, we analyzed the nucleotide composition across exons. Cassette exons that were down-regulated upon TCERG1 depletion exhibited GC content similar to intronic sequences, which was lower than the GC content of the up-regulated or control sequences (P < 0.05) (Fig. S2D). The higher GC content in exons relative to introns has been proposed to regulate exon inclusion [39], and higher GC content within the gene body has been associated with lower elongation rates [40, 41]. Given that TCERG1 has been shown to regulate alternative splicing by modulating the rate of RNAPII [22], these data may indicate a transcription-related mechanism that regulates the down-regulated cassette exons.
To characterize the differences between control and TCERG1-depleted cells, we assessed the genes related to the biological processes, cellular components, and molecular functions that were over-represented among the identified TCERG1-target genes using the DAVID bioinformatics resources webserver [33, 34]. Gene ontology (GO) analysis indicated that the most significantly enriched terms for the biological process category among the transcript-level targets are associated with the regulation of metabolic processes, morphogenesis and development, microtubule-based processes, and cell cycle arrest (Table 1). The exon-level targets showed changes in genes involved in processes such as metabolic regulation, neuron apoptosis, negative regulation of cellular component organization, cell cycle, and intracellular transport (Table 2). Analysis of the cellular components showed significant enrichment of the following: genes annotated as part of the z disc, a region of the sarcomere considered to primarily be important for mechanical stability, components of the contractile fiber, the basolateral plasma membrane, and the cell-substrate junction, among others (Tables 1 and 2). Finally, molecular function analysis revealed that transcription and RNA processing were significantly enriched terms among the TCERG1 exon-level targets (Table 2), supporting the hypothesis of the existence of a complex regulatory RNA network regulated and/or organized by transcription/splicing regulators [42].
To investigate the tissue specificity of TCERG1-regulated targets, we extracted tissue-specific profiles based on data for the 128 and 356 TCERG1 transcript- and alternative splicing-regulated events across 126 primary human tissues in a reference microarray dataset. The enrichment score was comparable between genes, allowing the genes in each tissue profile to be ranked. The software was described previously [43] and is available at http://xavierlab2.mgh.harvard.edu/EnrichmentProfiler/index.html. A non-supervised hierarchical clustering analysis using this tool revealed that cells derived from the nervous lineage possess a larger proportion of TCERG1-dependent differentially expressed genes (Fig. 3a). We evaluated the types of alternative transcriptional or splicing events within this population of targets specific for the CNS and found the cassette exon and alternative last exon to be over-represented patterns (Fig. 3b). To identify the most promising genes of interest in the dataset, we significantly increased the threshold value and observed that many of the targets in the central nervous system (CNS) were associated with cellular morphology and cytoskeleton dynamics (Fig. 3c and Table S2).
a Hierarchical clustering representation of enrichment scores for the TCERG1 transcript- and exon-level targets across 126 human primary tissues. Tissues are shown on the left of the panel, and each vertical line refers to a gene. Red indicates genes with increased expression, and green indicates genes with reduced expression relative to the geometrical mean. b Alternative processing events regulated by TCERG1 in the central nervous system. A total of 32 exon-level targets (28 genes) were manually inspected for known alternative splicing patterns and then categorized. c Gene ontology (GO) categories over-represented among the TCERG1 targets in the central nervous system. The ordinate is the name of the pathway, and the abscissa is the number of genes belonging to this pathway. Color scale representing the P values of the GO categories. Brown and purple correspond to values of 4.5 × 10−2 and 7.4 × 10−5, respectively, shown below the diagram
TCERG1 Depletion Leads to Alterations in Neurite Outgrowth
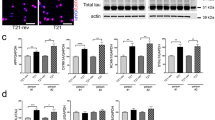
The transcriptome analysis of TCERG1-depleted HEK293T cells showed most of the alternative transcriptional or splicing events to be in genes predominantly expressed in the CNS and related to morphogenesis and cytoskeleton dynamics (Fig. 3 and Table S2). HEK293 cells were originally generated in the 1970s from normal human embryonic kidney cells through Adenovirus 5 transfection [44]. Despite their origin, HEK293 cells were later shown to possess characteristics of neurons (e.g., expressing more than 60 neuronal genes), suggesting that the original kidney culture was transformed into a neuronal lineage cell by the adenovirus [45]. As such, this cell line and derivatives have been extensively used in the study of neuron-specific voltage-gated ion channels ([46] and references therein), neuronal signaling [47], synaptogenesis [48–51], and propagation of highly neurotropic viruses [52–54]. This cell type, however, is not the best experimental model for studying neuronal function. Thus, to further investigate the role of TCERG1 in neurons, we