Abstract
Cyclin-dependent kinase 5 regulatory subunit 1 (CDK5R1) encodes p35, the main activatory subunit of cyclin-dependent kinase 5 (CDK5). The p35/CDK5 active complex plays a fundamental role in brain development and functioning, but its deregulated activity has also been implicated in various neurodegenerative disorders, including Alzheimer’s disease (AD). CDK5R1 displays a large and highly evolutionarily conserved 3′-untranslated region (3′-UTR), a fact that has suggested a role for this region in the post-transcriptional control of CDK5R1 expression. Our group has recently demonstrated that two miRNAs, miR-103 and miR-107, regulate CDK5R1 expression and affect the levels of p35. MiR-103 and miR-107 belong to the miR-15/107 family, a group of evolutionarily conserved miRNAs highly expressed in human cerebral cortex. In this work, we tested the hypothesis that other members of this group of miRNAs, in addition to miR-103 and miR-107, were able to modulate CDK5R1 expression. We provide evidence that several miRNAs belonging to the miR-15/107 family regulate p35 levels. BACE1 expression levels were also found to be modulated by different members of this family. Furthermore, overexpression of these miRNAs led to reduced APP phosphorylation levels at the CDK5-specific Thr668 residue. We also show that miR-15/107 miRNAs display reduced expression levels in hippocampus and temporal cortex, but not in cerebellum, of AD brains. Moreover, increased CDK5R1 mRNA levels were observed in AD hippocampus tissues. Our results suggest that the downregulation of the miR-15/107 family might have a role in the pathogenesis of AD by increasing the levels of CDK5R1/p35 and consequently enhancing CDK5 activity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cyclin-dependent kinase 5 regulatory subunit 1 (CDK5R1) encodes p35, a protein required for the activation of the serine/threonine kinase cyclin-dependent kinase 5 (CDK5). Through the activation of CDK5, p35 plays a critical role in several physiological processes in both embryonic and adult brain, including neuron maintenance and migration, cytoskeletal dynamics, neurite outgrowth, synaptogenesis, synaptic plasticity, behavior, and cognition [1]. In particular, the CDK5/p35 complex is essential for proper radial migration of neurons during the development of the mammalian cortex. In fact, mice lacking Cdk5 or p35 display severe defects in the migration and positioning of neurons and axonal fiber tracts [2, 3].
If the p35/CDK5 complex is essential for the correct development and functioning of the central nervous system (CNS) under normal physiology, CDK5-deregulated activity under pathological conditions has been implicated in various neurodegenerative disorders, particularly Alzheimer’s disease (AD). AD is a disorder causing a severe and permanent impairment of multiple cognitive faculties [4] which is having a huge social and economic impact on families and society. The incidence rate of AD among people 65 years of age and older is 19.4 and 15.0 per 1000 person-years in Europe and USA, respectively, but the incidence rate increases almost exponentially with increasing senility until 85 years of age [5]. There are two types of the pathology: familial and sporadic AD. The first one is a genetic condition with early onset, inherited in an autosomal dominant manner, which is caused by mutations in amyloid precursor protein (APP), presenilin 1 (PSEN1), or presenilin 2 (PSEN2) gene [6]. The second one is a late-onset complex condition whose molecular mechanisms are still poorly understood; some evidence shows a modest family clustering and interplay of genetic and environmental factors. The major genetic risk factor for this sporadic form is the presence of the epsilon 4 allele in the apolipoprotein E (APOE) gene, but recent genome-wide association studies and next-generation sequencing studies have successfully identified additional common genetic variations associated with late-onset AD [6].
Two main pathological hallmarks characterize AD: extracellular senile plaques, formed by the fibrillary β-amyloid (Aβ), and neurofibrillary tangles (NFTs), composed of the microtubule-associated protein Tau (encoded by the MAPT gene) in a hyperphosphorylated status [7]. CDK5 deregulation has been suggested to have a major role in the onset of these two neuropathological hallmarks. Under different stress conditions, including Aβ exposure, the regulatory subunit p35 is cleaved by the calcium-dependent protease calpain, generating a C-terminal truncated protein, p25, that retains the ability to bind the kinase and causes CDK5 hyperactivation and mislocalization [8, 9]. Noteworthy, mice overexpressing p25 show increased CDK5 activity and hyperphosphorylation of Tau and APP proteins [10, 11]. Furthermore, the silencing of CDK5 reduces the phosphorylation of Tau in primary neurons and the brain of wild-type mice and reduces neurofibrillary tangles in AD transgenic mice, indicating the involvement of CDK5 in Tau hyperphosphorylation [12]. In addition, APP phosphorylation by hyperactivated CDK5 at Thr668 makes it more prone to Aβ formation and has been proposed to represent an early event in AD [13, 14].
CDK5R1 (the gene encoding p35) displays a large and highly evolutionarily conserved 3′-untranslated region (3′-UTR), which has suggested a role for this region in the post-transcriptional control of CDK5R1 expression [15]. 3′-UTRs are known to function as post-transcriptional cis-regulatory elements that allow a precise spatio-temporal control of gene expression by modulating mRNA stability and translation efficiency through the binding of proteins and non-coding RNAs [16], notably miRNAs. MiRNAs are small regulatory RNAs with established roles in neurodevelopment, neuroplasticity, apoptosis, and other neurobiological processes [17, 18]. Accumulating evidence implicates miRNAs in the pathogenesis of neurodegenerative diseases, particularly AD [19–22].
Our group has recently demonstrated that two miRNAs, miR-103 and miR-107, regulate CDK5R1 expression and affect the levels of p35 [23], suggesting that miRNA-mediated mechanisms may influence CDK5 activity and the phosphorylation of its substrates, notably APP and Tau proteins. MiR-103 and miR-107 belong to the miR-15/107 family, a group of ten paralogous and evolutionarily conserved miRNAs sharing the identical AGCAGC “seed” sequence and known to regulate the expression of genes involved in cell division, metabolism, stress response, and angiogenesis [24]. The members of this family are highly expressed in human cerebral cortex [24, 25]. Recent evidence supports the hypothesis that miRNAs belonging to the miR-15/107 group are involved in AD pathogenesis. Expression levels of miR-15a and miR-107 are significantly reduced in AD brains compared to control brain samples [26–29]. Changes in miR-15a and miR-16 levels have been shown to affect Tau phosphorylation in primary cortical neurons [28]. Moreover, miR-16 directly targets APP [30], and miR-195 has been shown to regulate APP and BACE1 levels, as well as Aβ production [31, 32].
Therefore, besides the well-known paradigm of p25 production by calpain leading to CDK5 hyperactivity, other mechanisms might be evoked to explain CDK5 hyperactivation in AD, among them, the dysregulation of miRNAs that post-transcriptionally control CDK5R1 expression.
