Abstract
Circular RNA (circRNA) influences on the pathological process of osteoarthritis (OA) and may be a potential marker for disease diagnosis. The study was to scrutinize the association of circ_0045474 with OA. Clinical samples of OA patients were collected, and 12 circRNAs derived from KPNA2 gene were examined. CHON-001 cells were stimulated with IL-1β to construct an OA chondrocyte model. miR-485-3p, transcription factor 4 (TCF4) and circ_0045474, type II procollagen (COL2A1), and human collagenase-3 (MMP13) were tested. Furthermore, cell activities were analyzed. The relationship between miR-485-3p, TCF4, and circ_0045474 was determined. The role of circ_0045474 in vivo was further confirmed by constructing an OA mouse model by anterior cruciate ligament transection. circ_0045474 expression was elevated in OA patients. Suppressing circ_0045474 restrained IL-1β-stimulated extracellular matrix degradation, inflammatory cytokine secretion, and chondrocyte apoptosis. Circ_0045474 competitively combined with miR-485-3p, while TCF4 was the target of miR-485-3p. Circ_0045474 modulated IL-1β-stimulated extracellular matrix degradation, inflammatory cytokine secretion, and chondrocyte apoptosis via miR-485-3p/TCF4 axis. Suppressing circ 0045474 was effective to alleviate OA in mice. Silenced circ_0045474 suppresses OA progression in vitro and vivo via miR-485-3p/TCF4 axis. In short, circ_0045474 can be considered a novel therapeutic target for OA.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Osteoarthritis (OA) is a prevalent chronic degenerative joint disease in the elderly [1]. Histologically, OA is characterized by chondrocyte cloning, invasion of cartilage blood vessels into vertical fissures, early fragmentation of cartilage surfaces, and variable crystal deposition of tidal markers [2, 3]. OA can be identified by progressive function impairment and structure of joint components, particularly cartilage [4]. The anabolic and catabolic imbalance of the extracellular matrix (ECM) can lead to cartilage damage and deterioration of mechanical properties [5]. ECM degradation initially occurs on the surface of cartilage, accompanied by changes in matrix degrading enzymes such as collagenase-3 (MMP13). With the extension of degradation time, the cartilage area gradually forms calcification. Interleukin (IL)-1β stimulates chondrocyte inflammation, thus accelerating OA progression in a mouse model [6]. The pathogenesis of OA is still a mystery, so the existent treatment is basically to relieve symptoms [7]. Therefore, clarifying the molecular mechanism of OA can help to identify suitable therapeutic targets.
Circular RNA (circRNA) has been studied to be associated with various diseases. Stable circRNAs are endowed in cells by unique ring structures formed by intergenic regions, introns or exons [8]. Recent research has demonstrated that the interaction network between circRNA, microRNA (miRNA), and messenger RNA (mRNA) is closely linked to the development and advancement of osteoarthritis [9, 10]. In OA, circ_0008956 modulates Nicotinamide phosphoribosyltransferase via miR-149-5p sponge to boost ECM degradation [11]. Circ_0045474, located on chromosome 17, is produced by cyclization of the nuclear cell protein α2 (KPNA2). KPNA2, a member of the input protein alpha family, transports P65 into the nucleus by recognizing classical cargo protein nuclear localization signals. Meanwhile, KPNA2 is elevated in OA patients’ cartilage and regulates catabolism in OA [12]. Nevertheless, the regulatory network of KPNA2-derived circRNA with miRNA in OA is ill-informed.
MiRNA is extremely conserved noncoding RNA with 19 to 25 nucleotides in length and is involved in the OA process [13]. For instance, miR-485-3p is downregulated in the anterior cruciate ligament (ACL) tissue of knee in OA [14]. miR-485-3p accelerates OA chondrocyte progression via Notch2 and NF-κB pathways [15]. Transcription factor 4 (TCF4) on human chromosome 18 is a prevalent risk gene for human diseases [16]. TCF4 has been implicated in the progression of OA in the knee and is considered a potential target for OA treatment [17]. Additionally, TCF4 boosts chondrocyte progression via controlling adenosine monophosphate-activated protein kinase (AMPK)/NF-κB pathway [18], and TCF4 accelerates cartilage destruction by regulating osteopontin (OPN) [19].
In our study, we have confirmed that circ_004547 is upregulated in the serum of OA patients. In vitro experiments have demonstrated that circle_0045474 exerts a downregulatory effect on the expression of TCF4 through its interaction with miR-485-3p. This interaction results in the degradation of the extracellular matrix, secretion of inflammatory cytokines, and apoptosis of cells in osteoarthritis model cell. These research findings make a significant contribution to the understanding of potential mechanisms involved in the alleviation of osteoarthritis. Furthermore, in OA mouse, the downregulation of circ_0045474 can inhibit the expression of TCF4 through competitive binding with miRNA. This study describes a novel regulatory mechanism of circ_0045474 and provides a potential therapeutic target for the treatment of osteoarthritis.
