Abstract
Southern blight, stem rot, and root rot are serious soil-borne fungal diseases of peanut, which are caused by Sclerotium rolfsii, Lasiodiplodia theobromae, and Fusarium oxysporum, respectively. These diseases are difficult to be diagnosed in early stage of infection, causing the optimal treatment period was often missed. Therefore, establishing a rapid detection system is of great significance for early prevention of peanut soil-borne fungal diseases. Here, we have invented a multiplex PCR detection system to detect fungal pathogens of peanut southern blight, stem rot, and root rot at the same time. The quarantine fungal pathogen primer pairs were amplified to the specific number of base pairs in each of the following fungal pathogens: 1005-bp (F. oxysporum), 238-bp (L. theobromae), and 638-bp (S. rolfsii). The detection limit for the single and multiplex PCR primer sets was 1 ng of template DNA under in vitro conditions. Amplification of fungi of non-target species yielded no non-specific products. The validation showed that the multiplex PCR could effectively detect single and mixed infections in field samples. Overview, this study proved that this mPCR assay was a rapid, reliable, and simple tool for the simultaneous detection of three important peanut soil-borne diseases, which facilitated prompt treatment and prevention of peanut root diseases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Peanut is a vital oil crop and cash crop in the world [1], and China is the largest peanut producer [2]. In China, Shandong Province ranked second in peanut planting area in 2019, with a planting area of 650,000 hectares. Peanut soil-borne fungal diseases are one of the disastrous factors limiting peanut yield, which seriously threaten the quality of peanut and the safety of agricultural products [3]. The main causes of the serious soil-borne disease of peanut were the peanut continuous cropping barrier and the lack of disease resistant varieties [4].
At present, southern blight, stem rot, and root rot are three typical soil-borne fungal diseases of peanut [5], which have caused huge economic losses to the development of peanut-related industries. Peanut southern blight, caused by Sclerotium rolfsii, is a critical fungal disease in peanut worldwide [6,7,8]. Peanut stem rot is a vital soil-borne disease [9], which is caused by Lasiodiplodia theobromae that is a necrotic phytopathogen with a wide host range and widespread distribution [10]. Additionally, peanut root rot is caused by Fusarium oxysporum. The incidence rate of peanut root rot has greatly increased and already become a devastating disease [11,12,13]. These three diseases mainly damage the root and base of the stem, which were often difficult to diagnose in the early stage, so the best prevention period was easily missed. Therefore, establishment of rapid detection system of these three diseases is significant for protection of peanut cultivation.
Although individual PCR assay method that detects only single pathogen at a time is effective, PCR technology is time-consuming and expensive when applied to a large number of pathogen species leading to a variety of diseases [14, 15]. However, the emergence of multiplex PCR technology provides an effective method, which adds multiple pairs of primers in a reaction system at the same time and amplifies a large number of target fragments while using multiple DNA templates [16]. In recent years, based on the advantage of faster speed, higher sensitivity, and stronger specificity, multiplex PCR has been widely used [17,18,19,20].
In addition, fungal parasitism on seeds is an important factor in the spread of plant diseases. Thus, it is very important to establish a system to quickly detect and distinguish these three diseases. However, a multiplex PCR rapid detection system for detecting the main rhizome mycoses of peanuts has not been established. The contributions of this study were as follows: (1) designed the specific primers to detecting S. rolfsii and L. theobromae and used the universal primers for detection of Fusarium spp. and (2) established a multiplex PCR rapid detection system for southern blight, stem rot, and root rot of peanuts, which provided technical support for the early and rapid diagnosis of the three diseases, and this system also could be used for the rapid detection of diseased plants and seeds of peanut.