Since the members of the miR-15/107 family share an identical seed sequence, in this work we tested the hypothesis that other members of this group of miRNAs, in addition to miR-103 and miR-107, were able to modulate CDK5R1 expression. We provide evidence that the main and most conserved members of the miR-15/107 group regulate p35 levels. BACE1 expression levels were also found to be modulated by several members of this family. Furthermore, overexpression of these miRNAs led to reduced APP phosphorylation levels at the CDK5-specific Thr668 residue without affecting CDK5 cellular levels. We also show that miR-15/107 miRNAs display reduced expression levels in the hippocampus and temporal cortex, but not in the cerebellum, of AD brains compared to control samples. Moreover, increased CDK5R1 mRNA levels were observed in AD hippocampi, compared to controls. Our results suggest that increased levels of CDK5R1/p35 and consequent enhanced CDK5 activity caused by downregulation of the miR-15/107 family of miRNAs might be a further contributor to the pathogenesis of AD.
Materials and Methods
Cell Cultures
Human neuroblastoma SK-N-BE (2) cells (ATCC code CRL-271) were cultured in RPMI-1640 medium with 10 % fetal bovine serum (FBS), 100 U/ml penicillin-streptomycin, 0.01 mM L-glutamine, sodium pyruvate 11 g/l, and glucose 4.5 g/l. Human embryonic kidney HEK-293 cells (ATCC code CRL-1573) were maintained in DMEM medium with 10 % FBS, 100 U/ml penicillin-streptomycin, and 0.01 mM L-glutamine. Cultures were maintained at 37 °C in a 5 % CO2 incubator.
Brain Tissues
Post-mortem frozen brain tissue samples of AD patients (n = 12, age = 83 ± 8, m/f (%): 60/40) and age- and sex-matched non-demented individuals (n = 12) were obtained from MRC London Neurodegenerative Diseases Brain Bank (King’s College London), Newcastle Brain Tissue Resource (Newcastle University), and South West Dementia Brain Bank (University of Bristol). Tissue blocks were from hippocampus, anterior temporal cortex, and cerebellum, which are differently affected by AD pathology. All samples had a post-mortem interval (PMI) < 25 h (mean = 10, median = 10.5). The approval of the Ethics Committee of the University of Milan was obtained for the use of post-mortem tissues for research purposes (19.05.2011).
Pre-miRs and LNA Anti-miRs Transfection
The dsRNA miRNA precursors (pre-miRs) that mimic endogenous mature miRNAs (AM 17100: PM 10632 hsa-mir-103a-3p; PM 10339 hsa-mir-16-5p; PM 10235 hsa-mir-15a-5p) and the PremiR miRNA Precursor Molecules-Negative Control#1 (AM 17110) were purchased from Thermo Fisher Scientific (Waltham, MA, USA). LNA anti-miR oligonucleotides (mircury LNA inhibitors) were purchased from Exiqon (Vedbæk, Denmark). Each molecule inhibits one or two miRNAs: anti-miR-103/7 (hsa-miR-103 miRCURY LNA™ microRNA family, EX450028), anti-miR-15a/16, anti-miR-15b, anti-miR-195 (all three oligonucleotides included in hsa-miR-15 miRCURY LNA™ microRNA family, EX450022). Single-LNA anti-miRs were used at 5- or 25-nM concentration. LNA mixes were made using equal amounts of each anti-miR, to get a total concentration of 5 or 25 nM. The 125 × 103 SK-N-BE cells and 100 × 103 HEK-293 cells were seeded in 12-well plates, in order to extract total RNA (double quantity of cells was used in 6-well plates to extract proteins). The cells were transfected 24 h after seeding with pre-miRs (100 nM) or LNA anti-miR molecules (5 or 25 nM), using Lipofectamine 2000 (Thermo Fisher Scientific) transfection reagent according to the manufacturer’s instruction. Cell extracts were prepared for analysis 48 h after the transfection.
Real-Time PCR
Total RNA from transfected/nontransfected cells and from brain tissues (100 mg of each sample tissue was used) was isolated using TRIzol reagent (Thermo Fisher Scientific), according to the manufacturer’s instructions. Concentration and purity of RNA were measured using the Nanodrop spectrophotometer (Thermo Fisher Scientific). All RNA samples had an A260/A280 value of 1.8–2.1.
For the quantitative analysis of mature miRNAs, a two-step Taq-Man real-time PCR assay was performed, using primers and probes obtained from Thermo Fisher Scientific. cDNA was synthesized from total RNA (50 ng) in 15-μl reactions, using reverse transcriptase and the stem–loop primer for miR-15a (ID000389), miR-15b (ID000390), miR-16 (ID000391), miR-103 (ID000439), miR-107 (ID000443), miR-195 (ID000494), miR-34a (ID000426), or RNU6B (endogenous control as recommended by Thermo Fisher Scientific; ID001093) contained in the TaqMan MicroRNA Reverse Transcription kit (Thermo Fisher Scientific). The reverse transcriptase reaction was performed by incubating the samples at 16 °C for 30 min, 42 °C for 30 min, and 85 °C for 5 min. The PCR reaction (20 μl) contained 1.3 μl of reverse transcriptase product, 10 μl of Taq-Man Universal PCR Master Mix, and 1 μl of the appropriate TaqMan MicroRNA Assay (20×) containing primers and probes for the miR of interest. The PCR mixtures were incubated at 95 °C for 10 min, and this was followed by 40 cycles of 95 °C for 15 s and 60 °C for 60 s. The expression of miRs was based on the ΔCT method, using RNU6B as an endogenous control.
For the measurement of CDK5R1 and BACE1 mRNA, cDNA was synthesized from total RNA (1 μg) in 20-μl reactions using the High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). Real-Time PCR was performed using the SYBR Green PCR Master mixture (Promega, Madison, WI, USA) and the following primers: CDK5R1 fw: TGAGCGGGTCTAGTGGAAAG; CDK5R1 rev: AGCAGCAGACAAGGGGGTAG; BACE1 fw: GCAGGGCTACTACGTGGAGA; BACE1 rev: GTATCCACCAGGATGTTGAGC; CYC1 fw: TAGAGTTTGACGATGGCACCC, CYC1 rev: CGTTTTCGATGGTCGTGCTC; EIF4A2 fw: GGTCAGGGTCAAGTCGTGTT, EIF4A2 rev: CCCCCTCTGCCAATTCTGTG; SYP fw: CTTCGCCATCTTCGCCTTTG; SYP rev: TACACTTGGTGCAGCCTGAAG; GAPDH fw: TGCACCACCAACTGCTTAGC; GAPDH rev: GGCATGGACTGTGGTCATGAG. The PCR reactions (20 μl) were prepared with 2× SYBR Green mix containing 1 μl of reverse transcriptase product and 0.2 μl of each primer. The PCR mixtures were incubated at 95 °C for 3 min, followed by 40 cycles of 95 °C for 10 s, 60 °C for 20 s, and 72 °C for 10 s. The expression of CDK5R1 and BACE1 was based on the ΔΔCT method. The housekeeping gene GAPDH was used as internal control in pre-miR and anti-miR transfection experiments, while the housekeeping genes CYC1 and EIF4A2 and the neuronal marker SYP were used to normalize gene expression levels in brain tissues. All PCRs were performed in triplicate using an iQ5 Real-Time PCR detection system (Bio-Rad, Hercules, CA, USA).
