Abstract
H2A.Z-containing nucleosomes have been found to function in various developmental programs in Arabidopsis (e.g., floral transition, warm ambient temperature, and drought stress responses). The SWI2/SNF2-Related 1 Chromatin Remodeling (SWR1) complex is known to control the deposition of H2A.Z, and it has been unraveled that ACTIN-RELATED PROTEIN 6 (ARP6) is one component of this SWR1 complex. Previous studies showed that the arp6 mutant exhibited some distinguished phenotypes such as early flowering, leaf serration, elongated hypocotyl, and reduced seed germination rate in response to osmotic stress. In this study, we aimed to investigate the changes of arp6 mutant when the plants were grown in salt stress condition. The phenotypic observation showed that the arp6 mutant was more sensitive to salt stress than the wild type. Upon salt stress condition, this mutant exhibited attenuated root phenotypes such as shorter primary root length and fewer lateral root numbers. The transcript levels of stress-responsive genes, ABA INSENSITIVE 1 (ABI1) and ABI2, were found to be impaired in the arp6 mutant in comparison with wild-type plants in response to salt stress. In addition, a meta-analysis of published data indicated a number of genes involved in auxin response were induced in arp6 mutant grown in non-stress condition. These imply that the loss of H2A.Z balance (in arp6 mutant) may lead to change stress and auxin responses resulting in alternative root morphogenesis upon both normal and salinity stress conditions.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
H2A.Z-containing nucleosomes have been suggested to play a dual role in the regulation of gene transcription [10, 25]. Hence, these H2A.Z-containing nucleosomes have been documented to define the transcription of various genes involved in development and environmental responses such as floral transition, anthocyanin biosynthesis, thermomorphogenesis, drought stress responses, and shade avoidance syndrome [3, 16, 4, 10, 25, 2, 19, 28]. It is known that the deposition and eviction of H2A.Z are, respectively, controlled by two major chromatin-remodeling complexes including SWR1 and INO80 [21, 1, 11]. ACTIN-RELATED PROTEIN 6 (ARP6) has been characterized as a component of the Arabidopsis SWR1 chromatin-remodeling complex [3, 16, 4]. The Arabidopsis arp6 mutant has been investigated to exhibit several different phenotypes (e.g., early flowering, leaf serration, multiple phosphate-starvation-related phenotypes, high anthocyanins accumulation, and elongated hypocotyl) from wild-type plants [3, 4, 10, 23, 2].
In Arabidopsis, ABA INSENSITIVE 1 (ABI1) and ABI2 genes encode for two different clade A type 2C protein phosphatases (PP2Cs) [12]. These PP2Cs (ABI1 and ABI2) function as important factors in the abscisic acid (ABA) signaling pathway [8]. Previously, abi1 and abi2 mutants were shown to be more sensitive to ABA or abiotic stress (e.g., salt and heat stress conditions) [26, 9]. Auxin is one critical phytohormone that regulates root growth and development [8]. Besides, auxin was also found to contribute to control the root growth in response to salt stress [14]. In this study, the arp6 mutant is found to show retarded root phenotypes such as shorter primary root length and fewer lateral root (LR) numbers in response to salt stress conditions. To support the phenotypic data, the transcript levels of ABI1 and ABI2 were also tested in wild-type and arp6 plants grown under both normal and salt stress conditions. A meta-analysis of published data on arp6 mutant indicated that a set of auxin-related genes was upregulated in this mutant compared with wild type. These imply that the loss of H2A.Z balance (in the arp6 genome) can lead to changes in auxin signaling as well as salt stress response and eventually alterations in root morphogenesis in Arabidopsis thaliana.
Materials and Methods
Plant Materials and Growth Conditions
In this study, the used arp6 mutant is originally named as suppressor of FRIGIDA3 (suf3) which was isolated and characterized by Choi et al. [3]. Arabidopsis wild-type (Columbia, Col-0) and arp6 mutant seeds were surface-sterilized and sown on half-strength Murashige and Skoog (1/2 × MS) (2% sucrose). The whole medium plates containing seeds were then stored at 4 ℃ for 3 days. Afterward, these plates were transferred to a growth chamber with the following conditions: long day (16 h light/8 h dark), light intensity (~ 100 μmol photons m−2 s−1, white-light), and temperature (23 ± 1 ℃).
Salt Stress Treatments
Five-day-old seedlings were transferred to 1/2 × MS (2% sucrose) medium supplemented with 0 or 100 mM NaCl. Next, these seedlings were placed back to the same growth chamber and grown for phenotyping.