Materials and Methods
Tested Strains
The numbers and sources of all isolates are listed in Table 1. Sclerotium rolfsii, L. theobromae, and F. oxysporum were used as target strains for multiplex PCR detection. The specificity of primers was verified by Rhizoctonia cerealis, F. graminearum, F. pseudograminearum, R. solani, Sclerotinia sclerotiorum, Phytophthora parasitica var. nicotianae Tucker, and Pythium myriotylum. The strains were obtained from plant pathology laboratory of Shandong Agricultural University. These fungi were incubated in Petri dishes containing potato dextrose agar (PDA) medium for 3–5 days at 28 °C [21].
Primer Design
Specific primers were designed using Primer 5 software based on the specific gene beta-tubulin gene (GenBank accession no. MN078927.1) of S. rolfsii, the cytb-specific gene (GenBank accession no. MH880818.1) of L. theobromae, and the universal primer of F. oxysporum [22, 23]. As shown in Table 2, all primers were synthesized by Biochemical and Biological Engineering (Shanghai) Co., Ltd.
PCR Amplification
The single PCR system is shown in Table 3, and the multiplex PCR mixture system is shown in Table 4. Single PCR conditions: pre-denaturation at 94 °C for 5 min, denaturation at 94 °C for 40 s, annealing at 63 °C for 40 s, extension at 72 °C for 1 min, followed by 30 cycles, and final extension at 72 °C for 10 min [24]. Similar conditions were used for multiplex PCR, except for the annealing temperature.
Optimization of the Annealing Temperature for Multiplex PCR
The temperature gradient PCR procedure was designed between 58.6 and 63.4 °C, and temperatures were 58.6 °C, 59.2 °C, 59.9 °C, 60.6 °C, 61.3 °C, 62 °C, 62.8 °C, and 63.4 °C [25]. After the reactions, 5 μL of each product was used for 1% agarose gel electrophoresis (200 V, constant for 20 min), after ethidium bromide staining and then observed under UV light. All tests were repeated thrice.
Multiple PCR Primer-Specific Amplification
The mixed DNA of S. rolfsii, L. theobromae, and F. oxysporum was used as the positive control and ddH2O was used as the negative control. The single DNA of S. rolfsii, L. theobromae, and F. oxysporum and the genomic DNA of R. cerealis, R. solani, S. sclerotiorum, P. nicotianae, and P. fulvum were used as templates for triple PCR detection at the determined multiplex PCR annealing temperature to test the specificity of triple PCR primers [26]. All experiments were repeated thrice.
Sensitivity Analysis of Single and Multiplex PCR
First, dilute the initial template concentration of DAN of S. rolfsii, L. theobromae, and F. oxysporum to 10 ng/μL and then serially dilute tenfold (10 ng to 10 fg) with distilled water [27]. In addition, the initial template concentration of the mixed DAN of the three pathogenic fungus genomes was diluted to 10 ng/μL and serially diluted 10 times to evaluate the sensitivity of mPCR detection. Single multiplex and multiplex PCR were performed under optimized conditions [28]. All experiments were repeated thrice.
Multiple PCR Vaccination Testing
Sclerotium rolfsii, L. theobromae, and F. oxysporum were cultured in an incubator at 25 °C for 2 days. Then, five fungal culture blocks were taken out from those and transplanted into the wheat kernel medium, incubated at a constant temperature at 25 °C for 4 days, and shaken four times a day. Fungi used for inoculation experiment were cultured with wheat grains. Two wheat grains with different fungi were placed at the base of the peanut stems after the seedling emergence and cultivated in a biochemical incubator at 25 °C and a humidity of 80%. Drying-weighing method is used for soil moisture measurement [29]. The disease incidence of plants at 12 h, 16 h, 20 h, 24 h, and 48 h was observed and recorded. Total genomic DNA of the rhizome was extracted by the improved CTAB method [30], which was used as a template for PCR detection. The established triple PCR system was used for detection of pathogenic fungi in peanut.
Detection of Pathogenic Fungi in Peanut
One hundred and two peanut plant samples were collected from peanut fields in Linyi, Tai’an, and Jinan in Shandong Province in August 2020; 216 peanut seed samples from peanut fields in Linyi, Tai’an, and Jinan in Shandong Province. The total DNA of rhizomes and seeds was extracted and detected by the established triple PCR system.