examined the effects of TCERG1 depletion in the neuroblastoma cell line SH-SY5Y. Regulation of cytoskeleton dynamics in SH-SY5Y cells is fundamental to the outgrowth of neurites. To ensure long-term, reproducible, and defined silencing effects, we generated SH-SY5Y cells with stable TCERG1 mRNA interference using specific shRNAs delivered by lentiviruses. SH-SY5Y-sh9 cells were generated by infecting the cells with lentiviruses expressing a shRNA against TCERG1. The control SH-SY5Y cell line was generated using pLKO.1 shRNA, which does not target any known mammalian genes. shRNA-mediated TCERG1 depletion was verified by Western blot analysis (Fig. 4a). The control shRNA and other non-related shRNAs did not have an effect on TCERG1 expression (Fig. S3). We also verified TCERG1 depletion by RT-qPCR (Fig. 4b) and validated the change in the alternative first exon isoform of TCERG1 upon TCERG1 silencing in these cells (Fig. 4b). Depletion of TCERG1 in SH-SY5Y cells also led to increases in VIM protein (Fig. S4), suggesting that the gene expression changes observed in HEK293T cells are relevant to neuronal cells. Then, we analyzed the effects of TCERG1 depletion on cell morphology by bright-field microscopy. We observed that the TCERG1 knockdown cells became more rounded and had shorter neurites compared to the uninfected and shControl cells (Fig. 4c). To quantify the effects of TCERG1 depletion, we performed morphometric measurements on shTCERG1 and control neurons. A significant decrease in the length of the neurites and an increase in the size of the soma were observed in the TCERG1-depleted cells (Fig. 4d). The same results were obtained upon neuronal differentiation induced by retinoic acid treatment [55] (Fig. 4c, d). We did not observe clear differences between undifferentiated and retinoic-differentiated cells, which suggested that TCERG1 does not have a role in the differentiation process itself. Overall, these data indicate that TCERG1 is required for proper neurite outgrowth of SH-SY5Y cells.
Depletion of TCERG1 affects the morphology and neurite size of SH-SY5Y cells. a Western blot analysis of TCERG1 expression in the SH-SY5Y cell lines compared with endogenous CDK9 expression. b RT-qPCR analysis for TCERG1 transcript levels predicted to be differentially expressed. GAPDH was selected as a reference gene. Relative expression levels and fold changes were log2 transformed for visual evaluation. The data are from three independent experiments (means ± SEM). **P = 0.001–0.01; ***P < 0.001. c The cell morphology of uninfected, control, and TCERG1 (sh9) knockdown cells was evaluated by bright-field microscopy in the absence or presence of retinoic acid. Scale bar, 20 μm. A higher magnification of individual cells is also shown. Scale bar, 10 μm. d The graphs show the quantification of the total neurite length and soma area from three independent experiments (n = 100 for each condition) (means ± SEM). **P = 0.001–0.01; ***P < 0.001
We sought to corroborate these data by assessing the cytoskeletal organization in neurons by immunostaining with phalloidin, which binds specifically between F-actin polymer subunits. Alterations in cytoskeleton structural components, such as actin filaments and microtubules, affect neuronal morphology and neurite elongation and guidance [56]. Consistent with our previous results, the cells were more rounded and had shorter neurites (Fig. 5a–f). We corroborated these data upon neuronal differentiation induced by retinoic acid treatment (Fig. 5g–l). A different shRNA against TCERG1 (sh4) led to similar defects in neurite morphology (Fig. S5). These results suggest that TCERG1 is important for the organization of the actin cytoskeleton, which is required for neurite projection and outgrowth during neuron development.
TCERG1 depletion results in severe defects in the actin cytoskeleton and neurite outgrowth of SH-SY5Y neuroblastoma cells. Uninfected, shControl and shTCERG1 cells were stained with phalloidin-rhodamine to visualize the actin cytoskeleton in the absence (a to f) or presence (g to l) of retinoic acid. DAPI labeling was used to stain the nuclei. Scale bar, 50 μm
To confirm that the observed dendritic outgrowth defect was a direct effect of TCERG1 knockdown, we examined whether the cell alteration could be rescued by introducing an epitope-tagged TCERG1 expression vector into the depleted cells. shRNA-resistant, EGFP-tagged TCERG1 (Fig. 6a) was coexpressed in the SH-SY5Y cell lines. Western blot analysis confirmed expression of the tagged TCERG1 protein (Fig. 6b). The band corresponding to the ectopically expressed protein appears very faint when using TCERG1-specific antibodies because of the low transfection efficiency (~20%) obtained with these constructs (Fig. 6b). We observed that EGFP expression did not change the cellular structure of the TCERG1-depleted cells, which remained rounded and had shorter neurites (Fig. 6c, EGFP). Interestingly, a significant restoration of cellular morphology was observed in the TCERG1-depleted cells upon TCERG1 re-expression, resulting in significantly longer dendrites (Fig. 6c, TCERG1). Quantification of the data confirmed the restoration of the dendrite length in wild-type TCERG1-rescued cells compared to the control cells (Fig. 6d). These results show that TCERG1 expression can compensate for the functional deficits caused by TCERG1 depletion. Thus, endogenous TCERG1 regulates neurite outgrowth and elongation in these cells.