Quantitative Reverse Transcription PCR (RT-qPCR)
Two-week-old plants were transferred to 1/2 × MS (2% sucrose) medium supplemented with 0 (Control) or 250 mM NaCl (Salt stress) and were then placed back to the same growth chamber and grown for 6 h. Next, the plants were used for RNA extraction using TRIzol™ Reagent (Invitrogen™, Waltham, Massachusetts, USA). The RNA samples were subsequently subjected to cDNA synthesis and qPCR. The relative transcript levels of two genes, ABI1 (F: 5′-GCCTACCCATTTCCTCCTTCTT-3′ and R: 5′-GGGTTTCCTGGATTGTGGGTA-3′) and ABI2 (F: 5′-TTGCCCAGAATCCAGGAAAC-3′ and R: 5′-AGACAACCTTAGCTAGCACATGA-3′), were calculated by the 2(-Delta Delta C(T)) method [15] and ACTIN2 (F: 5′-GATCTCCAAGGCCGAGTATGAT-3′ and R: 5′-CCCATTCATAAAACCCCAGC-3′) was employed as the internal control.
Results
arp6 Mutant is Sensitive to Salt Stress
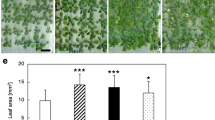
Salt stress was shown to increase H2A.Z-containing nucleosomes eviction resulting in activation of AtMYB44 transcription [18]. This implies that histone variant H2A.Z also involves in the regulation of salt stress responses. In the present work, arp6 mutant was used to test the growth performance upon salt stress condition. Five days after transferring to 1/2 × MS medium supplemented with NaCl (100 mM), both wild-type and arp6 seedlings exhibited shorted primary roots than those on control conditions (Fig. 1A). Upon salt stress, the primary roots of arp6 seedlings were significantly shorter than those of wild type (Fig. 1A). Besides, arp6 mutant also showed fewer LR numbers than wild type on both control and salt stress conditions (Fig. 1B). When the plants (wild-type and arp6) were grown longer on the medium supplemented with NaCl (100 mM) (15 days), it was clearer to see that the arp6 root system was much more sensitive to salt stress than wild type’s one (Fig. 2A). The quantification of primary root length showed that salt stress condition dramatically reduced the root elongation in arp6 in comparison with wild-type plants (Fig. 2B). These results indicate that loss-of-function mutation of ARP6 increases the salt sensitivity, especially in root growth, in Arabidopsis.
Root phenotypes of arp6 mutant in response to salt stress. Primary root length (A) and lateral root (LR) number (B) wild-type (Col-0) and arp6 plants on the 5th day after exposure to salt stress condition (growth medium supplemented with 100 mM NaCl). Data were obtained from three independent experiments. Columns marked with an asterisk (*) indicate significant differences (p < 0.05)
Whole-plant pictures (A) and primary root length (B) of wild-type (Col-0) and arp6 plants on the 15th day after exposure to salt stress condition (growth medium supplemented with 100 mM NaCl). Data were obtained from three independent experiments. Columns marked with an asterisk (*) indicate significant differences (p < 0.05)
ARP6 Positively Regulates Salt Stress-Responsive Genes
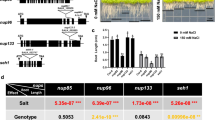
Next, we tested the transcript levels of stress-responsive genes in arp6 mutant in response to salt stress. As shown in Fig. 3, salt stress dramatically induced the expression of ABI1 and ABI2 genes. However, the salt stress-mediated induction of ABI1 and ABI2 was significantly impaired in arp6 mutant (Fig. 3). These molecular data support to explain why the arp6 mutant is more sensitive to salt stress than wild-type plants. A prior study revealed that salt stress inhibited root growth via negative influences on auxin signaling and transport [14]. As indicated in Fig. 1B, arp6 also exhibited fewer LR numbers even in normal growth condition. It implies that the lack of ARP6 may also influence the auxin signaling or response in Arabidopsis. In fact, a meta-analysis of the published data from a previous study [25] showed that many genes involved in auxin response were upregulated in arp6 mutant compared with wild-type when these plants were grown in normal condition (Fig. 4).