Results
Optimization of the Annealing Temperature for mPCR
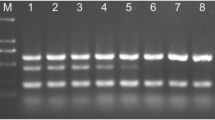
As shown in Fig. 1, all genes were amplified between 58.6 and 63.4 °C, with the highest amplification at 60.6 °C. Therefore, 54.9 °C was selected as the optimal annealing temperature.
Sensitivity of the Single and Multiplex PCR
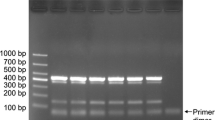
Genomic DNA was sequentially diluted for the sensitivity assay of single and multiplex PCR. A single PCR amplification sensitivity test detected 1-ng, 10-pg, and 10-pg DNA for S. rolfsii, L. theobromae, and F. oxysporum, respectively (Fig. 2), while the mPCR detected 1 ng, 10 pg, and 100 pg for S. rolfsii, L. theobromae, and F. oxysporum, respectively (Fig. 3). In addition, for the detection sensitivity of stem rot and southern blight, the results of mPCR were similar to that of the single PCR. However, as for root rot, the detection sensitivity of mPCR was decreased compared with that of the single PCR.
mPCR Primer Specificity Amplification
The mPCR assay yielded fragments of the expected size using S. rolfsii, L. theobromae, and F. oxysporum as genomic DNA with specific primers, while no products were obtained with non-target species (Fig. 4), thereby validating the specificity of the mPCR assay.
Specificity of multiplex PCR. Lane M: DL2000 marker; Lane 1: positive control; Lane 2: negative control; Lane 3–5: F. oxysporum; Lane 6–9: L. theobromae; Lane 10–14: S. rolfsii; Lane 15: R. solani; Lane 16: R. cerealis; Lane 17: S. sclerotiorum; Lane 18: P. parasitica var. nicotianae Tucker; and Lane 19: P. myriotylum
Evaluation of Multiple PCR in Vaccination Testing
The three fungi were inoculated in a biochemical incubator with 25 °C (soil temperature 23.9 °C) and 80% humidity (soil humidity 53.6%) and collected inoculated plants and control (uninoculated) plant. The DNA of the rhizome was extracted for multiplex PCR detection. After inoculation for 20 h, 24 h, and 48 h, the southern blight fungus, stalk rot fungus, and root rot fungus were detected, respectively (Fig. 5). Moreover, plants exhibited no obvious symptoms, indicating that the system could detect these three pathogens in plants that have not yet shown disease symptoms and could be used for early detection of peanut diseases in the field.
Application of mPCR Detection of Peanut
For the collected 102 peanut samples, DNA was extracted from the rhizomes and subjected to mPCR detection. As shown in Fig. 6, a total of 24 plants with peanut southern blight, 15 plants with peanut root rot, and 6 plants with peanut stem rot were detected. Among them, 4 peanut plants were co-infected with stem rot fungus (L. theobromae) and southern blight fungus (S. rolfsii), which proved that this multiplex PCR system could be used to detect the plants with lesions caused by these three diseases (Table 5).
For 216 peanut seeds collected in Shandong Province, mPCR detection was performed after extracting the seed DNA. As shown in Fig. 7, a total of 71 peanut seeds carrying southern blight, 16 peanut seeds carrying root rot, and 12 peanut seeds carrying stem rot pathogen were detected. Among them, 3 seeds had the phenomenon of co-infection, 1 seed was detected with southern blight fungus and stem rot fungus at the same time and 2 seeds were detected by southern blight fungus and root rot fungus at the same time (Table 6). This result indicated that the established multiplex PCR system can be used to detect seeds and then effective control measures can be taken to reduce the damage of seed-borne diseases and minimize the losses from diseases caused by seeds with fungus.