Western Blotting
SK-N-BE and HEK-293 cells were homogenized in lysis buffer (150 mM NaCl, 20 mM Tris–HCl, pH 7.4, 1 % Triton X-100, protease inhibitor mixture). Samples were centrifuged at 12,000g for 15 min at 4 °C, and supernatants were collected.
Western blot assay was performed under standard conditions. In brief, equal amounts of proteins were analyzed by 12 % SDS-PAGE at 100 V for 90 min and blotted onto nitrocellulose membrane Hybond-enhanced chemiluminescence (ECL; Amersham Biosciences, Buckinghamshire, UK) in a Bio-Rad Trans-blot apparatus at 350 mA for 3 h. The blots were probed with anti-p35 (C-19, Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-BACE1 (D10E5, Cell Signaling, Beverly, MA), anti-CDK5 antibody (C-8, Santa Cruz Biotechnology), anti-pAPP (Thr668) (Cell Signaling, Danvers, CA, USA), anti-APP (Millipore, Billerica, MA, USA), and anti GAPDH (Novus biologicals, Littleton, CO, USA) antibodies followed by an alkaline phosphatase-conjugated IgG secondary antibody. Blots were processed by an ECL Plus detection kit as instructed by the supplier (Amersham Biosciences). All experiments were performed at least three times, and the quantization was made using the ImageJ program (National Institute of Health, Bethesda, MD, USA).
3′-UTR and “miR-sensor” Constructs
The Renilla luciferase pGL4.71P construct (Promega) containing the CDK5R1 3′-UTR was previously described [23]. For transfection efficiency normalization, the pGL3 firefly luciferase plasmid (Promega) was used.
For the generation of “miR-sensor” constructs, oligonucleotide pairs were designed to contain the desired mature miRNA sequence and, when annealed, overhangs complementary to those generated by XbaI digestion (Table S1). Once ligated into the pmirGLO Vector (Promega), the result is a region perfectly complementary to the mature miRNA in the 3′-UTR of the hRluc reporter gene. Both oligonucleotides were diluted to 1 μg/μl, and 2 μl of each oligonucleotide was combined with 46 μl of Oligo Annealing Buffer, heated at 90 °C for 3 min, and then transferred to a 37 °C water bath for 15 min. Annealed oligonucleotides were diluted 1:10 in nuclease-free water to a final concentration of 4 ng/μl per oligonucleotide, and 4 ng of annealed oligonucleotides were ligated with 50 ng of XbaI-linearized vector using a standard ligation protocol. The correct orientation of the insertion was verified by direct sequencing. This method was used to generate “sensor” luciferase constructs specific for each member of the miR-15/107 family (sensor-103, sensor-107, sensor-15a, sensor-15b, sensor-16, and sensor-195) and miR-34a (sensor-34a). The sensor-34a was chosen as a negative control because miR-34a is not expressed in SK-N-BE cells [23]. The same sensor constructs with a 4-base mutation inside the seed sequence were produced by direct mutagenesis, using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA) with specific oligonucleotides (Table S1), according to the protocol provided with the kit.
Luciferase Assay
For transient transfection of SK-N-BE cells with pre-miRs and the luciferase construct containing the entire human CDK5R1 3′-UTR, 125 × 103 cells were seeded in 12-well plates and then transfected with 100 nM pre-miRs, 24 h after seeding, and with the luciferase reporter construct (150 ng), 48 h after seeding, using Lipofectamine 2000 transfection reagent according to the manufacturer’s instruction. To normalize the value of Renilla luciferase activity for transfection efficiency and cell viability after transfection, we cotransfected the pGL3-Promoter Firefly luciferase reporter gene (150 ng). Seventy-two hours after seeding, cells were washed with PBS and lysed with passive lysis buffer (Promega), the Renilla and firefly luciferase activities were measured using the Dual-Glo Luciferase Assay System (Promega) in a single channel luminometer. Each experiment was performed at least three times.
For transient transfection of SK-N-BE with wild-type and mutated sensor constructs, 125 × 103 cells were seeded in 12-well plates and then transfected with 150 ng of the specific sensor construct, 24 h after seeding using Lipofectamine 2000 transfection reagent. pmiRGLO contains both Renilla and firefly reporter genes. Forty-eight hours after seeding, the luciferase activity was determined as described above.
Finally, 125 × 103 SK-N-BE cells were seeded in 12-well plates and then transfected with 150 ng of the specific wt sensor construct only, or cotransfected with 5 nM of the corresponding LNA anti-miR molecule. Forty-eight hours after anti-miR transfection and 24 h after sensor construct transfection, cellular lysates were subjected to luciferase assays, as described above.
Statistical Analysis
Each experiment was carried out three times, at least. Histograms represent the mean values, and bars indicate the standard deviation of the mean. The box plots show median, 25 and 75 percentile values, and whiskers to the minimum and maximum value. The statistical significance of the results was determined using Student’s t test, with data considered significant when p < 0.05. The degree of linear relationship between miRNA and gene expression levels was calculated using Pearson’s correlation coefficient (r value) using the mean values of three experiments. The p value is calculated from an extra sum-of-squares F test.
Results
miR-15/107 Family Modulates CDK5R1 Expression
We recently demonstrated that two miRNAs, miR-103 and miR-107, both belonging to the miR-15/107 family, regulate CDK5R1 expression through the binding to its 3′-UTR and affect the levels of p35 [23]. To verify whether the CDK5R1 mRNA and p35 cellular levels are also regulated by other members of the miR-15/107 group, we transiently transfected SK-N-BE and HEK-293 cells with the precursors of other two representative members of the miR-15/107 family: miR-15a and miR-16.