Loss-of-function mutation of ARP6 impaired the expression of stress-responsive genes upon salt stress condition. Two-week-old plants [wild-type (Col-0) and arp6] were either grown in normal condition (Control) or treated with NaCl (250 mM) for 6 h (Salt stress). Subsequently, the plants were subjected to RNA extraction and RT-qPCR to test the transcript levels of two stress-responsive genes, ABI1 and ABI2. Data were obtained from three independent experiments. Columns marked with an asterisk (*) indicate significant differences (p < 0.05)
Meta-analysis of upregulated genes in arp6 plants grown in normal conditions. The data published by Sura et al. [25] were obtained and used for gene ontology (GO) analysis. The list of upregulated genes in arp6 mutant when compared to wild-type plants grown in non-stress conditions was extracted from a prior published data [25]. Next, this list was used for GO analysis by DAVID Gene Functional Classification Tool [6]. Ten Biological Process (BP) categories which had highest [−log10 (p-value)] were shown
Discussion
A previous study reported that phosphate-starvation stress also strongly reduced the primary root length of arp6 mutant when compared with wild type [23]. It is well known that different severe abiotic stress conditions (such as drought, salinity, and cold stresses) can significantly inhibit primary root elongation [29, 7]. A high concentration of NaCl (100 mM) in growth media was also found to reduce the lateral root numbers [30]. The cytological analysis showed that salt stress conditions can cause a reduction in cell production and cell division as well as smaller mature cell length leading to unhealthy root morphogenesis [27, 13]. In the present work, we show that loss-of-function mutation of ARP6 increased the salt sensitivity, and this was especially indicated as strongly attenuated root growth of arp6 mutant upon salt stress (Fig. 1 and 2). To further explore the underlying mechanisms, expressions of stress-responsive genes were tested in wild-type and arp6 plants grown under both normal and salt stress conditions. The salt stress significantly triggered the mRNA levels of ABI1 and ABI2 genes in both wild-type and arp6 plants (Fig. 3). However, the mRNA levels of these genes were found to be lower in arp6 than those in wild-type plants (Fig. 3). Previous studies did show that abi1 and abi2 mutants were more sensitive to ABA or abiotic stress (e.g., salt and heat stress conditions) in comparison with wild-type plants [26, 9]. These indicate that ARP6 may control the plant response to salinity stress via the regulation of different PP2C genes and these may consequently influence the ABA signaling pathway. ABA has been known to play as a negative regulator of root growth and branching [20, 24]. In the ABA signaling pathway, the PP2Cs (e.g., ABI1 and ABI2) proteins function as the brake which negatively influences the signal transduction [8]. As shown in Fig. 3, the loss-of-function mutation in ARP6 resulted in the reduction of transcript levels of ABI1 and ABI2 genes upon salt stress treatment. Thus, it is possible that the ABA signaling transduction was highly activated in arp6 mutant grown on salt-supplemented media leading to retarding root growth.
Auxin is known as a critical phytohormone in the regulation of root growth in both normal growth as well as salt stress conditions [5, 22, 14, 17]. Here, we found that the arp6 mutant not only exhibited retarded root growth upon salt stress condition, but it also reduced the primary root growth under normal condition (Fig. 1 and 2). As shown in Fig. 4, the meta-analysis of published data on arp6 mutant indicated that a set of auxin-related genes was upregulated in this mutant compared with wild type. Overall, these data suggest that the loss of balance in H2A.Z in the genome of arp6 mutant may alter auxin signaling in plants resulting in inhibition of root growth upon both normal and salt stress conditions.
Conclusion
In this study, arp6 mutant was found to exhibit impaired root growth such as shorter primary root length and fewer lateral root numbers in response to salt stress. The RT-qPCR analysis revealed that ABI1 and ABI2 transcript levels were significantly lower in arp6 mutant in comparison with wild type upon salt stress treatment. Based on the phenotypic and gene expression analyses of arp6 mutant, this study provides an additional role of H2A.Z histone variant in the regulation of plant response to salt stress.
Data Availability
The data published by Sura et al. [25] were obtained and used for gene ontology (GO) analysis and are available at https://doi.org/10.1105/tpc.16.00573.