Experimental Methods
Clinical Specimens
From January 2019 to January 2021, patients diagnosed as OA were enrolled at Haining People’s Hospital with informed consent in this study. Inclusion criteria: People who voluntarily participated in the study; People who met the diagnostic criteria of OA; 18 years old or more; Patients were conscious, communicated normally, and had no serious physical diseases such as tumors. Exclusion criteria: Less than 18 years old; Nervous system damage disease; Acute and chronic pancreatitis; Liver and kidney dysfunction; Psychoactive substance abuse history.
A total of 38 OA patients (19 males and 19 females) were involved. The mean age of male OA subjects was 71.14 ± 5.31 years (ranged from 63 to 84 years), and the mean course of disease was 7.3 ± 0.49 years. The mean age of female OA subjects was 72.21 ± 3.42 years (ranged from 67 to 77 years), and the mean course of disease was 6.8 ± 0.53 years. Healthy people (19 males and 19 females) undergoing physical examination in Haining People’s Hospital were controls. The mean age of male and female was 67.45 ± 3.54 years and 68.68 ± 4.27 years, respectively. The study was approved by the Medical Ethics Committee of Haining People’s Hospital. All patients provided written informed consent. From each OA patient and healthy control, 3 mL of peripheral blood was collected and centrifuged. The supernatant was collected and stored in a − 80 °C refrigerator.
Cell Culture
Human chondrocytes CHON-001 were purchased from the American Type Culture Collection (ATCC, MD, USA). Cells were cultured in Dulbecco modified eagle medium (Gibco) replenished with 10% fetal bovine serum. CHON-001 cells were stimulated with 10 ng/mL 1L-1β to establish an OA cell model (OA-like chondrocytes) in vitro [20].
Cell Transfection
miR-485-3p mimic/inhibitor, circ_0045474 plasmids (si-circ_0045474#1, si-circ_0045474#2, si-circ_0045474 #3, oe-circ_0045474), TCF4 low expression plasmid (si-TCF4), and its corresponding negative control (NC) were provided by RiboBio (Guangzhou, China). These oligonucleotides and plasmids were transfected into CHON-001 chondrocytes using Lipofectamine 2000 (Invitrogen, CA, USA) in line with the manufacturer’s protocol. Follow-up tests were conducted after transfection (sequences were shown supplementary Table 1).
Cell Counting Kit (CCK)-8 Assay
Chondrocyte proliferation was analyzed with CCK-8 kit (Sigma–Aldrich, MO, USA). Chondrocytes were cultured at 1 × 105 cells/well on a 96-well plate. After 24, 48, and 72 h of transfection, 10 μL CCK-8 was added to chondrocytes and incubated. Subsequently, optical density was read at 450 nm on a microplate reader (Bio-Rad, CA, USA).
Flow Cytometry
Measurements of apoptosis rates of CHON-001 cells were done in line with the protocol of annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) (BD Biosciences, CA, USA). In short, CHON-001 cells were incubated with annexin V-FITC and PI.
Nuclear/Cytoplasmic RNA Separation Test
Nuclear and cytoplasmic RNA were isolated from CHON-001 cells using PARIS ™ kit (Thermo Fisher Scientific, MA, USA) in line with the protocol. Cells (107) were suspended in 300 μL cold buffer, incubated on ice, and centrifuged. The cytoplasm was separated from the nuclear precipitate. Subsequently, approximately 400 μL of cold cell lysis buffer and equal volume of 2 × lysis/binding solution were added to the nuclear precipitate. Then, 400 μL 100% ethanol was added to the mixture. After rinse, centrifugation, and filtration, RNA was eluted and stored for use.
Luciferase Reporter Gene Assay
Potential binding sites of miR-485-3p with TCF4 and circ_0045474 were predicted by Starbase website (http://starbase.sysu.edu.cn). The 3′-untranslated region (3′-UTR) sequence of circ_0045474 or TCF4 containing miR-485-3p wild-type or mutant binding sites was inserted into the pmirGLO reporter (Promega, WI, USA) to form WT-circ_0045474 or MUT-circ_0045474 and WT-TCF4 or MUT-TCF4. miR-485-3p or mimic-NC was transfected into 293T cells using Lipofectamine 2000 (Invitrogen) in line with the manufacturer’s protocol. Luciferase activity was examined using a luciferase reporting kit (Promega).
RNA Pull-Down Assay
Biotin-labeled miR-140-5p wild-type and mutant plasmid (50 nM each) were transfected, respectively, into cells. After transfection, cells were incubated with specific cell lysis buffer (Ambion, Austin, Texas, USA). Cell lysates were incubated with M-280 streptavidin magnetic beads precoated with RNase-free and yeast tRNA (all Sigma, St. Louis, MO, USA). After elution with salt buffer, total RNA was isolated to test circ_0045474. An antagonistic miR-140-5p probe was as a negative control.
RNA Immunoprecipitation (RIP)
Magna RNA Binding Protein Immunoprecipitation Kit (Merck KGaA, Darmstadt, Germany) was designed for RIP analysis. CHON-001 cells were lysed with RIP lysis buffer (Beyotime, Shanghai, China) and incubated with magnetic beads conjugating with Argonaute-2 (Ago2, Millipore, MA, USA) or control immunoglobulin G (IgG, Millipore). The compound was digested with protease K (Sigma-Aldrich), and the binding RNA was analyzed.