Discussion
In general, staining, culture, and biochemical identification are major traditional methods to detect pathogenic fungi, which are time-consuming and possess low sensitivity [31, 32]. With the rapid development of molecular technology, molecular detection technology based on PCR technology is increasingly valued and favored [32,33,34,35]. Among them, a single PCR is suitable for detecting a single disease [36]. However, the peanut soil-borne diseases often mixing occurs, thus the single PCR is usually difficult to screen a large number of peanut samples for the presence of pathogens. Although the detection sensitivity of multiplex PCR is slightly lower or close to that of single PCR [37], multiplex PCR can simultaneously amplify multiple fragments and the characteristics of simplicity and convenience in practice and relatively low cost [38,39,40,41], which satisfies the needs of rapid detection of peanut root diseases.
In addition, it was the first time to detect the peanut soil-borne diseases by multiplex PCR. In this study, the multiple PCR assays established for the detection of pathogenic fungi of peanut stem rot, root rot, and southern blight with 10 pg/L, 100 pg/L, and 1 ng/L, respectively (Fig. 3). Similar findings have been reported before [42,43,44]. Compared with the single PCR sensitivity test results (Fig. 2), the sensitivity results of stem rot pathogen and southern blight pathogen were the same and the sensitivity of root rot fungus decreased from 10 to 100 pg/μL (Fig. 3). Presumably, the decreased sensitivity is due to competition between the three pairs for Taq DNA enzymes and dNTPs, decreasing the number of amplification products [20, 45], another reason may be that the three pairs of primers interact with each other so that the binding of the primers to the DNA template is not tight and does not occur sufficiently [14, 46].
According to the relevant research, the phenomenon of mixed occurrence of diseases on the same plant is common [47, 48]. At present, a variety of diseases have been detected in strawberries, grape, and mango [49,50,51]. Since the stem rot, root rot, and southern blight disease of peanut can infect the seeds and make the seeds carry bacteria, the established multiplex PCR system is able to detect and verify not only the field peanut plant samples, but also the peanut seed samples. It has been reported that, in the seeds of rapeseed (Brassica campestris L.) [52] and sunflower (Solanum tuberosum L.) [53], multiplex PCR has been successfully used in detecting multiple seed-borne pathogens. Our results showed that three pathogens could be detected in peanut and peanut seeds with a relative high positive detection rate. The relative high rate of positive detection of peanut plants may be the collection of peanut plant samples with more typical diseases (Table 5) [54,55,56]. The positive detection rate of peanut seeds was not higher than that of plants (Table 6), may be due to the low amount of seed epidermis with fungi.
The results of the testing of field samples showed that it was found that there was a phenomenon of co-infection of peanut southern blight and peanut stem rot on peanut plants. There are co-infection of peanut southern blight fungus and peanut stem rot on peanut seeds, as well as peanut southern blight fungus and peanut root rot fungus in practice, indicating that these three diseases are indeed mixed. Studies have shown that this co-infection occurs not only in peanuts but has also been found in sugarcane, watermelon, and wheat. [20, 57, 58]. In this study, the peanut mPCR system enables pathogen detection of co-infected peanut soil-borne diseases, which contributed to effectively controlling peanut diseases.
In conclusion, the mPCR system established in our work for the detection of peanut southern blight, stem rot, and root rot is simple, rapid, and sensitive. The system has important practical value for the control of peanut root and stem diseases to ensure the safety of peanut crops.
Data Availability
The data that support the findings of this study are available on request from the corresponding author, upon reasonable request.
References
Yang, H., Ni, J., Gao, J., Han, Z., & Luan, T. (2021). A novel method for peanut variety identification and classification by improved VGG16. Scientific Reports, 11(1), 15756.
Ahmad, R., Hussain, B., & Ahmad, T. (2021). Fresh and dry fruit production in Himalayan Kashmir, Sub-Himalayan Jammu and Trans-Himalayan Ladakh, India. Heliyon, 7(1), e05835.