Total mRNA was extracted from SK-N-BE and HEK-293 cells 48 h after transfection with 100 nM pre-miR-15a, pre-miR-16, pre-miR-103 (as a positive control), or a scramble control. qRT-PCR analysis showed a decrease in CDK5R1 transcript levels after transfection with the miR-103 precursor in both cell lines, as expected [23], and after transfection with the other two precursors only in HEK-293 cells (Fig. 1a). Forty-eight hours after pre-miRNA transfection, a significant reduction of the p35 levels was also observed by Western blot analysis in both cell lines, compared to the scramble control (Fig. 1b).
miR-15/107 precursors reduce p35 levels. a SK-N-BE and HEK-293 cells were treated with 100 nM pre-miR-103, pre-miR-15a, pre-miR16, or a scramble control. After 48 h, total RNA was extracted, and the levels of CDK5R1 mRNA were measured with qRT-PCR and normalized on the housekeeping GAPDH gene. Results are compared with the cells treated with the scramble control. n = 5, mean ± s.d. b SK-N-BE or HEK-293 cells were treated as in A, and Western blot on p35 and GAPDH was performed on protein lysates. n = 5, mean ± s.d. c A pGL4.11 firefly luciferase construct containing the entire human CDK5R1 3′-UTR and a pGL3 Renilla luciferase plasmid as normalizer [23] were cotransfected with 100 nM miR-103/107, miR-15a, and miR-16 precursors in SK-N-BE cells, and the luciferase activity was measured by a dual luciferase assay after 72 h. Results are represented as firefly/Renilla luciferase activity. n = 5, mean ± s.d., *p < 0.05; **p < 0.01, Student’s t test
To investigate whether miR-15/107 miRNAs can act in a direct or indirect manner on CDK5R1 3′-UTR, luciferase assays were performed. A luciferase construct containing the entire human CDK5R1 3′-UTR [23] and a pGL3 firefly luciferase plasmid were co-transfected with 100 nM pre-miR-103, pre-miR-15a, or pre-miR-16 in SK-N-BE cells, and the luciferase activity was measured by a dual luciferase assay. The luciferase activity decreased following the co-transfection with all three precursors compared to the untransfected cells or to the cells transfected with pre-miR-34a, which has no target sites in CDK5R1 3′-UTR (Fig. 1c), demonstrating a direct action of these miRNAs on CDK5R1 3′-UTR.
To further analyze the effect of miR-15/107 family on CDK5R1 expression, we used locked nucleic acid (LNA) antisense oligonucleotides with perfect sequence complementarity to the main members of the miR-15/107 group to inhibit their endogenous activity. qRT-PCR on mature miRNAs is not a reliable method for measuring the level of miRNA knockdown by miRNA inhibitors, as they do not degrade their targets but rather form stable complexes with their targets causing an accumulation of the miRNA due to reduced turnover. For this reason, to test the efficacy of the LNA oligonucleotides in inhibiting endogenous miR-15/107 activity, we decided to generate sensor luciferase constructs by cloning a sequence perfectly complementary to the mature miRNA of interest downstream the firefly luciferase of the pmiR-GLO plasmid. The binding of the endogenous miRNA to the corresponding target sequence is expected to result in decreased luciferase activity, due to the repressive action of the miRNA. Indeed, we measured a significant luciferase activity reduction in SK-N-BE cells for every transfected pmiR-GLO-sensor, compared to the empty pmiR-GLO control vector, indicating that all the miRNAs of the miR-15/107 family tested have a significant endogenous activity in this cell line (Fig. S1a). On the contrary, no reduction in luciferase activity was observed for the miR-34a sensor, as expected, given that miR-34a is not expressed in SK-N-BE cells [23]. To ensure the specificity of miRNA binding to the sensors, we generated sensor constructs mutated in the seed region. The luciferase activity of the mutant constructs was increased in all cases (Fig. S1b).
To verify the efficacy of miRNA inhibition by antisense LNA molecules, we co-transfected each sensor with 5 nM of the corresponding LNA miRNA inhibitor (anti-miR) in SK-N-BE cells. In each case, we observed a significant increase of the luciferase activity (Fig. S1c), indicating an effective reduction of the miRNA repressive activity caused by the inhibitors.
Following the transfection of the miR-15/107 LNA inhibitors in SK-N-BE and HEK-293 cell lines, we extracted total RNA and proteins and performed qRT-PCR and Western blot experiments, to assess the levels of CDK5R1 mRNA and p35 after the treatment. In SK-N-BE cells, significantly augmented CDK5R1 mRNA levels were measured by qRT-PCR after the transfection with 5 nM anti-miR-103/107 and with 25 nM anti-miR-103/107, anti-miR-15a/16, and a mix including all the inhibitors (anti-miR-103/7-15a/16-15b-195), compared to an equal dose of control anti-miR (anti-miR-C-) (Fig. 2a). In HEK-293 cells, CDK5R1 transcript levels significantly increased following the transfection of 5 nM anti-miR-103/107, anti-miR-15a/16, a mix containing all the inhibitors except anti-miR-103/107 (anti-miR-15a/16-15b-195) and the mix containing all the inhibitors. At 25 nM dose, a significant increase of CDK5R1 mRNA levels was observed after the transfection with all single anti-miRs and mixes (Fig. 2a). Western blot analysis showed that the transfection of 5 nM anti-miR-103/107, anti-miR-15a/16, anti-miR-15b, or anti-miR-195, as well as of the inhibitor mixes, leads to a significant increase (up to two-fold) of p35 levels, compared to the control, in both cell lines (Fig. 2b).
miR-15/107 inhibition increases p35 levels. a SK-N-BE and HEK-293 cells were treated with 5 nM (dark gray) or 25 nM (light gray) anti-miR-C-scramble control, anti-miR-103/7, anti-miR-15a/16, anti-miR-15b, anti-miR-195, or a combination of anti-miRs. After 48 h, total RNA was extracted, and the levels of CDK5R1 mRNA were measured with qRT-PCR and normalized on the housekeeping GAPDH gene. Results are compared with the cells treated with the scramble control. n = 5, mean ± s.d. b SK-N-BE or HEK-293 cells were treated with 5 nM anti-miR as in A, and Western blot on p35 and GAPDH was performed on protein lysates. n = 5, mean ± s.d., *p < 0.05; **p < 0.01, Student’s t test
Taken together, these data support the hypothesis that several members of the miR-15/107 group of miRNAs regulate CDK5R1 expression.
miR-15/107 Family Modulates BACE1 Expression
BACE1 is a known target of miR-107 and miR-195 which is involved in the pathogenesis of AD [27, 31]. We tested the hypothesis that other members of the miR-15/107 family can regulate BACE1 expression. A reduction of the levels of BACE1 was observed by Western blot analysis following the transfection of SK-N-BE cells with 100 nM pre-miR-15a, pre-miR-16, and pre-miR-103, compared to the scramble control (Fig. 3a). Moreover, BACE1 protein levels increased after the transfection of the same cell line with 5 nM anti-miR-103/107, anti-miR-15a/16, anti-miR-15b, or anti-miR-195 inhibitors and inhibitor mixes (Fig. 3b). These experiments demonstrated that, in addition to miR-107 and miR-195, also other members of the miR-15/107 group of miRNAs regulate BACE1 expression.