TCERG1 rescues the neuron morphological defects. a Schematic representation of the silent mutations in mutant TCERG1 (mut-TCERG1) that prevented its knockdown by sh9. b Western blot of samples of cells transiently transfected with control (EGFP) or sh-resistant TCERG1-EGFP expression vectors. The antibodies used are listed on the right. The asterisk indicates the position of the transiently overexpressed protein. Lanes from the original blot were removed for presentation purposes. c Immunofluorescence of TCERG1-knockdown SH-SY5Y cells transiently transfected with the control (EGFP) and sh-resistant TCERG1-EGFP expression vectors. White arrowheads indicate examples of longer dendrites observed in the rescued cells. Green, EGFP fluorescence; red, phalloidin-rhodamine staining; blue, DAPI staining. Images representative of triplicate experiments are shown. Scale bar, 50 μm. d The graph shows the quantification of the total neurite length from three independent experiments (green channel, EGFP; n = 60, cells for each condition) (means ± SEM). ***P < 0.001
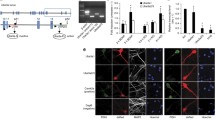
According to The Human Protein Atlas (http://www.proteinatlas.org/), human TCERG1 is widely expressed in the brain (cerebral cortex, hippocampus, lateral ventricle, and cerebellum). Murine TCERG1 is also expressed in the brain and hippocampal neurons as assessed by retrieval of its profile from quantitative expression databases of the adult mouse brain (http://brainstars.org/ and http://mouse.brain-map.org/). As further confirmation of the effects of TCERG1 on dendritic outgrowth, we knocked down TCERG1 expression in primary neuronal cultures derived from mouse hippocampi. Although the sequence of shRNA (sh9) is conserved between humans and mice, we first confirmed its capacity to knock down expression of TCERG1 in C2C12 murine cells prior to transfection into primary cultures (data not shown). Next, hippocampal neurons were transfected with a shRNA targeting TCERG1 or the control shRNA together with a GFP vector. RT-qPCR analysis confirmed inhibition of the TCERG1 transcript upon lentiviral transduction (Fig. S6). We observed that neurons with reduced TCERG1 levels showed a general alteration in cellular morphology, with substantially diminished dendrite branching (Fig. 7a). Quantification of the changes in neurite outgrowth confirmed the reduced total neurite length in the shTCERG1-treated cells compared with cells expressing the control shRNA and a reduction in the complexity of the dendrite ramification and branching near the soma (Fig. 7b–d). These results confirmed our previous data and indicate that TCERG1 is essential for proper neurite projection and outgrowth during neuritogenesis in cell culture. Thus, the absence of TCERG1 may lead to defects in neuronal functions.
TCERG1 affects dendritic outgrowth and branching in mouse hippocampal neurons. a Immunofluorescence of individual neurons that were transiently transfected with either the non-targeting control shRNA (shControl) or the TCERG1-targeting shRNA (sh9) together with a GFP vector. The morphology of the transfected cells was detected based on the GFP signal (green). Scale bar, 25 μm. b The graph shows the quantification of the total neurite length and total branch number from three independent experiments (n = 24 and 22 for shControl and sh9-transfected neurons, respectively); hippocampal neurons were transfected at 3 DIV and fixed at 7 DIV (means ± SEM). **P = 0.001–0.01; ***P < 0.001. c Sholl analysis of dendritic branching. Concentric circles were drawn at intervals of 5 μm. The number of dendritic branch interactions within each concentric circle was counted. The abscissa depicts the distance of the concentric circle from the neuron soma center, and the ordinate depicts the number of branch points. Mean values between 15 and 90 μm are highlighted with a line. ***P < 0.001. d Automated centrifugal labeling of neurites transiently transfected with the shControl and shTCERG1 was performed using NeuronStudio software. The graph shows the number of branches for each order of a branching structure. Note the marked decrease in branching complexity between branch order 3 and 12, which is labeled with a line. ***P < 0.001
Discussion
In this study, we showed that the transcription/splicing and disease-associated factor TCERG1 plays a role in cytoskeleton dynamics and neurite outgrowth. Following down-regulation of TCERG1, we observed that cells derived from the nervous lineage possess a greater proportion of TCERG1-dependent differentially expressed genes. Specifically, mRNA profiling of TCERG1-depleted cells revealed preferential misregulation of alternative transcriptional or splicing in factors that are important for proper morphogenesis and neurite outgrowth. Accordingly, defective morphogenesis and neurite outgrowth were observed in SH-SY5Y cells and primary neuronal cultures upon TCERG1 depletion. Collectively, our data indicate a role for TCERG1 in the synthesis and processing of pre-mRNA of important neuronal genes and suggest that alterations in alternative transcriptional or splicing events regulated by TCERG1 are involved in the neurological disorders associated with this factor.