Minarni, Warman, I., & Yuhendra. (2019). Implementation of case-based reasoning and nearest neighbor similarity for peanut disease diagnosis. Journal of Physics: Conference Series, 1196, 012053.
Ahmad, A., Attia, A. G., Mohamed, M. S., & Elsayed, H. E. (2019). Fermentation, formulation and evaluation of PGPR Bacillus subtilis isolate as a bioagent for reducing occurrence of peanut soil-borne diseases. Journal of Integrative Agriculture, 18(9), 140–152.
Liu, J., Li, X., Jia, Z., Zhang, T., & Wang, X. (2016). Effect of benzoic acid on soil microbial communities associated with soilborne peanut diseases. Applied Soil Ecology, 110, 34–42.
Jogi, A., Kerry, J. W., Brenneman, T. B., Leebens-Mack, J. H., & Gold, S. E. (2016). Identification of genes differentially expressed during early interactions between the stem rot fungus (Sclerotium rolfsii) and peanut (Arachis hypogaea) cultivars with increasing disease resistance levels. Microbiological Research, 184, 1–12.
Jacob, S., Sajjalaguddam, R. R., & Sudini, H. K. (2018). Streptomyces sp. RP1A-12 mediated control of peanut stem rot caused by Sclerotium rolfsii. Journal of Integrative Agriculture, 17(04), 892–900.
Standish, J. R., Culbreath, A. K., Branch, W. D., & Brenneman, T. B. (2019). Disease and yield response of a stem-rot-resistant and -susceptible peanut cultivar under varying fungicide inputs. Plant Disease, 103(11), 2781–2785.
Liu, F., Yan, S., Xu, F., Zhang, Z., Lu, Y., Zhang, J., & Wang, G. (2022). Characteristics of biological control and mechanisms of Pseudomonas chlororaphis zm-1 against peanut stem rot. BMC Microbiology, 22(1), 9.
Xie, X. G., Zhang, F. M., Wang, X. X., Li, X. G., & Dai, C. C. (2019). Phomopsis liquidambari colonization promotes continuous cropping peanut growth by improving the rhizosphere microenvironment, nutrient uptake and disease incidence. Journal of the Science of Food and Agriculture, 99(4), 1898–1907.
Palacios, S., Casasnovas, F., Ramirez, M. L., Reynoso, M. M., & Torres, A. M. (2014). Impact of water potential on growth and germination of Fusarium solani soilborne pathogen of peanut. Brazilian Journal of Microbiology, 45(3), 1105–1112.
Sun, K., Xie, X. G., Lu, F., Zhang, F. M., & Dai, C. C. (2021). Peanut preinoculation with a root endophyte induces plant resistance to soil-borne pathogen Fusarium oxysporum via activation of salicylic acid-dependent signaling. Plant and Soil, 460, 297–312.
Zhang, F. M., He, W., Wu, C. Y., Sun, K., Zhang, W., & Dai, C. C. (2020). Phomopsis liquidambaris inoculation induces resistance in peanut to leaf spot and root rot. BioControl, 65, 475–488.
Bilgiç, H. B., Karagenç, T., Simuunza, M., Shiels, B., Tait, A., Eren, H., & Weir, W. (2013). Development of a multiplex PCR assay for simultaneous detection of Theileria annulata, Babesia bovis and Anaplasma marginale in cattle. Experimental Parasitology, 133(2), 222–229.
Adachi, N., & Oku, T. (2000). PCR-mediated detection of Xanthomonas oryzae pv. oryzae by amplification of the 16S–23S rDNA spacer region sequence. Journal of General Plant Pathology, 66, 303–309.
Henegariu, O., Heerema, N. A., Dlouhy, S. R., Vance, G. H., & Vogt, P. H. (1997). Multiplex PCR: Critical parameters and step-by-step protocol. BioTechniques, 23(3), 504–511.
Chen, S., Cao, Y. Y., Li, T. Y., & Wu, X. X. (2015). Simultaneous detection of three wheat pathogenic fungal species by multiplex PCR. Phytoparasitica, 43, 449–460.