miR-15/107 regulates BACE1 levels. a SK-N-BE cells were treated with 100 nM pre-miR-103, pre-miR-15a, pre-miR16, or a scramble control. After 48 h, total Western blot on BACE1 and GAPDH was performed on protein lysates. n = 5, mean ± s.d. b SK-N-BE cells were treated with 5 nM anti-miR-C-scramble control, anti-miR-103/7, anti-miR-15a/16, anti-miR-15b, anti-miR-195, or a combination of anti-miRs. After 48 h, total Western blot on BACE1 and GAPDH was performed on protein lysates. n = 5, mean ± s.d. *p < 0.05 **p < 0.01, Student’s t test
miR-15/107 miRNAs Do Not Influence CDK5 Expression But Its Kinase Activity
We also wanted to investigate whether miR-15/107 miRNAs can affect the levels of the CDK5 kinase. To test this hypothesis, we transfected SK-N-BE cells with 100 nM of the miR-15/107 precursors (pre-miR-15a, pre-miR-16, and pre-miR-103), extracted total proteins, and measured CDK5 levels by Western blot. We observed no change in CDK5 levels after the treatments, compared to the scramble control (Fig. 4a).
miR-15/107 effects on CDK5 level and activity. a SK-N-BE cells were treated with 100 nM pre-miR-103, pre-miR-15a, pre-miR16, or a scramble control. After 48 h, total Western blot on CDK5 and GAPDH was performed on protein lysates. n = 3, mean ± s.d. b SK-N-BE cells were treated with 100 nM pre-miR-103, pre-miR-15a, pre-miR16, or a scramble control. After 48 h, total Western blot on pAPP-Thr668 and total APP was performed on protein lysates. n = 3, mean ± s.d., *p < 0.05, Student’s t test
We then tested the effect of the miR-15/107 family on CDK5 activity by studying one of the known targets of CDK5: the amyloid precursor protein (APP), which is specifically phosphorylated by CDK5 at the threonine 668 residue [13]. We transfected SK-N-BE cells with the miR-15/107 precursors and measured the levels of pAPP-Thr668 and total APP in Western blot analysis. A reduction of APP phosphorylation levels, measured as pAPP-Thr668/APP ratio, was observed after 48 h treatment with 100 nM pre-miR-103 and pre-miR-15a (Fig. 4b), suggesting that miRNAs belonging to the miR-15/107 group can control CDK5 activity acting on the levels of the regulatory subunit p35 but leaving the levels of the kinase unchanged.
miR-15/107 Family Is Downregulated in AD Temporal Cortex and Hippocampus
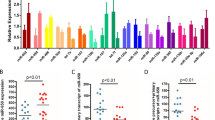
CDK5 deregulation has been suggested to have a major role in the onset of AD [14]. While p35 cleavage by calpain, to generate the truncated p25 form, leading to CDK5 hyperactivity, has been proposed since a long time as a possible pathogenic mechanism related to AD [8, 9], CDK5 aberrant activity could also be caused, at least partially, by altered miRNA-mediated regulation of its activator p35. To evaluate whether an altered expression of miR-15/107 miRNAs is associated to AD, we measured their levels by qRT-PCR in post-mortem tissue samples from the temporal cortex, hippocampus, and cerebellum of 12 AD patients and 12 age-matched healthy controls. These experiments showed that the most expressed miRNA is miR-16, followed by miR-195 and miR-103, in both AD patients and control-derived tissue samples (Fig. S2). A significant reduction of miRNA expression levels was observed in the temporal cortex and hippocampus of AD patients, compared to controls, for all analyzed miR-15/107 miRNAs, except miR-15a. On the contrary, no statistically significant difference in miRNA levels between patients and controls was detected in the cerebellum (Fig. 5).
Comparison between the levels of miR-15/107 miRNA expression in AD and control brain tissues. Box plot of the levels of miRNA expression in three different brain areas (temporal cortex, hippocampus, and cerebellum) of AD patients and controls. Dark horizontal lines represent the median, with the box representing the 25th and 75th percentiles, the whiskers the 5th and 95th percentiles. For each miR, the average of control values was set to 1, and all values were calculated relatively. *p < 0.05, **p < 0.01, Student’s t test
CDK5R1 mRNA Levels Are Increased in AD Hippocampus
Since we observed reduced levels of members of the miR-15/107 family in AD brains, we also performed qRT-PCR experiments in the same tissue samples, to verify if a corresponding CDK5R1 mRNA upregulation could be detected. The mRNA levels of BACE1, which has been reported to be upregulated in AD [27], were also measured. The transcript levels of the two genes were normalized against the median expression levels of the Cytochrome c1 (CYC1) and eukaryotic translation initiation factor 4A, isoform 2 (EIF4A2) housekeeping genes, as suggested by Penna and colleagues [33], as well as against the levels of the neuronal marker SYP, in order to normalize gene expression taking into account the neuronal population only [34].
Our qRT-PCR experiments showed no significant differences in CDK5R1 and BACE1 mRNA levels between AD and control tissues when normalized to the CYC1 and EIF4A2 housekeeping genes in temporal cortex and hippocampus (Fig. 6). Instead, reduced transcript levels were detected for both genes in the cerebellum of AD patients, compared to controls. On the contrary, when normalizing to SYP expression, we evidenced an increase of CDK5R1 mRNA levels in the hippocampus of AD samples, compared to the controls (Fig. 6); BACE1 mRNA levels were also increased in AD hippocampus, even if the data were slightly above the statistical significance (p = 0.078). A trend of increased levels was also observed in temporal cortex for both genes, but the results were not statistically significant (CDK5R1, p = 0.19; BACE1, p = 0.23). Reduced CDK5R1 and BACE1 mRNA levels were still displayed in AD cerebellum tissues. Moreover, the expression levels of CDK5R1 were significantly higher in temporal cortex and hippocampus compared to cerebellum in AD patients but not in control individuals (Fig. S3). BACE1 levels were comparable in AD tissues while they were significantly reduced in temporal cortex and hippocampus compared to cerebellum in control tissue samples. These findings suggest that an altered expression of CDK5R1 and BACE1 may specifically occur in the neuronal component of AD tissues. We also calculated the Pearson’s correlation between the expression levels of miR-15/107 miRNAs and CDK5R1 and BACE1 genes (normalized on SYP). We found a significant inverse correlation of expression between every miRNA (except miR-15a) and CDK5R1 in hippocampus samples (Fig. 7). We also observed an inverse correlation between miRNA and CDK5R1 expression levels in temporal cortex and cerebellum, as well as between miRNA and BACE1 expression levels in all three tissues, but these data did not reach the statistical significance (data not shown).