References
Brahma, S., Udugama, M. I., Kim, J., Hada, A., Bhardwaj, S. K., Hailu, S. G., Lee, T. H., & Bartholomew, B. (2017). INO80 exchanges H2A.Z for H2A by translocating on DNA proximal to histone dimers. Nat Commun, 8, 15616. https://doi.org/10.1038/ncomms15616
Cai, H., Zhang, M., Chai, M., He, Q., Huang, X., Zhao, L., & Qin, Y. (2019). Epigenetic regulation of anthocyanin biosynthesis by an antagonistic interaction between H2A.Z and H3K4me3. New Phytologist, 221, 295–308. https://doi.org/10.1111/nph.15306
Choi, K., Kim, S., Kim, S. Y., Kim, M., Hyun, Y., Lee, H., Choe, S., Kim, S. G., Michaels, S., & Lee, I. (2005). SUPPRESSOR OF FRIGIDA3 encodes a nuclear ACTIN-RELATED PROTEIN6 required for floral repression in Arabidopsis. The Plant Cell, 17, 2647–2660. https://doi.org/10.1105/tpc.105.035485
Deal, R. B., Topp, C. N., McKinney, E. C., & Meagher, R. B. (2007). Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A.Z. The Plant Cell, 19, 74–83. https://doi.org/10.1105/tpc.106.048447
Fukaki, H., & Tasaka, M. (2009). Hormone interactions during lateral root formation. Plant Molecular Biology, 69, 437–449. https://doi.org/10.1007/s11103-008-9417-2
Huang, D. W., Sherman, B. T., Tan, Q., Collins, J. R., Alvord, W. G., Roayaei, J., Stephens, R., Baseler, M. W., Lane, H. C., & Lempicki, R. A. (2007). The DAVID Gene Functional Classification Tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biology, 8, R183. https://doi.org/10.1186/gb-2007-8-9-r183
Julkowska, M. M., Hoefsloot, H. C., Mol, S., Feron, R., de Boer, G. J., Haring, M. A., & Testerink, C. (2014). Capturing Arabidopsis root architecture dynamics with ROOT-FIT reveals diversity in responses to salinity. Plant Physiology, 166, 1387–1402. https://doi.org/10.1104/pp.114.248963
Jung, C., Nguyen, N. H., & Cheong, J.-J. (2020). Transcriptional regulation of protein phosphatase 2C genes to modulate abscisic acid signaling. International Journal of Molecular Sciences, 21, 9517.
Khan, I. U., Ali, A., Khan, H. A., Baek, D., Park, J., Lim, C. J., Zareen, S., Jan, M., Lee, S. Y., Pardo, J. M., et al. (2020). PWR/HDA9/ABI4 complex epigenetically regulates ABA dependent drought stress tolerance in Arabidopsis. Frontiers in Plant Science, 11, 623. https://doi.org/10.3389/fpls.2020.00623
Kumar, S. V., & Wigge, P. A. (2010). H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell, 140, 136–147. https://doi.org/10.1016/j.cell.2009.11.006
Lademann, C. A., Renkawitz, J., Pfander, B., & Jentsch, S. (2017). The INO80 complex removes H2A.Z to promote presynaptic filament formation during homologous recombination. Cell Reports, 19, 1294–1303. https://doi.org/10.1016/j.celrep.2017.04.051
Leung, J., Merlot, S., & Giraudat, J. (1997). The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. The Plant Cell, 9, 759–771. https://doi.org/10.1105/tpc.9.5.759
Li, H., Yan, S., Zhao, L., Tan, J., Zhang, Q., Gao, F., Wang, P., Hou, H., & Li, L. (2014). Histone acetylation associated up-regulation of the cell wall related genes is involved in salt stress induced maize root swelling. BMC Plant Biology, 14, 105. https://doi.org/10.1186/1471-2229-14-105
Liu, W., Li, R. J., Han, T. T., Cai, W., Fu, Z. W., & Lu, Y. T. (2015). Salt stress reduces root meristem size by nitric oxide-mediated modulation of auxin accumulation and signaling in Arabidopsis. Plant Physiology, 168, 343–356. https://doi.org/10.1104/pp.15.00030
Livak, K. J., & Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods, 25, 402–408. https://doi.org/10.1006/meth.2001.1262
Martin-Trillo, M., Lazaro, A., Poethig, R. S., Gomez-Mena, C., Pineiro, M. A., Martinez-Zapater, J. M., & Jarillo, J. A. (2006). Early in short days 1 (ESD1) encodes ACTIN-RELATED PROTEIN 6 (AtARP6), a putative component of chromatin remodelling complexes that positively regulates FLC accumulation in Arabidopsis. Development, 133, 1241–1252. https://doi.org/10.1242/dev.02301
Nguyen, C. T., Tran, G. B., & Nguyen, N. H. (2020). Homeostasis of histone acetylation is critical for auxin signaling and root morphogenesis. Plant Molecular Biology, 103, 1–7. https://doi.org/10.1007/s11103-020-00985-1
Nguyen, N. H., & Cheong, J. J. (2018). H2A.Z-containing nucleosomes are evicted to activate AtMYB44 transcription in response to salt stress. Biochemical and Biophysical Research Communications, 499, 1039–1043. https://doi.org/10.1016/j.bbrc.2018.04.048