Cho, H. J., Hong, S. W., Kim, H. J., & Kwak, Y. S. (2016). Development of a multiplex PCR method to detect fungal pathogens for quarantine on exported cacti. The Plant Pathology Journal, 32(1), 53–57.
Yu, J., Zhao, Y., Ai, G., Xu, H., Dou, D., & Shen, D. (2019). Development of multiplex PCR assay for simultaneous detection of five cucumber pathogens based on comparative genomics. Australasian Plant Pathology, 48, 369–372.
Sun, X., Zhang, L., Meng, C., Zhang, D., Xu, N., & Yu, J. (2020). Establishment and application of a multiplex PCR assay for detection of Rhizoctonia cerealis, Bipolaris sorokiniana, and Fusarium spp. in winter wheat. Journal of Plant Pathology, 102, 19–27.
Youssef, N. A. (2015). Efficacy of the entomopathogenic nematodes and fungi for controlling the tomato leaf miner, Tuta absoluta (Meyrick) (Lepidoptera: Gelechiidae). Arab Universities Journal of Agricultural Sciences, 23(2), 591–598.
Murray, M. G., & Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research, 8(19), 4321–4326.
Michielse, C. B., & Rep, M. (2010). Pathogen profile update: Fusarium oxysporum. Molecular Plant Pathology, 10(3), 311–324.
Waters, D. L., & Shapter, F. M. (2014). The polymerase chain reaction (PCR): General methods. Methods in Molecular Biology, 1099, 65–75.
Zhang, H. W., Liu, J., Dong, Y. L., Hu, J., Lamour, K., & Yang, Z. M. (2022). A one-step multiplex PCR assay for the detection and differentiation of four species of Clarireedia causing dollar spot on turfgrass. Pest Management Science. https://doi.org/10.1002/ps.7276
Bangratz, M., Wonni, I., Kini, K., Sondo, M., Brugidou, C., Béna, G., Gnacko, F., Barro, M., Koebnik, R., Silué, D., & Tollenaere, C. (2020). Design of a new multiplex PCR assay for rice pathogenic bacteria detection and its application to infer disease incidence and detect co-infection in rice fields in Burkina Faso. PLoS ONE, 15(4), e0232115.
Lee, G. Y., Suh, S. M., Lee, Y. M., & Kim, H. Y. (2022). Multiplex PCR assay for simultaneous identification of five types of tuna (Katsuwonus pelamis, Thunnus alalonga, T. albacares, T. obesus and T. thynnus). Foods, 11(3), 280.
Lee, O. M., Lee, H. J., Kang, S. I., Jeong, J. Y., Kwon, Y. K., & Kang, M. S. (2022). A multiplex real-time PCR assay for differential identification of avian Chlamydia. Avian Pathology: Journal of the W.V.P.A., 51(2), 164–170.
Vauclin, M., Haverkamp, R., & Vachaud, G. (1984). Error analysis in estimating soil water content from neutron probe measurements: 2. Spatial standpoint1. Soil Science, 137, 78–90.
Nicholson, P., & Parry, D. W. (1996). Development and use of a PCR assay to detect Rhizoctonia cerealis, the cause of sharp eyespot in wheat. Plant Pathology, 45, 872–883.
Atkins, S. D., & Clark, I. M. (2004). Fungal molecular diagnostics: A mini review. Journal of Applied Genetics, 45(1), 3–15.
McCartney, H. A., Foster, S. J., Fraaije, B. A., & Ward, E. (2003). Molecular diagnostics for fungal plant pathogens. Pest Management Science, 59(2), 129–142.
Zhou, J., Liao, Y., Li, H., Lu, X., Han, X., Tian, Y., Chen, S., & Yang, R. (2015). Development of a loop-mediated isothermal amplification assay for rapid detection of Trichosporon asahii in experimental and clinical samples. Biomed Research International, 2015, 732573.