Anterior Cruciate Ligament Transection (ACLT) to Construct OA Mouse Model
Animal experimental procedures were approved by the Animal Care and Use Committee of Haining People’s Hospital. C57BL/6 J mice (12 weeks; 8 mice per group) were purchased from Shanghai SLAC Laboratory Animal Co., Ltd. (Shanghai, China). Mice were randomly into the following experimental groups: (1) sham group; (2) OA group; (3) OA + sh circ_0045474 group; (4) OA + sh-NC. Mice were anesthetized with ketamine at 75 mg/kg and thiazide at 10 mg/kg for unilateral ACLT surgery. A skin incision was made to expose the patella, which was laterally dislocated under a microscope with tweezers. ACL was exposed by complete knee flexion and then transected with a needle. The incision was sutured with 3–0 silk thread. The sham group underwent skin incision and suture without patellar dislocation and ligament transection. One week following surgery, the joint was promptly administered with an injection of 100 μL control or adenovirus interference vectors (sh-NC, scrambling sh-circ_0045274 (RiboBio, Guangzhou, China) with a viral titer of 2 × 108 TU/mL 7. After 8 weeks, thermophyperalgesia was assessed by hot plate test and joint pain was evaluated by plate test [21]. All of the mice were euthanized after completing behavioral tests.
Safranin O/Fast Green and Alcian Blue Staining
Knee joint was fixed with 4% paraformaldehyde, decalcified with 10% Ethylene Diamine Tetraacetic Acid, embedded in paraffin, and cut into 5 μm. The sections were stained with 0.1% Safranin O and 0.001% fast green solution or Alcian blue solution (pH 2.5). Glass slide images were observed under a microscope (Olympus, Tokyo, Japan). OA severity/cartilage injury was assessed using Osteoarthritis Research Society International (OARSI) score (Grade 0–6) [22]. The concentrations of inflammatory cytokines tumor necrosis factor-α (TNF-α) and interleukin 6 (IL-6) in tissue and cell culture supernatant were measured in line with the manufacturer’s protocol of ELISA Kit (Sigma-Aldrich).
Reverse Transcription Quantitative Polymerase Chain Reaction (RT-qPCR)
Extraction of RNA was done with TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Subsequently, the RNA quality was assessed using the Nanodrop 2000 and the RNA concentration was adjusted accordingly. The miRNA reverse transcription kit (TaKaRa, Japan) was employed for the synthesis of cDNA for miRNA, while the PrimeScript ™ RT Reagent kit with gDNA Eraser (TaKaRa, Japan) was utilized for the synthesis of cDNA for mRNA and circRNA. Subsequently, three replicates of RT-qPCR were conducted on each sample using the Miscript SYBR Green PCR kit (Qiagen, Dusseldorf, Germany) with cDNA as the template. The primer list for the target gene can be found in Tables 1 and 2. U6 or β-Actin was employed as a control for the experiment. Gene expression was determined using the 2−ΔΔCT method for calculation.
Western Blot
Chondrocytes and tissues were lysed with a radio-immunoprecipitation assay lysis buffer. Measurements of protein concentration were performed with the bicinchoninic acid protein kit. Then, 20 μg protein was separated with 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis, and electroblotted to polyvinylidene fluoride membranes. The membranes were incubated with primary antibodies anti-TCF4 (1:10000), anti-COL2A1 (1:1000), anti-MMP13 (1:1000), and anti-β-actin (1 µg/mL). The membranes were further detected with a horseradish peroxidase-conjugated secondary antibody. All antibodies were provided by Abcam (MA, USA). ECL was used to detect protein bands, and the protein gray value was calculated by Image J (NIH Image J system, Bethesda, MD).
Statistical Analysis
SPSS 22.0 software was used to process all experimental data. Measurement data were shown in the form of mean ± standard deviation (SD). The comparison between two groups was done by t test, while that among multiple groups was done by one-way analysis of variance (ANOVA) and Tukey’s multiple comparisons test. Graphs were drawn using GraphPad Prism 8.0.1 (GraphPad Software, Inc., La Jolla, CA). P < 0.05 was accepted as indicative of statistical differences.
Results
KPNA2-Derived circRNAs in the Serum of OA Patients
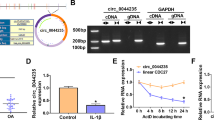
A total of 12 circRNAs derived from KPNA2 gene were detected in human on the bioinformatics website (http://www.circbase.org/). The detailed information is shown in Table 3. Serum samples were obtained from 5 OA patients and 5 healthy controls to test KPNA2-derived circRNAs. Four circRNAs (hsa_ circ_0045474, hsa_circ_0045482, hsa_circ_0045478, and hsa_circ_0045477) were elevated in the serum of OA patients (Fig. 1A). The 4 circRNAs were further analyzed in a larger sample size. Serum hsa_circ_0045474 (circ_KPNA2) was higher in OA patients than in healthy controls (Fig. 1B), while other 3 circRNAs represented no differences (Fig. 1C–E). The above results clarified that KPNA2-derived hsa_circ_0045474 was upregulated in OA patients.