Comparison between the levels of CDK5R1 and BACE1 expression in AD and control brain tissues. Box plots of the levels of CDK5R1 and BACE1 mRNA expression in three different brain areas (temporal cortex, hippocampus, and cerebellum) of AD patients and controls. Results are shown normalized on housekeeping genes EIF4A2 and CYC1 (top panels) or SYP (bottom panels). Dark horizontal lines represent the median, with the box representing the 25th and 75th percentiles, the whiskers the 5th and 95th percentiles. For each mRNA, the average of control values was set to 1, and all values were calculated relatively. *p < 0.05, **p < 0.01, Student’s t test
Overall, these data suggest a role of the miR-15/107 family of miRNAs in the pathogenesis of AD through upregulation of CDK5R1 and BACE1.
Discussion
In the last two decades, post-transcriptional regulation has decisively emerged as an additional level of gene expression control whose importance is, at least, equal to that exerted by transcriptional regulation, providing the cell an efficient way to induce rapid changes in protein synthesis without the need for transcriptional activation and subsequent mRNA processing steps. Importantly, it is now recognized that alterations of post-transcriptional mechanisms can contribute to a variety of disorders in humans [35]. MicroRNAs (miRNAs) are small, 21–23nt-long, non-coding RNAs with a key post-transcriptional regulatory function. These small RNAs play a role in neurodevelopment, neuroplasticity, apoptosis, and other fundamental neurobiological processes [17, 18], acting as negative regulators of the expression of hundreds of target genes, either by triggering mRNA degradation or by blocking translation [36]. It is therefore not surprising that dysregulation of miRNAs has been recently implicated in neurodegenerative disorders [19, 21], including AD [20, 22]. Altered expression of several miRNAs has been found in AD brains, a relevant number of which have been involved in the regulation of genes with a key role in AD pathogenesis, notably APP and BACE1 (reviewed in [22]). In particular, different members of the miR-15/107 group of miRNAs have been recently involved in AD pathogenesis in various ways, including alteration of APP, BACE1 and Cofilin levels, Aβ production, and Tau phosphorylation levels [26–32, 37]. We previously demonstrated that two miRNAs belonging to this family, miR-103 and miR-107, regulate CDK5R1 expression, thereby affecting the levels of the encoded protein, p35 [23]. p35 represents the main activatory subunit of the CDK5 kinase, whose deregulation has been implicated in AD pathogenesis [10–14].
The present work provides evidence that all the main and most conserved members of the miR-15/107 group, not only miR-103 and miR-107, modulate CDK5R1 expression and p35 levels, through the binding to CDK5R1 3′-UTR, as shown by luciferase assays. The expression of another protein relevant to AD, BACE1, was also found to be regulated by several members of this family. Moreover, APP phosphorylation at threonin 668, a residue specifically phosphorylated by CDK5 [13], was significantly reduced upon overexpression of different miR-15/107 miRNAs, suggesting that CDK5 activity can be modulated by the cellular levels of these miRNAs. We also found that miR-15/107 miRNAs are downregulated in the hippocampus and temporal cortex, but not in cerebellum, of AD patients and that CDK5R1 mRNA levels are upregulated in the AD hippocampus. Importantly, an inverse correlation between miR-15/107 miRNAs and CDK5R1 mRNA levels was detected in hippocampus of AD patients.
In our study, we analyzed the main and most conserved miRNAs of the miR-15/107 group, i.e., miR-103, miR-107, miR-15a, miR-15b, miR-16, and miR-195, which were also found to be the most highly expressed in human brain by microarray experiments, while the remaining members of the family—miR-497, miR-503, miR-424, and miR-646—showed low expression levels [24]. However, miR-497 was recently found to be highly expressed in brain using another platform for expression analysis, deep sequencing RNA-seq [25]. Therefore, we cannot exclude a relevant role in the regulation of CDK5R1 expression also for this member of the family.
Our findings are in accordance with evidence regarding one member of the miR-15/107 family, miR-195, which was recently shown to bind CDK5R1 3′-UTR, thereby inhibiting p35 expression and reducing CDK5 activity and Tau phosphorylation in rat hippocampi and cultured neurons [38].
Despite CDK5 3′-UTR does not contain any miR-15/107 predicted binding sites, an indirect effect on CDK5 levels by regulation of other targets which are in turn able to influence CDK5 cellular abundance cannot be excluded. Our experiments showed that CDK5 levels are not affected by miR-15/107 overexpression, suggesting that these miRNAs can control CDK5 activity exclusively modulating the levels of the regulatory subunit p35. This evidence is also supported by the study of Sun and collaborators who did not observe changes in CDK5 levels after miR-195 knockdown or overexpression in rat hippocampi and neurons [38].
It is worth noting that, in our set of data, the overexpression of miR-16 does not reduce BACE1 levels as efficiently as the other miRNAs. This is at variance with what recently found by Parsi and colleagues, who reported that miR-16 is the most efficient in repressing human BACE1 protein among its family members. On the other hand, differently from our results, their data did not support the action of miR-103 on BACE1 levels [39]. These discrepancies may be ascribed to the different cell lines and/or to the different concentrations of miRNA precursors/mimics used in the two studies. However, miR-16 reduces APP phosphorylation to a level similar to that of miR-103 and miR-15a, albeit lacking statistical significance due to slightly higher variability in the three replicates. This evidence is consistent with the ability of miR-16 to reduce p35 to levels comparable to that of the other two miRNAs.
Our expression data confirm prior evidence of downmodulation of miR-15/107 miRNAs in AD brains [26–29]. In particular, we observed reduced expression levels in hippocampus and temporal cortex of AD patients for all analyzed miRNAs except miR-15a. These results on miR-15a are not consistent with those obtained by Hébert and colleagues [28], who observed miR-15a downregulation in AD brains. On the contrary, they did not detect a decrease of miR-195, as shown by our expression analysis. A bias due to the low sample size might be a possible explanation of these discrepancies. It is also interesting to note that we did not observe downmodulation of miR-15/107 miRNAs in AD cerebellum, which has been shown to be largely spared by the neurodegenerative process.
The results obtained by our and other groups ultimately show that different genes (APP, BACE1, CDK5R1) and molecular processes (amyloidogenesis, APP, and Tau phosphorylation) involved in the pathogenesis of AD are regulated by the miR-15/107 family of miRNAs, which is dysregulated in AD. Taken together, these findings point to these miRNAs as true “AD-miRs,” whose altered expression can simultaneously affect different AD-related pathways, contributing to trigger the neurodegenerative process. As such, the miR-15/107 family of miRNAs could constitute an optimal target for the development of drugs that can simultaneously act on different pathways involved in AD neurodegeneration [39].
Interestingly, the miR-15/107 microRNA gene group was also shown to cause dysregulation of Granulin (GRN) [40, 41], a gene implicated in frontotemporal dementia (FTD) [42], suggesting a very broad involvement of this miRNA family in the pathogenesis of dementias.