We used the Affymetrix Human Exon 1.0 ST Array, which contains approximately 5.4 million probes grouped into 1.4 million probe sets for interrogating over 1 million exon clusters, to identify genes that are differentially expressed in normal TCERG1-depleted HEK293T cells. Through combining the exon array with a powerful commercial software for gene expression analysis, we persuasively identified expression variations in 414 human genes. We observed that TCERG1 depletion changed the overall transcript levels of 128 genes. Eighty-four percent of these genes were down-regulated, which was consistent with the positive regulation of the transcription of these endogenous targets by TCERG1. Initially, TCERG1 was identified as an elongation factor that represses RNAPII transcription, although in a specific fashion, i.e., it does not repress the transcription of all genes [30]. Consistently, TCERG1 acts as a co-repressor that inhibits the transcriptional activities of the CEBPA protein [18]. TCERG1 also acts as an activator of transcription [21]. These data suggest that TCERG1 is able to repress or activate genes, depending on the cell type context and/or gene sequence. Nevertheless, our study establishes that TCERG1 is not a general elongation factor because most genes are not transcriptionally sensitive to its depletion. Another finding from our analysis corroborates the functional link between TCERG1 and splicing in vivo . We observed that TCERG1 knockdown changed the alternative transcriptional or splicing patterns of 314 genes, most of which were up-regulated, particularly in the cases of alternative last and first exons and alternative 5′ splice sites (72, 80, and 94%, respectively). Furthermore, our data suggest that TCERG1 alters the use of alternative last exons (approximately 23% of neuron-specific genes compared to a prevalence of approximately 7% of this splicing event in the genome as assessed by retrieving alternatively splicing events from the ENCODE data at UCSC), which might lead to shorter or longer UTRs that potentially influence mRNA stability by changing miRNA and/or polyadenylation binding sites, and/or modify the fate of the nascent transcript by nonsense-mediated decay [57, 58]. Finally, previous studies concluded that TCERG1 regulates alternative splicing by changing the transcriptional capabilities of RNAPII [22]; however, our data indicate that only a limited number of genes are regulated through coupling between transcription and splicing, at least in the HEK293T cells used here. A previous report used classical GeneChip arrays to identify TCERG1 targets in HEK293T and HeLa cells [20]. A relatively small number of the targets (31 genes) identified in this previous work were also targets in our study. Possible explanations for these discrepancies include the reduced gene content coverage of this earlier study that was not able to detect a wide range of alternative splicing events [59], the different experimental silencing conditions, and/or the use of different bioinformatics approaches. Interestingly, a significant number of genes from this previous genome-wide analysis related to cytoskeleton dynamics were also observed in this previous study [20].
Abnormalities in the dendritic tree and in the synaptic spine morphology and density are the origin of several neurodegenerative disorders [8, 60, 61]. Cytoskeletal organization is widely believed to be key to the formation and loss of dendrites and dendrite spines [56, 62, 63]. In our analysis, we found many TCERG1-regulated genes with enriched CNS expression to be related to morphogenesis, cytoskeleton, and cell dynamics: actin-related genes, such as the actin-binding protein ACTN2 (Actinin), ABLIM1 (Actin-Binding LIM Protein 1), and PAK3 and SRGAP3 (Rho-GTPase signaling); microtubule-related proteins, such as TUBB2B (Tubulin, Beta 2B Class IIb), MAP2 and MAP7 (Microtubule-Associated Proteins 2 and 7, respectively); and linker proteins between the inner cytoskeleton and the extracellular matrix, such as ANK2 (Ankyrin 2), DMD (Dystrophin), CLASP2 (Cytoplasmic Linker Associated Protein 2), and EFEMP1 (EGF Containing Fibulin-Like Extracellular Matrix). Notably, more than half of the endogenous CNS gene targets (56%, 23 genes) affected by TCERG1 depletion were cytoskeleton components. Based on our data, a possible mechanism for the TCERG1 knockdown-mediated disruption of neurite outgrowth is reliant on alterations in the cytoskeletal organization, e.g., in the polymerization and/or stability of actin filaments, which are critical for the regulation of neuronal morphogenesis and structural plasticity ([63] and references herein). Microtubule cytoskeleton organization and regulation of cell substrate adhesion are also potential targets contributing to the observed phenotype. Synaptic plasticity through cytoskeletal remodeling can also be achieved by the action of factors regulating other important neuronal functions, such as the transport, localization, and translation of mRNA granules. UHMK1 (KIS) is a protein kinase with a unique RNA binding domain that localizes to RNA granules and enhances local mRNA translation [64]. UHMK1 plays important roles in dendritic spine morphology and dendritic arborization by affecting the tubulin cytoskeleton [29], which controls actin polymerization within the spines [65]. Interestingly, TCERG1 depletion down-regulated UHMK1 transcript levels and provoked an increase in the inclusion of its second exon, which possesses a longer alternative 5′UTR. Our initial efforts to identify the target genes responsible for the phenotype observed upon TCERG1 depletion failed, and further experimentation involving a genome-wide complementation assay is required (data not shown). However, we are aware that TCERG1 could be affecting gene targets direct or indirectly. We observed that TCERG1 depletion affects the expression of several transcription and splicing factors as well as critical regulators of the miRNA pathway, which may change the expression levels of other downstream gene targets. Further work involving each of the endogenous genes that was affected by the depletion of TCERG1 and that was detected in our assay will distinguish between both effects. Nevertheless, coexpression of TCERG1 in the knockdown cells compensates the functional deficits caused by TCERG1 depletion, strongly suggesting that endogenous TCERG1 regulates neurite outgrowth and elongation.