Nguyen, N. H. (2020). Histone variant H2A.Z and transcriptional activators may antagonistically regulate flavonoid biosynthesis. AIMS Bioengineering, 7, 55–59. https://doi.org/10.3934/bioeng.2020005
Nguyen, H. N., Kim, J. H., Hyun, W. Y., Nguyen, N. T., Hong, S. W., & Lee, H. (2013). TTG1-mediated flavonols biosynthesis alleviates root growth inhibition in response to ABA. Plant Cell Reports, 32, 503–514. https://doi.org/10.1007/s00299-012-1382-1
Papamichos-Chronakis, M., Watanabe, S., Rando, O. J., & Peterson, C. L. (2011). Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell, 144, 200–213. https://doi.org/10.1016/j.cell.2010.12.021
Petricka, J. J., Winter, C. M., & Benfey, P. N. (2012). Control of Arabidopsis root development. Annual Review of Plant Biology, 63, 563–590. https://doi.org/10.1146/annurev-arplant-042811-105501
Smith, A. P., Jain, A., Deal, R. B., Nagarajan, V. K., Poling, M. D., Raghothama, K. G., & Meagher, R. B. (2010). Histone H2A.Z regulates the expression of several classes of phosphate starvation response genes but not as a transcriptional activator. Plant Physiology, 152, 217–225. https://doi.org/10.1104/pp.109.145532
Sun, L. R., Wang, Y. B., He, S. B., & Hao, F. S. (2018). Mechanisms for abscisic acid inhibition of primary root growth. Plant Signaling & Behavior, 13, e1500069. https://doi.org/10.1080/15592324.2018.1500069
Sura, W., Kabza, M., Karlowski, W. M., Bieluszewski, T., Kus-Slowinska, M., Paweloszek, L., Sadowski, J., & Ziolkowski, P. A. (2017). Dual role of the histone variant H2A.Z in transcriptional regulation of stress-response genes. The Plant Cell, 29, 791–807. https://doi.org/10.1105/tpc.16.00573
Suzuki, N., Bassil, E., Hamilton, J. S., Inupakutika, M. A., Zandalinas, S. I., Tripathy, D., Luo, Y., Dion, E., Fukui, G., Kumazaki, A., et al. (2016). ABA is required for plant acclimation to a combination of salt and heat stress. PLoS One, 11, e0147625. https://doi.org/10.1371/journal.pone.0147625
West, G., Inze, D., & Beemster, G. T. (2004). Cell cycle modulation in the response of the primary root of Arabidopsis to salt stress. Plant Physiology, 135, 1050–1058. https://doi.org/10.1104/pp.104.040022
Willige, B. C., Zander, M., Yoo, C. Y., Phan, A., Garza, R. M., Trigg, S. A., He, Y., Nery, J. R., Chen, H., Chen, M., Ecker, J. R., & Chory, J. (2021). PHYTOCHROME-INTERACTING FACTORs trigger environmentally responsive chromatin dynamics in plants. Nature Genetics, 53, 955–961. https://doi.org/10.1038/s41588-021-00882-3
Xiong, L., Wang, R. G., Mao, G., & Koczan, J. M. (2006). Identification of drought tolerance determinants by genetic analysis of root response to drought stress and abscisic acid. Plant Physiology, 142, 1065–1074. https://doi.org/10.1104/pp.106.084632
Zolla, G., Heimer, Y. M., & Barak, S. (2010). Mild salinity stimulates a stress-induced morphogenic response in Arabidopsis thaliana roots. Journal of Experimental Botany, 61, 211–224. https://doi.org/10.1093/jxb/erp290
Acknowledgements
We would like to thank Prof. Ilha Lee (Seoul National University, South Korea) for his kindness in providing the arp6 mutant. We sincerely appreciate Dr. Nguyen Minh Tan (UCLA, USA) for his critical reading and comments on the manuscript.
Funding
This work was supported by Ho Chi Minh City Open University.
Author information
Authors and Affiliations
Contributions
NHN designed and supervised the research. BHD and NHN performed the experiments. BHD and NHN wrote the manuscript. NTH and TDL read and commented on the manuscript. All authors gave final approval for publication and agree to be held accountable for the work performed therein.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical Approval
This study was performed on the model plant (Arabidopsis thaliana) and was not involved in human participants and/or animals.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original article has been corrected to update affiliation 1.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Do, B.H., Hiep, N.T., Lao, T.D. et al. Loss-of-Function Mutation of ACTIN-RELATED PROTEIN 6 (ARP6) Impairs Root Growth in Response to Salinity Stress. Mol Biotechnol 65, 1414–1420 (2023). https://doi.org/10.1007/s12033-023-00653-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-023-00653-x