We recently demonstrated that, besides microRNAs, CDK5R1 3′-UTR is also bound by two different classes of RNA-binding proteins (RBPs), nELAVs (neuronal ELAV-like proteins), and hnRNPA2/B1, which showed an opposite effect on CDK5R1 transcript stability [43]. These findings highlight the complexity of the regulation of CDK5R1 expression at the post-transcriptional level, where different trans-acting factors, both miRNAs and RNA-binding proteins, can contribute to the determination of p35 levels. It is worth noting that CDK5R1 3′-UTR is characterized by the presence of several AU-rich elements (ARE) [15], composed by a core sequence of AUUUA within a U-rich contest. Since both nELAVs, that are well-known ARE-binding proteins, and miR-16, which contains the UAAAU complementary sequence, recognize these cis-regulatory elements, possible competition for binding to the same ARE sequences may be suggested. Moreover, given that, besides miR-15/107 miRNAs, both nELAVs and hnRNPA2/B1 RBPs were also shown to display altered expression in AD brains [44, 45], we hypothesize that deregulation of post-transcriptional control of CDK5R1 expression might constitute a pathomechanism in AD.
Collectively, our data support a relevant role of the miR-15/107 group of miRNAs in the pathogenesis of AD, affecting the levels of genes with a key involvement in the molecular events leading to neurodegeneration, such as CDK5R1 and BACE1. Our findings ultimately confirm that the miR-15/107 family is a promising candidate pharmacological target for AD drug development.
References
Su SC, Tsai LH (2011) Cyclin-dependent kinases in brain development and disease. Annu Rev Cell Dev Biol 27:465–491
Ohshima T, Ward JM, Huh CG, Longenecker G, Veeranna, Pant HC, Brady RO, Martin LJ et al (1996) Targeted disruption of the cyclin-dependent kinase 5 gene results in abnormal corticogenesis, neuronal pathology and perinatal death. Proc Natl Acad Sci U S A 93:11173–11178
Chae T, Kwon YT, Bronson R, Dikkes P, Li E, Tsai LH (1997) Mice lacking p35, a neuronal specific activator of Cdk5, display cortical lamination defects, seizures, and adult lethality. Neuron 18:29–42
McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM (1984) Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology 34:939–944
Qiu C, Kivipelto M, von Strauss E (2009) Epidemiology of Alzheimer’s disease: occurrence, determinants, and strategies toward intervention. Dialogues Clin Neurosci 11:111–128
Chouraki V, Seshadri S (2014) Genetics of Alzheimer’s disease. Adv Genet 87:245–294
Jiang T, Chang RC, Rosenmann H, Yu JT (2015) Advances in Alzheimer’s disease: from bench to bedside. Biomed Res Int 2015:202676
Patrick GN, Zukerberg L, Nikolic M, de la Monte S, Dikkes P, Tsai LH (1999) Conversion of p35 to p25 deregulates Cdk5 activity and promotes neurodegeneration. Nature 402:615–622
Kusakawa G, Saito T, Onuki R, Ishiguro K, Kishimoto T, Hisanaga S (2000) Calpain-dependent proteolytic cleavage of the p35 cyclin-dependent kinase 5 activator to p25. J Biol Chem 275:17166–17172
Ahlijanian MK, Barrezueta NX, Williams RD, Jakowski A, Kowsz KP, McCarthy S, Coskran T, Carlo A et al (2000) Hyperphosphorylated tau and neurofilament and cytoskeletal disruptions in mice overexpressing human p25, an activator of cdk5. Proc Natl Acad Sci U S A 97:2910–2915
Cruz JC, Tseng HC, Goldman JA, Shih H, Tsai LH (2003) Aberrant Cdk5 activation by p25 triggers pathological events leading to neurodegeneration and neurofibrillary tangles. Neuron 40:471–483
Piedrahita D, Hernández I, López-Tobón A, Fedorov D, Obara B, Manjunath BS, Boudreau RL, Davidson B et al (2010) Silencing of CDK5 reduces neurofibrillary tangles in transgenic alzheimer’s mice. J Neurosci 30:13966–13976
Iijima K, Ando K, Takeda S, Satoh Y, Seki T, Itohara S, Greengard P, Kirino Y et al (2000) Neuron-specific phosphorylation of Alzheimer’s beta-amyloid precursor protein by cyclin-dependent kinase 5. J Neurochem 75:1085–1091
Shukla V, Skuntz S, Pant HC (2012) Deregulated Cdk5 activity is involved in inducing Alzheimer’s disease. Arch Med Res 43:655–662
Moncini S, Bevilacqua A, Venturin M, Fallini C, Ratti A, Nicolin A, Riva P (2007) The 3′ untranslated region of human Cyclin-Dependent Kinase 5 Regulatory subunit 1 contains regulatory elements affecting transcript stability. BMC Mol Biol 8:111
Mignone F, Gissi C, Liuni S, Pesole G (2002) Untranslated regions of mRNAs. Genome Biol 3:REVIEWS0004
Nelson P, Kiriakidou M, Sharma A, Maniataki E, Mourelatos Z (2003) The microRNA world: small is mighty. Trends Biochem Sci 28:534–540
Kosik KS, Krichevsky AM (2005) The elegance of the MicroRNAs: a neuronal perspective. Neuron 47:779–782
Nelson PT, Wang WX, Rajeev BW (2008) MicroRNAs (miRNAs) in neurodegenerative diseases. Brain Pathol 18:130–138
Satoh J (2010) MicroRNAs and their therapeutic potential for human diseases: aberrant microRNA expression in Alzheimer’s disease brains. J Pharmacol Sci 114:269–275
Salta E, De Strooper B (2012) Non-coding RNAs with essential roles in neurodegenerative disorders. Lancet Neurol 11:189–200
Tan L, Yu JT, Hu N, Tan L (2013) Non-coding RNAs in Alzheimer’s disease. Mol Neurobiol 47:382–393
Moncini S, Salvi A, Zuccotti P, Viero G, Quattrone A, Barlati S, De Petro G, Venturin M et al (2011) The role of miR-103 and miR-107 in regulation of CDK5R1 expression and in cellular migration. PLoS One 6, e20038
Finnerty JR, Wang WX, Hébert SS, Wilfred BR, Mao G, Nelson PT (2010) The miR-15/107 group of microRNA genes: evolutionary biology, cellular functions, and roles inhuman diseases. J Mol Biol 402:491–509
Wang WX, Danaher RJ, Miller CS, Berger JR, Nubia VG, Wilfred BS, Neltner JH, Norris CM et al (2014) Expression of miR-15/107 family microRNAs in human tissues and cultured rat brain cells. Genomics Proteomics Bioinformatics 12:19–30
Hébert SS, Horré K, Nicolaï L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, Kauppinen S, Delacourte A et al (2008) Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci U S A 105:6415–6420