Analysis of the morphological parameters of the shTCERG1 primary cultures showed a decrease in total neurite length, the number of dendritic ramifications up to approximately 40 μm from the soma, and less complex ramifications than control cells; similarly, we did not observe significant differences in neurite branches at longer distances. Although our analysis was not separately performed on dendrites and axons, these results suggest that TCERG1 regulates overall dendrite ramification and branching complexity near the soma in hippocampus neurons. Axon length and branching at long distances do not seem to be affected by TCERG1 depletion. Importantly, changes in the dendritic arborization of pyramidal neurons affect their physiological properties and may lead to many brain disorders [8].
Several findings suggest that TCERG1 may be involved in HD pathogenesis: It binds to HTT, accumulates in striatal and cortical neurons as the disease grade increases, rescues striatal cell death for mutant HTT neurotoxicity, and may modify the age of HD onset [25, 26, 66, 67]. Abnormal interaction of misfolded mutant HTT species with TCERG1, as is the case for CREB binding protein (CREB), Sp1, and TAFII 130 [68–70], could affect the expression of important TCERG1-regulated genes, and this may be central to HD pathogenesis. Such rescue by TCERG1 is consistent with the restoration of transcriptional/splicing activation. There are also several reports showing the amyloidogenic properties of the second WW domain of human TCERG1 [71–74]. These comprehensive structural data may explain the deposits observed during HD and other neurodegenerative pathologies, such as Parkinson’s disease and Alzheimer’s disease. Finally, the article by Gau et al. [75] suggested a relationship between TCERG1 deficiency and cognitive impairment. These authors identified a microdeletion at the TCERG1 chromosomal location together with a microduplication on a different chromosome in a male patient with autism. Although more genes are present within the altered chromosomal regions and because of the previous studies linking TCERG1 with HD, the authors proposed that TCERG1 might play an important role in neurons, and that haploinsufficiency of this gene might have relevance to the clinical phenotypes of the patient.
Gaining insights into the molecular mechanisms by which factors such as TCERG1 regulate the morphology and morphological plasticity of dendrites may help to elucidate their role and function in normal and disease states.
Abbreviations
- TCERG1:
-
Transcription elongation regulator 1
- HD:
-
Huntington’s disease
- RNAPII:
-
RNA polymerase II;
- qPCR:
-
Quantitative PCR
References
Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ (2008) Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40:1413–1415
Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C et al (2008) Alternative isoform regulation in human tissue transcriptomes. Nature 456:470–476
Calarco JA, Zhen M, Blencowe BJ (2011) Networking in a global world: establishing functional connections between neural splicing regulators and their target transcripts. RNA 17:775–791
Kelemen O, Convertini P, Zhang Z, Wen Y, Shen M, Falaleeva M et al (2013) Function of alternative splicing. Gene 514:1–30
Licatalosi DD, Darnell RBRNA Processing and its regulation: global insights into biological networks. Nat Rev Genet 11:75–87
Singh RK, Cooper TA (2012) Pre-mRNA splicing in disease and therapeutics. Trends Mol Med 18:472–482
Cooper TA, Wan L, Dreyfuss G (2009) RNA and disease. Cell 136:777–793
Kulkarni VA, Firestein BL (2012) The dendritic tree and brain disorders. Mol Cell Neurosci 50:10–20
Lenzken SC, Achsel T, Carri MT, Barabino SM (2014) Neuronal RNA-binding proteins in health and disease. Wiley Interdiscip Rev RNA 5:565–576
Ward AJ, Cooper TA (2010) The pathobiology of splicing. J Pathol 220:152–163
Suñé C, Hayashi T, Liu Y, Lane WS, Young RA, Garcia-Blanco MA (1997) CA150, a nuclear protein associated with the RNA polymerase II holoenzyme, is involved in tat-activated human immunodeficiency virus type 1 transcription. Mol Cell Biol 17:6029–6039
Goldstrohm AC, Albrecht TR, Suñé C, Bedford MT, Garcia-Blanco MA (2001) The transcription elongation factor CA150 interacts with RNA polymerase II and the pre-mRNA splicing factor SF1. Mol Cell Biol 21:7617–7628