KpNA2-derived circRNA in serum of OA patients. A Detection of among 12 circRNA molecules stemmed from KPNA2 by RT-qPCR, n = 5; B–E Examination of 4 circRNAs (hsa_circ_0045474, hsa_circ_0045482, hsa_circ_0045478 and hsa_circ_0045477) in healthy controls and OA patients, n = 38. **P < 0.01, ***P < 0.001. The data in the figure were shown in the form of mean ± standard deviation (SD)
Silenced circ_0045474 Suppresses IL-1β-Induced Inflammatory Cytokine Secretion, ECM Degradation, and Chondrocyte Apoptosis
To further elucidate the function of circ_0045474 in OA, chondrocytes were treated with IL-1β (5 ng/mL), and circ_0045474 was found to be elevated in IL-1β-stimulated chondrocytes (Fig. 2A). Additionally, si-circ_0045474 was transfected into chondrocytes, with si-circ_0045474#2 showed the greatest knockdown effect (Fig. 2B). Therefore, si-circ_0045474#2 was selected for subsequent experiments. circ_0045474 knockdown restrained IL-1β-induced chondrocyte apoptosis and promoted proliferation (Fig. 2C, D). IL-1β treatment induced TNF-α and IL-6 secretion, while silencing circ_0045474 reduced TNF-α and IL-6 secretion (Fig. 2E). Additionally, IL-1β treatment repressed COL2A1 and elevated MMP13, while this effect was reversed by downregulating circ_0045474 (Fig. 2F). In short, silenced circ_0045474 suppresses IL-1β-induced inflammatory cytokine secretion, ECM degradation, and chondrocyte apoptosis.
Repressed circ_0045474 suppresses IL-1β-stimulated inflammatory factor secretion, ECM degradation, and chondrocyte apoptosis. A RT-qPCR to test circ_0045474 in IL-1β-stimulated chondrocytes; B RT-qPCR to examine the transfection efficiency of circ_0045474; C CCK-8 to test chondrocyte proliferation; D FCM to examine chondrocyte apoptosis; E ELISA to test inflammatory cytokines TNF-α and IL-6; F WB to test ECM degradation-correlated markers COL2A1 and MMP13; *P < 0.05, **P < 0.01. N = 3. The data in the figure were shown in the form of mean ± SD
Hsa_circ_0045474 Adsorbs miR-485-3p
The subcellular localization of hsa_circ_0045474 (circ KPNA2) was predicted by the lncLocator website (http://www.csbio.sjtu.edu.cn/bioinf/lncLocator/). It was found that circ_0045474 was mainly distributed in the cytoplasm (Fig. 3A, B). Hsa_circ_0045474 at chr17:66042637–66042657 locus had a binding site with miR-485-3p, which was predicted by the Starbase website (http://starbase.sysu.edu.cn/) (Fig. 3C). Accordingly, miR-485-3p mimic and wild-type circ_0045474 co-transfection reduced the luciferase activity of cells (Fig. 3D). RNA pull-down assay further found that the enrichment of circ_0045474 in the Bio-miR-485-3p-WT group was higher than that in the Bio-miR-485-3p-MUT and Bio-probe NC group (Fig. 3E). miR-485-3p expression was negatively regulated by circ_0045474 (Fig. 3F). In brief, circ_0045474 negatively modulates miR-485-3p via competing with miR-485-3p.
Hsa_circ_0045474 adsorbs miR-485-3p and negatively modulates miR-485-3p expression. A lncLocator to predict the subcellular localization of circ_0045474; B Nucleo-cytoplasmic separation assay to testify that circ_0045474 was distributed in the cytoplasm. C Starbase to predict the binding site of circ_0045474 and miR-485-3p; D Luciferase reporter assay to test the relationship between circ_0045474 and miR-485-3p; E RNA pull-down to detect the enrichment of circ_0045474 and miR-485-3p; F RT-qPCR to examine miR-485-3p; **P < 0.01; N = 3. The data in the figure were shown in the form of mean ± SD
Hsa_circ_0045474 Controls IL-1β-Stimulated Inflammatory Factor Secretion, ECM Degradation, and Chondrocyte Apoptosis Via Adsorbing miR-485-3p
MiR-485-3p was downregulated in OA-like chondrocytes, and co-transfection with miR-485-3p mimic and oe-circ_0045474 resulted in miR-485-3p downregulation (Fig. 4A). The results clarified that miR-485-3p upregulation restrained OA-like chondrocyte apoptosis and enhanced proliferation, while upregulation of miR-485-3p and circ_0045474 led to the opposite results (Fig. 4B, C). miR-485-3p upregulation reduced the secretion of inflammatory factors TNF-α and IL-6, while the inflammatory factors were restored after co-transfection of miR-485-3p mimic and oe-circ_0045474 (Fig. 4D). miR-485-3p upregulation repressed IL-1β-stimulated ECM degradation in chondrocytes, while co-transfection of miR-485-3p mimic and oe-circ_0045474 enhanced ECM degradation (Fig. 4E). In short, miR-485-3p inhibits extracellular matrix degradation, inflammatory cytokine secretion, and cell apoptosis of OA-like chondrocytes, circ_40045474 promotes the adverse behavior of chondrocytes by inhibiting miR-485-3p.