Wang WX, Rajeev BW, Stromberg AJ, Ren N, Tang G, Huang Q, Rigoutsos I, Nelson PT (2008) The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci 28:1213–1223
Hébert SS, Papadopoulou AS, Smith P, Galas MC, Planel E, Silahtaroglu AN, Sergeant N, Buée L et al (2010) Genetic ablation of Dicer in adult forebrain neurons results in abnormal tau hyperphosphorylation and neurodegeneration. Hum Mol Genet 19:3959–3969
Nunez-Iglesias J, Liu CC, Morgan TE, Finch CE, Zhou XJ (2010) Joint genome-wide profiling of miRNA and mRNA expression in Alzheimer’s disease cortex reveals altered miRNA regulation. PLoS One 5, e8898
Liu W, Liu C, Zhu J, Shu P, Yin B, Gong Y, Qiang B, Yuan J et al (2012) MicroRNA-16 targets amyloid precursor protein to potentially modulate Alzheimer’s-associated pathogenesis in SAMP8 mice. Neurobiol Aging 33:522–534
Zhu HC, Wang LM, Wang M, Song B, Tan S, Teng JF, Duan DX (2012) MicroRNA-195 downregulates Alzheimer’s disease amyloid-β production by targeting BACE1. Brain Res Bull 88:596–601
Ai J, Sun LH, Che H, Zhang R, Zhang TZ, Wu WC, Su XL, Chen X et al (2013) MicroRNA-195 protects against dementia induced by chronic brain hypoperfusion via its anti-amyloidogenic effect in rats. J Neurosci 33:3989–4001
Penna I, Vella S, Gigoni A, Russo C, Cancedda R, Pagano A (2011) Selection of candidate housekeeping genes for normalization in human postmortem brain samples. Int J Mol Sci 12:5461–5470
Linnertz C, Saucier L, Ge D, Cronin KD, Burke JR, Browndyke JN, Hulette CM, Welsh-Bohmer KA et al (2009) Genetic regulation of alpha-synuclein mRNA expression in various human brain tissues. PLoS One 4, e7480
Michalova E, Vojtesek B, Hrstka R (2013) Impaired pre-mRNA processing and altered architecture of 3′ untranslated regions contribute to the development of human disorders. Int J Mol Sci 14:15681–15694
Pillai RS, Bhattacharyya SN, Filipowicz W (2007) Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol 17:118–126
Yao J, Hennessey T, Flynt A, Lai E, Beal MF, Lin MT (2010) MicroRNA-related cofilin abnormality in Alzheimer’s disease. PLoS One 5, e15546
Sun LH, Ban T, Liu CD, Chen QX, Wang X, Yan ML, Hu XL, Su XL et al (2015) Activation of Cdk5/p25 and tau phosphorylation following chronic brain hypoperfusion in rats involves microRNA-195 downregulation. J Neurochem 134:1139–1151
Parsi S, Smith PY, Goupil C, Dorval V, Hébert SS (2015) Preclinical evaluation of miR-15/107 family members as multifactorial drug targets for Alzheimer’s Disease. Mol Ther Nucleic Acids 4, e256
Wang WX, Wilfred BR, Madathil SK, Tang G, Hu Y, Dimayuga J, Stromberg AJ, Huang Q et al (2010) miR-107 regulates granulin/progranulin with implications for traumatic brain injury and neurodegenerative disease. Am J Pathol 177:334–345
Wang WX, Kyprianou N, Wang X, Nelson PT (2010) Dysregulation of the mitogen granulin in human cancer through the miR-15/107 microRNA gene group. Cancer Res 70:9137–9142
Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, Lindholm C, Snowden J, Adamson J et al (2006) Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 442:916–919
Zuccotti P, Colombrita C, Moncini S, Barbieri A, Lunghi M, Gelfi C, De Palma S, Nicolin A et al (2014) hnRNPA2/B1 and nELAV proteins bind to a specific U-rich element in CDK5R1 3′-UTR and oppositely regulate its expression. Biochim Biophys Acta 1839:506–516
Mizukami K, Ishikawa M, Iwakiri M, Ikonomovic MD, Dekosky ST, Kamma H, Asada T (2005) Immunohistochemical study of the hnRNP A2 and B1 in the hippocampal formations of brains with Alzheimer’s disease. Neurosci Lett 386:111–115
Amadio M, Pascale A, Wang J, Ho L, Quattrone A, Gandy S, Haroutunian V, Racchi M et al (2009) nELAV proteins alteration in Alzheimer’s disease brain: a novel putative target for amyloid-beta reverberating on AbetaPP processing. J Alzheimers Dis 16:409–419
Acknowledgments
The authors would like to thank the MRC London Neurodegenerative Diseases Brain Bank, the Newcastle Brain Tissue Resource, and the South West Dementia Brain Bank (SWDBB) for providing brain tissue for this study. The SWDBB is supported by BRACE (Bristol Research into Alzheimer’s and Care of the Elderly), Brains for Dementia Research and the Medical Research Council. This work was supported by Ministero dell’Istruzione, dell’Università e della Ricerca (RBFR0895DC_02, FIRB (Basic Research Investment Fund) 2008 Grant to MV and RBFR0895DC_01, FIRB (Basic Research Investment Fund) 2008 Grant to MAD).
Conflict of Interest
The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Table S1
Oligonucleotide pairs designed to generate wild-type and mutated miR-sensor constructs (XLSX 10 kb)
Fig. S1
Validation of miRNA sensors. a Luciferase activity of miRNA sensor constructs in SK-N-BE cells normalized on pmirGLO-empty vector. b Luciferase activity of representative miRNA sensors with a sequence perfectly complementary to the endogenous miRNA (wt) or with a 4-base mutation in the seed sequence (mut). c Luciferase activity of sensor constructs cotransfected with 5 nM of the specific miRNA antisense LNA molecule. *p < 0.05, **p < 0.01, Student’s t test (GIF 1 kb)
Fig. S2
Levels of miR-15/107 miRNA expression in AD and control brain tissues. The expression levels of miR-15/107 miRNAs in three different brain areas (temporal cortex, hippocampus, and cerebellum) of AD patients (n = 12) and controls (n = 12) are represented as relative expression to the small nucleolar RNA RNU6B (GIF 352 bytes)
Fig. S3
Levels of CDK5R1 and BACE1 expression in AD and control brain tissues. The expression levels of CDK5R1 and BACE1 genes in three different brain areas (temporal cortex, hippocampus, and cerebellum) of AD patients (n = 12) and controls (n = 12) are represented as relative expression to the SYP gene. *p < 0.05, Student’s t test (GIF 725 bytes)
Rights and permissions
About this article
Cite this article
Moncini, S., Lunghi, M., Valmadre, A. et al. The miR-15/107 Family of microRNA Genes Regulates CDK5R1/p35 with Implications for Alzheimer’s Disease Pathogenesis. Mol Neurobiol 54, 4329–4342 (2017). https://doi.org/10.1007/s12035-016-0002-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-016-0002-4