Lin KT, Lu RM, Tarn WY (2004) The WW domain-containing proteins interact with the early spliceosome and participate in pre-mRNA splicing in vivo. Mol Cell Biol 24:9176–9185
Sánchez-Álvarez M, Goldstrohm AC, Garcia-Blanco MA, Suñé C (2006) Human transcription elongation factor CA150 localizes to splicing factor-rich nuclear speckles and assembles transcription and splicing components into complexes through its amino and carboxyl regions. Mol Cell Biol 26:4998–5014
Carty SM, Goldstrohm AC, Sune C, Garcia-Blanco MA, Greenleaf AL (2000) Protein-interaction modules that organize nuclear function: FF domains of CA150 bind the phosphoCTD of RNA polymerase II. Proc Natl Acad Sci U S A 97:9015–9020
Smith MJ, Kulkarni S, Pawson T (2004) FF domains of CA150 bind transcription and splicing factors through multiple weak interactions. Mol Cell Biol 24:9274–9285
Sánchez-Hernández N, Ruiz L, Sánchez-Álvarez M, Montes M, Macias MJ, Hernández-Munain C et al (2012) The FF4 and FF5 domains of transcription elongation regulator 1 (TCERG1) target proteins to the periphery of speckles. J Biol Chem 287:17789–17800
McFie PJ, Wang GL, Timchenko NA, Wilson HL, Hu X, Roesler WJ (2006) Identification of a co-repressor that inhibits the transcriptional and growth-arrest activities of CCAAT/enhancer-binding protein alpha. J Biol Chem 281:18069–18080
Cheng D, Cote J, Shaaban S, Bedford MT (2007) The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol Cell 25:71–83
Pearson JL, Robinson TJ, Munoz MJ, Kornblihtt AR, Garcia-Blanco MA (2008) Identification of the cellular targets of the transcription factor TCERG1 reveals a prevalent role in mRNA processing. J Biol Chem 283:7949–7961
Sánchez-Álvarez M, Montes M, Sánchez-Hernández N, Hernández-Munain C, Suñé C (2010) Differential effects of sumoylation on transcription and alternative splicing by transcription elongation regulator 1 (TCERG1). J Biol Chem 285:15220–15233
Montes M, Cloutier A, Sánchez-Hernández N, Michelle L, Lemieux B, Blanchette M et al (2012) TCERG1 regulates alternative splicing of Bcl-x gene by modulating the rate of RNAPII transcription. Mol Cell Biol 32:751–762
Goldstrohm AC, Greenleaf AL, Garcia-Blanco MA (2001) Co-transcriptional splicing of pre-messenger RNAs: considerations for the mechanism of alternative splicing. Gene 277:31–47
Liu J, Fan S, Lee CJ, Greenleaf AL, Zhou P (2013) Specific interaction of the transcription elongation regulator TCERG1 with RNA polymerase II requires simultaneous phosphorylation at Ser2, Ser5, and Ser7 within the carboxyl-terminal domain repeat. J Biol Chem 288:10890–10901
Holbert S, Denghien I, Kiechle T, Rosenblatt A, Wellington C, Hayden MR et al (2001) The Gln-Ala repeat transcriptional activator CA150 interacts with huntingtin: neuropathologic and genetic evidence for a role in Huntington’s disease pathogenesis. Proc Natl Acad Sci U S A 98:1811–1816
Arango M, Holbert S, Zala D, Brouillet E, Pearson J, Regulier E et al (2006) CA150 expression delays striatal cell death in overexpression and knock-in conditions for mutant huntingtin neurotoxicity. J Neurosci 26:4649–4659
de la Grange P, Gratadou L, Delord M, Dutertre M, Auboeuf D (2010) Splicing factor and exon profiling across human tissues. Nucleic Acids Res 38:2825–2838
Coiras M, Montes M, Montanuy I, Lopez-Huertas MR, Mateos E, Le Sommer C et al (2013) Transcription elongation regulator 1 (TCERG1) regulates competent RNA polymerase II-mediated elongation of HIV-1 transcription and facilitates efficient viral replication. Retrovirology 10:124
Pedraza N, Ortiz R, Cornado A, Llobet A, Aldea M, Gallego C (2014) KIS, a kinase associated with microtubule regulators, enhances translation of AMPA receptors and stimulates dendritic spine remodeling. J Neurosci 34:13988–13997
Suñé C, Garcia-Blanco MA (1999) Transcriptional cofactor CA150 regulates RNA polymerase II elongation in a TATA-box-dependent manner. Mol Cell Biol 19:4719–4728
de la Grange P, Dutertre M, Correa M, Auboeuf D (2007) A new advance in alternative splicing databases: from catalogue to detailed analysis of regulation of expression and function of human alternative splicing variants. BMC Bioinformatics 8:180