Hsa_circ_0045474 exerts a role in IL-1β-stimulated chondrocytes via miR-485-3p. A RT-qPCR to test circ_0045474 and miR-485-3p in chondrocytes; B CCK-8 to detect chondrocyte proliferation; C FCM to examine chondrocyte apoptosis; D ELISA to test inflammatory cytokines TNF-α and IL-6; E WB to test ECM degradation-associated markers COL2A1 and MMP13; *P < 0.05, **P < 0.01. N = 3. The data in the figure were shown in the form of mean ± SD
TCF4 Performs as miR-485-3p Downstream to Target mRNA
MiR-485-3p at chr17:66042637–66042657 locus had a binding site with TCF4 as predicted by Starbase website (http://starbase.sysu.edu.cn/) (Fig. 5A). The targeting relationship between TCF4 and miR-485-3p was supported by luciferase reporter gene assay and RIP assay (Fig. 5B, C). Furthermore, TCF4 expression was negatively mediated by miR-485-3p in chondrocytes (Fig. 5D, E). In brief, miR-485-3p targets TCF4.
MiR-485-3p targets TCF4 3’UTR. A Starbase to predict the binding site of miR-485-3p and TCF4; B Luciferase reporter gene assay to test the combination of miR-485-3p and TCF4; C RIP to examine the combination of miR-485-3p and TCF4; D, E RT-qPCR and WB to analyze TCF4 expression; *P < 0.05, **P < 0.01. N = 3. The data in the figure were shown in the form of mean ± SD
MiR-485-3p Targets TCF4 to Mediate IL-1β-Stimulated ECM Degradation, Inflammatory Cytokine Secretion, and Chondrocyte Apoptosis
TCF4 expression was augmented in OA-like chondrocytes, while TCF4 expression was elevated after co-transfection with miR-485-3p inhibitor and TCF4 low expression vector (si-TCF4) (Fig. 6A, B). si-TCF4 restrained apoptosis and enhanced proliferation of OA-like chondrocytes, and si-TCF4 and miR-485-3p inhibitor increased apoptosis and inhibited proliferation (Fig. 6C, D). si-TCF4 suppressed the secretion of inflammatory factors TNF-α and IL-6, while inflammatory factors were restored after co-transfection with miR-485-3p inhibitor and si-TCF4 (Fig. 6E). Additionally, si-TCF4 restrained ECM degradation in OA-like chondrocytes, while si-TCF4 and miR-485-3p inhibitor enhanced ECM degradation (Fig. 6F). In short, miR-485-3p inhibits ECM degradation, inflammatory cytokine secretion, and cell apoptosis in OA-like chondrocytes by targeting TCF4.
MiR-485-3p exerts a role in IL-1β-stimulated chondrocytes via targeting TCF4. A, B RT-qPCR and WB to test miR-485-3p and TCF4 in chondrocytes; C CCK-8 to detect chondrocyte proliferation; D FCM to testify chondrocyte apoptosis; E ELISA to test inflammatory cytokines TNF-α and IL-6; F WB to test ECM degradation-correlated markers COL2A1 and MMP13; *P < 0.05, **P < 0.01. N = 3. The data in the figure were shown in the form of mean ± SD
Silenced circ_0045474 Alleviates OA Progression in the Mouse ACLT Model
ACLT mice were injected with sh circ_0045474 vector to downregulate circ_0045474 (Fig. 7A). After sh circ_0045474 injection, ACLT mice manifested higher tolerance levels on the hot plate (Fig. 7B). Furthermore, sh circ_0045474 reduced the number of shocks in mice, implying that silenced circ_0045474 rescued OA progression (Fig. 7C). Cartilage surface of ACLT-stimulated OA mice was ameliorated with the injection of sh circ_0045474 (Fig. 7D). Meanwhile, inflammatory factors TNF-α and IL-6 were elevated in OA tissue, while silenced circ_0045474 reduced TNF-α and IL-6 secretion (Fig. 7E). Additionally, COL2A1 was repressed and MMP13 was enhanced in OA mice, while the effect was turned around by silencing circ_0045474 (Fig. 7F). In brief, silenced circ_0045474 alleviates OA progression in the mouse ACLT model.
Silenced circ_0045474 mitigates OA progression in the mouse model of ACLT. A, B RT-qPCR and WB to examine circ_0045474, miR-485-3p, and TCF4 in OA cartilage tissues; B Tolerance level on the hot plate; C Number of shocks; D Alcian blue and Safranin O/fast green staining, Scale bar = 50–100 μm. E ELISA to test inflammatory cytokines TNF-α and IL-6; F WB to test ECM degradation-correlated markers COL2A1 and MMP13; **P < 0.01. N = 8. The data in the figure were shown in the form of mean ± SD
Discussion
This study presents novel findings regarding the overexpression of circ_0045474 in cartilage tissue of an OA-like chondrocytes and OA model mice, alongside a concurrent decrease in the expression level of miR-485-3p. The role of TCF4 in regulating the progression of osteoarthritis has been confirmed [23]. There have been reports indicating that miR-93-5p exerts inhibitory effects on TCF4 expression, thereby reducing chondrocyte apoptosis and partially impeding the progression of osteoarthritis [24]. Our subsequent experiments demonstrated that the inhibition of circ_0045474 or the overexpression of miR-485-3p resulted in a reduction of cell apoptosis, MMP13 deposition, and an increase in the expression of COL2A1 protein in vitro OA cells, thereby mitigating OA damage.