Law, J. W., Ab Mutalib, N. S., Chan, K. G., & Lee, L. H. (2014). Rapid methods for the detection of foodborne bacterial pathogens: Principles, applications, advantages and limitations. Frontiers in Microbiology, 5, 1–19.
Sharma, A., Chouhan, A., Bhatt, T., Kaur, A., & Minhas, A. P. (2022). Selectable markers to marker-free selection in rice. Molecular Biotechnology, 64, 841–851.
Huang, Y., Zhang, H., Wei, Y., Cao, Y., Zhu, Q., Li, X., Shan, T., Dai, X., & Zhang, J. (2023). Characterizing the amplification of STR markers in multiplex polymerase chain displacement reaction using massively parallel sequencing. Forensic Science International: Genetics, 62, 102802.
Moustacas, V. S., Silva, T. M., Costa, L. F., Xavier, M. N., Carvalho, C. A., Jr., Costa, É. A., Paixão, T. A., & Santos, R. L. (2013). Species-specific multiplex PCR for the diagnosis of Brucella ovis, Actinobacillus seminis, and Histophilus somni infection in rams. BMC Veterinary Research, 9, 51.
Sun, Q., Cheng, J., Lin, R., Li, J., Zhang, Y., Liang, X., Su, Y., Pang, R., Xue, L., Zeng, H., Gu, Q., Ding, Y., Wu, Q., Chen, M., & Zhang, J. (2022). A novel multiplex PCR method for simultaneous identification of hypervirulent Listeria monocytogenes clonal complex 87 and CC88 strains in China. International Journal of Food Microbiology, 366, 109558.
Korbie, D., & Trau, M. (2022). Multiplex PCR design for scalable resequencing. Methods in Molecular Biology, 2392, 143–158.
Liu, Z., Yu, Y., Fotina, T., Petrov, R., Klishchova, Z., Fotin, A., & Ma, J. (2022). Multiplex PCR assay based on the citE2 gene and intergenic sequence for the rapid detection of Salmonella Pullorum in chickens. Poultry Science, 101(8), 101981.
Hong, Y., Ji, R., Wang, Z., Gu, J., Jiao, X., & Li, Q. (2023). Development and application of a multiplex PCR method to differentiate Salmonella enterica serovar Typhimurium from its monophasic variants in pig farms. Food Microbiology, 109, 104135.
Kundave, V. R., Ram, H., Banerjee, P. S., Garg, R., Mahendran, K., Ravikumar, G., & Tiwari, A. K. (2018). Development of multiplex PCR assay for concurrent detection of tick borne haemoparasitic infections in bovines. Acta Parasitologica, 63(3), 759–765.
de Cássia-Pires, R., de Melo, M. F., Barbosa, R. D., & Roque, A. L. (2017). Multiplex PCR as a tool for the diagnosis of Leishmania spp. kDNA and the gapdh housekeeping gene of mammal hosts. PLoS ONE, 12, e0173922.
Peňázová, E., Dvořák, M., Ragasová, L., Kiss, T., Pečenka, J., Čechová, J., & Eichmeier, A. (2020). Multiplex real-time PCR for the detection of Clavibacter michiganensis subsp. michiganensis, Pseudomonas syringae pv. tomato and pathogenic Xanthomonas species on tomato plants. PLoS ONE, 15, e0227559.
Zhang, X. M., Li, D., Liu, C. B., Yu, N. J., Li, Y. J., & Xie, D. M. (2022). Multiplex PCR method for simultaneous identification of Xanthii Fructus and its adulterants. China Journal of Chinese Materia Medica, 47(10), 2605–2613.
Edwards, M. C., & Gibbs, R. A. (1994). Multiplex PCR: Advantages, development, and applications. PCR Methods and Applications, 3(4), 65–75.