de la Grange P, Dutertre M, Martin N, Auboeuf D (2005) FAST DB: a website resource for the study of the expression regulation of human gene products. Nucleic Acids Res 33:4276–4284
Huang da W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57
Huang da W, Sherman BT, Lempicki RA (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37:1–13
Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JA (2007) Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res 35:W71–W74
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108
Wearne SL, Rodriguez A, Ehlenberger DB, Rocher AB, Henderson SC, Hof PR (2005) New techniques for imaging, digitization and analysis of three-dimensional neural morphology on multiple scales. Neuroscience 136:661–680
Gardina PJ, Clark TA, Shimada B, Staples MK, Yang Q, Veitch J et al (2006) Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array. BMC Genomics 7:325
Amit M, Donyo M, Hollander D, Goren A, Kim E, Gelfman S et al (2012) Differential GC content between exons and introns establishes distinct strategies of splice-site recognition. Cell Rep 1:543–556
Jonkers I, Kwak H, Lis JT (2014) Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. Elife 3:e02407
Veloso A, Kirkconnell KS, Magnuson B, Biewen B, Paulsen MT, Wilson TE et al (2014) Rate of elongation by RNA polymerase II is associated with specific gene features and epigenetic modifications. Genome Res 24:896–905
Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH (2009) An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol 16:130–137
Benita Y, Cao Z, Giallourakis C, Li C, Gardet A, Xavier RJ (2010) Gene enrichment profiles reveal T-cell development, differentiation, and lineage-specific transcription factors including ZBTB25 as a novel NF-AT repressor. Blood 115:5376–5384
Graham FL, Smiley J, Russell WC, Nairn R (1977) Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol 36:59–74
Shaw G, Morse S, Ararat M, Graham FL (2002) Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J 16:869–871
He B, Soderlund DM (2010) Human embryonic kidney (HEK293) cells express endogenous voltage-gated sodium currents and Na v 1.7 sodium channels. Neurosci Lett 469:268–272
Vetter I, Lewis RJ (2010) Characterization of endogenous calcium responses in neuronal cell lines. Biochem Pharmacol 79:908–920
Fu Z, Washbourne P, Ortinski P, Vicini S (2003) Functional excitatory synapses in HEK293 cells expressing neuroligin and glutamate receptors. J Neurophysiol 90:3950–3957
Kim S, Burette A, Chung HS, Kwon SK, Woo J, Lee HW et al (2006) NGL family PSD-95-interacting adhesion molecules regulate excitatory synapse formation. Nat Neurosci 9:1294–1301
Mah W, Ko J, Nam J, Han K, Chung WS, Kim E (2010) Selected SALM (synaptic adhesion-like molecule) family proteins regulate synapse formation. J Neurosci 30:5559–5568
Woo J, Kwon SK, Choi S, Kim S, Lee JR, Dunah AW et al (2009) Trans-synaptic adhesion between NGL-3 and LAR regulates the formation of excitatory synapses. Nat Neurosci 12:428–437
Campbell SA, Lin J, Dobrikova EY, Gromeier M (2005) Genetic determinants of cell type-specific poliovirus propagation in HEK 293 cells. J Virol 79:6281–6290
Jahan N, Wimmer E, Mueller S (2011) A host-specific, temperature-sensitive translation defect determines the attenuation phenotype of a human rhinovirus/poliovirus chimera, PV1(RIPO). J Virol 85:7225–7235
Madhusudana SN, Sundaramoorthy S, Ullas PT (2010) Utility of human embryonic kidney cell line HEK-293 for rapid isolation of fixed and street rabies viruses: comparison with Neuro-2a and BHK-21 cell lines. Int J Infect Dis 14:e1067–e1071
Pahlman S, Hoehner JC, Nanberg E, Hedborg F, Fagerstrom S, Gestblom C et al (1995) Differentiation and survival influences of growth factors in human neuroblastoma. Eur J Cancer 31A:453–458
Hotulainen P, Hoogenraad CC (2010) Actin in dendritic spines: connecting dynamics to function. J Cell Biol 189:619–629
McGlincy NJ, Smith CW (2008) Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends Biochem Sci 33:385–393
Jia J, Yao P, Arif A, Fox PL (2013) Regulation and dysregulation of 3’UTR-mediated translational control. Curr Opin Genet Dev 23:29–34