Circular RNA can act as an activating or inhibitory factor and play an important role in the occurrence and development of osteoarthritis. Zhou et al. declare that 255 circRNAs are differentially expressed in IL-1β-treated mouse articular chondrocytes [25]. Liu et al. maintain that 71 circRNAs are dysregulated in human OA tissues [26]. Silenced circ_0136474 is available to mitigate IL-1β-stimulated chondrocyte apoptosis and oxidative stress [27]. However, the precise role of circ_0045474 in osteoarthritis (OA) remains uncertain. The data presented in our study indicate that the elevated expression of circ_0045474 in both OA-like chondrocytes and OA mouse models may play a significant role in facilitating chondrocyte apoptosis and functional impairment, thereby contributing to the initiation and progression of osteoarthritis.
The localization of circular RNA exhibits a strong correlation with the sponge effect exerted on miRNA [28, 29]. It is postulated that circ_0045474 is localized to cytoplasm and act as a miRNA molecular sponge to regulate the functions of miRNAs. Previous investigations have demonstrated that circ_0045474 predominantly localizes in the cytoplasm [30, 31]. Subsequently, we have identified miR-485-3p as a target of circ_0045474. We have substantiated that miR-485-3p is downregulated in OA-like chondrocytes and in cartilage tissue of OA mice. Moreover, its overexpression significantly mitigates cell apoptosis, extracellular matrix degradation, and exacerbation of inflammatory factors in OA-like chondrocytes. In summary, it is hypothesized that miR-485-3p exerts a negative regulatory influence on the progression of OA. The results of our study align with previous research, which demonstrated a decrease in miR-485-3p expression in the cartilage tissue of individuals with osteoarthritis [32]. Zhou et al. also confirmed the downregulation of miR-485-3p in OA-like chondrocytes induced with lipopolysaccharide, and observed that overexpression of miR-485-3p mitigated extracellular matrix degradation, inflammation, and oxidative stress in these chondrocytes [15]. These findings further substantiate the inhibitory role of miR-485-3p in osteoarthritis.
miRNAs regulate gene expression by inducing mRNA degradation or inhibiting mRNA translation [33].Bioinformatics analysis shows that miR-485-3p interacts with the 3’UTR of TCF4. Our study confirmed that TCF4 is highly expressed in OA-like chondrocytes and cartilage tissues of OA mice. TCF4 is an alkaline helix loop helix transcription factor that is highly expressed in various diseases [14]. The downregulation of TCF4 mediated by small interfering RNA or short hairpin RNA inhibits the occurrence and development of diseases [34, 35]. In a study conducted on osteoarthritis, the administration of PKF118-310 via intra-articular injection (an inhibitor of TCF4-β catenin interaction) exhibited a decrease in cartilage degradation and a reduction in MMP13 expression in mice with osteoarthritis [36]. Likewise, our investigation demonstrated that the suppression of TCF4 expression resulted in the inhibition of extracellular matrix degradation, secretion of inflammatory cytokines, and apoptosis in OA-like chondrocytes.
Finally, the present study highlights the significance of circ 0045474 as a regulatory element in modulating TCF4 expression via miR-485-3p. Through in vivo investigations, it was demonstrated that the lentivirus-mediated inhibition of circ 0045474 in OA mice resulted in an upregulation of miR-485-3p, a downregulation of TCF4 expression, and an amelioration of cartilage tissue damage associated with OA. However, the specific regulatory mechanism of TCF4 in regulating extracellular matrix degradation, inflammatory cytokine secretion, and cell apoptosis of OA-like chondrocytes is not yet clear, and further research is needed.
Conclusion
In brief, circ_0045474 and TCF4 are elevated and miR-485-3p is silenced in OA tissues and cells. Knockdown of circ_0045474 facilitates OA progression. circ_0045474 accelerates OA progression via interacting with the miR-485-3p/TCF4 axis. Circ_0045474 may become a potential clinical biomarker of OA, providing a new perspective on the molecular pathological mechanism of OA, and has important value for the specific targeted treatment strategy of OA.
Data Availability
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
References
Chen, D., Shen, J., Zhao, W., Wang, T., Han, L., Hamilton, J. L., & Im, H.-J. (2017). Osteoarthritis: toward a comprehensive understanding of pathological mechanism. Bone Research, 5, 16044.
Madry, H., Luyten, F. P., & Facchini, A. (2012). Biological aspects of early osteoarthritis. Knee Surgery, Sports Traumatology, Arthroscopy, 20, 407–422.
Xia, B., Di Chen, C., Zhang, J., Hu, S., Jin, H., & Tong, P. (2014). Osteoarthritis pathogenesis: A review of molecular mechanisms. Calcified Tissue International, 95, 495–505.
Masson, A. O., & Krawetz, R. J. (2020). Understanding cartilage protection in OA and injury: A spectrum of possibilities. BMC Musculoskeletal Disorders, 21, 432.
Han, L., Grodzinsky, A. J., & Ortiz, C. (2011). Nanomechanics of the cartilage extracellular matrix. Annual Review of Materials Research, 41, 133–168.
Yin, W., & Lei, Y. (2018). Leonurine inhibits IL-1beta induced inflammation in murine chondrocytes and ameliorates murine osteoarthritis. International Immunopharmacology, 65, 50–59.
Chen, Y., Guo, H., Li, L., Bao, D., Gao, F., Li, Q., Huang, Q., Duan, X., & Xiang, Z. (2020). Long non-coding RNA (lncRNA) small nucleolar RNA host gene 15 (SNHG15) alleviates osteoarthritis progression by regulation of extracellular matrix homeostasis. Medical Science Monitor, 26, e923868.
Qu, S., Yang, X., Li, X., Wang, J., Gao, Y., Shang, R., Sun, W., Dou, K., & Li, H. (2015). Circular RNA: A new star of noncoding RNAs. Cancer Letters, 365, 141–148.
Kong, H., Sun, M.-L., Zhang, X.-A., & Wang, X.-Q. (2021). Crosstalk among circRNA/lncRNA, miRNA, and mRNA in osteoarthritis. Frontiers in Cell and Developmental Biology, 9, 774370.
Zheng, Y.-L., Song, G., Guo, J.-B., Su, X., Chen, Y.-M., Yang, Z., Chen, P.-J., & Wang, X.-Q. (2021). Interactions among lncRNA/circRNA, miRNA, and mRNA in musculoskeletal degenerative diseases. Frontiers in Cell and Developmental Biology, 9, 753931.
Fu, S., Fan, Q., Xu, J., Yu, S., Sun, M., Ji, Y., & Liu, D. (2021). Circ_0008956 contributes to IL-1beta-induced osteoarthritis progression via miR-149-5p/NAMPT axis. International Immunopharmacology, 98, 107857.
Tao, R., Xu, X., Sun, C., Wang, Y., Wang, S., Liu, Z., Zhai, L., Cheng, H., Xiao, M., & Zhang, D. (2015). KPNA2 interacts with P65 to modulate catabolic events in osteoarthritis. Experimental and Molecular Pathology, 99, 245–252.
Huang, P.-y, Wu, J.-g, Gu, J., Zhang, T.-q, Li, L.-f, Wang, S.-q, & Wang, M. (2021). Bioinformatics analysis of miRNA and mRNA expression profiles to reveal the key miRNAs and genes in osteoarthritis. Journal of Orthopaedic Surgery and Research, 16, 63.
Li, B., Bai, L., Shen, P., Sun, Y., Chen, Z., & Wen, Y. (2017). Identification of differentially expressed microRNAs in knee anterior cruciate ligament tissues surgically removed from patients with osteoarthritis. International Journal of Molecular Medicine, 40, 1105–1113.
Zhou, Y., Zhao, Z., Yan, L., & Yang, J. (2021). MiR-485-3p promotes proliferation of osteoarthritis chondrocytes and inhibits apoptosis via Notch2 and the NF-kappaB pathway. Immunopharmacology and Immunotoxicology, 43, 370–379.
Xi, P., Zhang, C.-l, S-y, Wu., Liu, L., Li, W.-j, & Li, Y.-m. (2021). Circ circ-IQGAP1 Knockdown alleviates interleukin-1beta-induced osteoarthritis progression via targeting miR-671-5p/TCF4. Orthopaedic Surgery, 13, 1036–1046.
Wu, L., Guo, H., Sun, K., Zhao, X., Ma, T., & Jin, Q. (2016). Sclerostin expression in the subchondral bone of patients with knee osteoarthritis. International Journal of Molecular Medicine, 38, 1395–1402.
Wang, J., Fang, L., Ye, L., Ma, S., Huang, H., Lan, X., & Ma, J. (2020). miR-137 targets the inhibition of TCF4 to reverse the progression of osteoarthritis through the AMPK/NF-kappaB signaling pathway. Bioscience Reports. https://doi.org/10.1042/BSR20200466
Tian, J., Gao, S.-G., Li, Y.-S., Cheng, C., Deng, Z.-H., Luo, W., & Zhang, F.-J. (2020). The beta-catenin/TCF-4 pathway regulates the expression of OPN in human osteoarthritic chondrocytes. Journal of Orthopaedic Surgery and Research, 15, 344.
Wang, M., Sampson, E. R., Jin, H., Li, J., Ke, Q. H., Im, H.-J., & Chen, D. (2013). MMP13 is a critical target gene during the progression of osteoarthritis. Arthritis Research & Therapy. https://doi.org/10.1186/ar4133
Ni, W., Jiang, C., Wu, Y., Zhang, H., Wang, L., Yik, J. H., Haudenschild, D. R., Fan, S., Shen, S., & Hu, Z. (2021). CircSLC7A2 protects against osteoarthritis through inhibition of the miR-4498/TIMP3 axis. Cell Proliferation, 54, e13047.