Ha K, Coulombe-Huntington J, Majewski J (2009) Comparison of affymetrix gene array with the exon array shows potential application for detection of transcript isoform variation. BMC Genomics 10:519
Parrish JZ, Emoto K, Kim MD, Jan YN (2007) Mechanisms that regulate establishment, maintenance, and remodeling of dendritic fields. Annu Rev Neurosci 30:399–423
Kasai H, Fukuda M, Watanabe S, Hayashi-Takagi A, Noguchi J (2010) Structural dynamics of dendritic spines in memory and cognition. Trends Neurosci 33:121–129
Shirao T, Gonzalez-Billault C (2013) Actin filaments and microtubules in dendritic spines. J Neurochem 126:155–164
Luo L (2002) Actin cytoskeleton regulation in neuronal morphogenesis and structural plasticity. Annu Rev Cell Dev Biol 18:601–635
Cambray S, Pedraza N, Rafel M, Gari E, Aldea M, Gallego C (2009) Protein kinase KIS localizes to RNA granules and enhances local translation. Mol Cell Biol 29:726–735
Jaworski J, Kapitein LC, Gouveia SM, Dortland BR, Wulf PS, Grigoriev I et al (2009) Dynamic microtubules regulate dendritic spine morphology and synaptic plasticity. Neuron 61:85–100
Andresen JM, Gayan J, Cherny SS, Brocklebank D, Alkorta-Aranburu G, Addis EA et al (2007) Replication of twelve association studies for Huntington’s disease residual age of onset in large Venezuelan kindreds. J Med Genet 44:44–50
Chattopadhyay B, Ghosh S, Gangopadhyay PK, Das SK, Roy T, Sinha KK et al (2003) Modulation of age at onset in Huntington’s disease and spinocerebellar ataxia type 2 patients originated from eastern India. Neurosci Lett 345:93–96
Dunah AW, Jeong H, Griffin A, Kim YM, Standaert DG, Hersch SM et al (2002) Sp1 and TAFII130 transcriptional activity disrupted in early Huntington’s disease. Science 296:2238–2243
Steffan JS, Kazantsev A, Spasic-Boskovic O, Greenwald M, Zhu YZ, Gohler H et al (2000) The Huntington’s disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc Natl Acad Sci U S A 97:6763–6768
Shimohata T, Nakajima T, Yamada M, Uchida C, Onodera O, Naruse S et al (2000) Expanded polyglutamine stretches interact with TAFII130, interfering with CREB-dependent transcription. Nat Genet 26:29–36
Nguyen H, Jager M, Moretto A, Gruebele M, Kelly JW (2003) Tuning the free-energy landscape of a WW domain by temperature, mutation, and truncation. Proc Natl Acad Sci U S A 100:3948–3953
Ferguson N, Berriman J, Petrovich M, Sharpe TD, Finch JT, Fersht AR (2003) Rapid amyloid fiber formation from the fast-folding WW domain FBP28. Proc Natl Acad Sci U S A 100:9814–9819
Ferguson N, Becker J, Tidow H, Tremmel S, Sharpe TD, Krause G et al (2006) General structural motifs of amyloid protofilaments. Proc Natl Acad Sci U S A 103:16248–16253
Tartaglia GG, Pawar AP, Campioni S, Dobson CM, Chiti F, Vendruscolo M (2008) Prediction of aggregation-prone regions in structured proteins. J Mol Biol 380:425–436
Gau SS, Liao HM, Hong CC, Chien WH, Chen CH (2012) Identification of two inherited copy number variants in a male with autism supports two-hit and compound heterozygosity models of autism. Am J Med Genet B Neuropsychiatr Genet 159B:710–717
Acknowledgements
We are grateful to many colleagues for their helpful suggestions, critical discussions, and comments. The technical assistance of Laura Montosa and Eduardo Andrés-León during the confocal microscopy and bioinformatic studies is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Funding
This work was supported by grants from the Spanish Ministry of Economy and Competitiveness (grant number BFU2014–54660-R) and the Andalusian Government (Excellence Project BIO-2515/2012) to C.S. and from the Spanish Ministry of Economy and Competitiveness (grant number BFU2013–44660-R) and the Andalusian Government (Excellence Project CTS-6587) to C.H.M. Support from the European Region Development Fund (ERDF [FEDER]), is also acknowledged.
Rights and permissions
About this article
Cite this article
Muñoz-Cobo, J.P., Sánchez-Hernández, N., Gutiérrez, S. et al. Transcriptional Elongation Regulator 1 Affects Transcription and Splicing of Genes Associated with Cellular Morphology and Cytoskeleton Dynamics and Is Required for Neurite Outgrowth in Neuroblastoma Cells and Primary Neuronal Cultures. Mol Neurobiol 54, 7808–7823 (2017). https://doi.org/10.1007/s12035-016-0284-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-016-0284-6