Deng, Z., Chen, F., Liu, Y., Wang, J., Lu, W., Jiang, W., & Zhu, W. (2021). Losartan protects against osteoarthritis by repressing the TGF-beta1 signaling pathway via upregulation of PPARgamma. Journal of Orthopaedic Translation, 29, 30–41.
Luo, X., Wang, J., Wei, X., Wang, S., & Wang, A. (2020). Knockdown of lncRNA MFI2-AS1 inhibits lipopolysaccharide-induced osteoarthritis progression by miR-130a-3p/TCF4. Life Sciences, 240, 117019.
Xue, H., Tu, Y., Ma, T., Wen, T., Yang, T., Xue, L., Cai, M., Wang, F., & Guan, M. (2019). miR-93–5p attenuates IL-1β-induced chondrocyte apoptosis and cartilage degradation in osteoarthritis partially by targeting TCF4. Bone, 123, 129–136.
Zhou, Z., Du, D., Chen, A., & Zhu, L. (2018). Circular RNA expression profile of articular chondrocytes in an IL-1beta-induced mouse model of osteoarthritis. Gene, 644, 20–26.
Liu, Q., Zhang, X., Hu, X., Dai, L., Fu, X., Zhang, J., & Ao, Y. (2016). Circular RNA related to the chondrocyte ECM regulates MMP13 expression by functioning as a MiR-136 ‘sponge’ in human cartilage degradation. Science and Reports, 6, 22572.
Zhu, H., Zhu, S., Shang, X., Meng, X., Jing, S., Yu, L., & Deng, Y. (2021). Exhausting circ_0136474 and restoring miR-766-3p attenuate chondrocyte oxidative injury in IL-1beta-induced osteoarthritis progression through regulating DNMT3A. Frontiers in Genetics, 12, 648709.
Kulcheski, F. R., Christoff, A. P., & Margis, R. (2016). Circular RNAs are miRNA sponges and can be used as a new class of biomarker. Journal of Biotechnology, 238, 42–51.
Liao, H.-X., Zhang, Z.-H., Chen, H.-L., Huang, Y.-M., Liu, Z.-L., & Huang, J. (2021). CircHYBID regulates hyaluronan metabolism in chondrocytes via hsa-miR-29b-3p/TGF-beta1 axis. Molecular Medicine, 27, 56.
Wu, M., Liu, Z., & Zhang, S. (2022). Down-regulation of hsa_circ_0045474 induces macrophage autophagy in tuberculosis via miR-582–5p/TNKS2 axis. Innate Immunity, 28, 11–18.
Xu, J., & Ma, X. (2021). Hsa_circ_0032131 knockdown inhibits osteoarthritis progression via the miR-502-5p/PRDX3 axis. Aging (Albany NY), 13, 15100–15113.
da Silveira, W., Renaud, L., Simpson, J., Glen, W., Hazard, E., Chung, D., & Hardiman, G. (2018). miRmapper: A tool for interpretation of miRNA-mRNA interaction networks. Genes (Basel), 9, 458.
Forrest, M. P., Hill, M. J., Quantock, A. J., Martin-Rendon, E., & Blake, D. J. (2014). The emerging roles of TCF4 in disease and development. Trends in Molecular Medicine, 20, 322–331.
Xu, W., Du, M., Zhao, Y., Wang, Q., Sun, W., & Chen, B. (2012). γ-Tocotrienol inhibits cell viability through suppression of β-catenin/Tcf signaling in human colon carcinoma HT-29 cells. The Journal of Nutritional Biochemistry, 23, 800–807.
Jeong, J., Lee, J., & Lee, S.-H. (2015). TCF4 is a molecular target of resveratrol in the prevention of colorectal cancer. International Journal of Molecular Sciences, 16, 10411–10425.
Bouaziz, W., Sigaux, J., Modrowski, D., Devignes, C.-S., Funck-Brentano, T., Richette, P., Ea, H.-K., Provot, S., Cohen-Solal, M., & Haÿ, E. (2016). Interaction of HIF1α and β-catenin inhibits matrix metalloproteinase 13 expression and prevents cartilage damage in mice. Proceedings of the National Academy of Sciences USA, 113, 5453–5458.
Acknowledgements
Not applicable.
Funding
Not applicable.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing Interests
The authors have no conflict of interest to declare.
Ethical Approval
The present study was approved by the Ethics Committee of Haining People’s Hospital and written informed consent was provided by all patients prior to the study start. All procedures were performed in accordance with the ethical standards of the Institutional Review Board and The Declaration of Helsinki, and its later amendments or comparable ethical standards (Approval number: 201806HN01). And the experiments were approved by the Institutional Animal Care and Use Committee of Haining People’s Hospital and all procedures complied with the National Institutes of Health Guide for the Use of Laboratory Animals (Approval number: 201806HN20).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, Z., Yu, P. & Bai, L. Hsa_circular RNA_0045474 Facilitates Osteoarthritis Via Modulating microRNA-485-3p and Augmenting Transcription Factor 4. Mol Biotechnol 66, 1174–1187 (2024). https://doi.org/10.1007/s12033-023-01019-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-023-01019-z