Li, J. T., Fu, J. F., & Li, H. C. (2013). Epidemic processes and yield loss of northern leaf blight (Exserohilum turcicum), curvularia leaf spot (Curvularia lunata) and gray leaf spot (Cercospora zeae-maydis) of maize when they occurred together in the field. Acta Phytopathologica Sinica, 43(3), 301–309.
Dang, P. M., Lamb, M. C., & Chen, C. Y. (2021). Association of differentially expressed R-gene candidates with leaf spot resistance in peanut (Arachis hypogaea L.). Molecular Biology Reports, 48(1), 323–334.
Wang, N., Wang, J., Yin, D., Gao, G., & Wang, W. (2010). Triplex PCR detection of Botrytis cinerea, Colletotrichum gloeosporioides and Verticillium dahliae in infected strawberry plant tissues. Scientia Agricultura Sinica, 43(21), 4392–4400.
Ni, H. F., Yang, H. R., Chen, R. S., Hung, T. H., & Liou, R. F. (2012). A nested multiplex PCR for species-specific identification and detection of Botryosphaeriaceae species on mango. European Journal of Plant Pathology, 133, 819–828.
Catal, M., Jordan, S., Butterworth, S., & Schilder, A. (2007). Detection of Eutypa lata and Eutypella vitis in grapevine by nested multiplex polymerase chain reaction. Phytopathology, 97(6), 737–747.
Kiran, R., Kumar, P., Akhtar, J., Nair, K., & Dubey, S. C. (2022). Development of multiplex PCR assay for detection of Alternaria brassicae, A. brassicicola and Xanthomonas campestris pv. campestris in crucifers. Archives of Microbiology, 204(4), 224.
Iradukunda, L., Wang, Y. P., Nkurikiyimfura, O., Wang, T., Yang, L. N., & Zhan, J. (2022). Establishment and application of a multiplex PCR assay for the rapid detection of Rhizoctonia solani anastomosis group (AG)-3PT, the pathogen causing potato black scurf and stem canker. Pathogens, 11(6), 627.
Thiessen, L. D., & Woodward, J. E. (2012). Diseases of peanut caused by soilborne pathogens in the Southwestern United States. Isrn Agronomy, 2012, 1–9.
Li, H., Li, C., Song, X., Liu, Y., Gao, Q., Zheng, R., Li, J., Zhang, P., & Liu, X. (2022). Impacts of continuous and rotational cropping practices on soil chemical properties and microbial communities during peanut cultivation. Scientific Reports, 12(1), 2758.
Kong, X. Y., Zhou, X. D., Wu, H. S., Yan, S., Liu, X. X., & Wang, Z. H. (2010). Research advance in control of soil-borne diseases in continuously-cultivated peanut. Acta Agriculturae Jiangxi, 22(08), 12–15.
Cho, J. D., Kim, J. S., Park, H. S., La, Y. J., & Kim, K. S. (2000). Ultrastructural aspects of the mixed infections of watermelon mosaic potyvirus isolated from pumpkin and cucumber green mottle mosaic tobamovirus from watermelon. The Plant Pathology Journal, 16(4), 216–221.
Nithya, K., Parameswari, B., & Viswanathan, R. (2020). Mixed infection of sugarcane yellow leaf virus and grassy shoot phytoplasma in yellow leaf affected Indian sugarcane cultivars. The Plant Pathology Journal, 36(4), 364–377.
Acknowledgements
This study was supported by the National Natural Sciences Foundation of China (Grant No. 32202266), the Shandong Modern Agricultural Industry Technology System (Grant No. SDAIT-04-08), the Outstanding Youth Foundation of Shandong Province (Grant No. ZR2021YQ20), and the Key Research and Development Program of Shandong Province (Grant No. 2019JZZY020608).
Funding
Funding was provided by the National Natural Sciences Foundation of China (Grant No. 32202266), the Shandong Modern Agricultural Industry Technology System (Grant No. SDAIT-04-08), the Outstanding Youth Foundation of Shandong Province (Grant No. ZR2021YQ20), and the Key Research and Development Program of Shandong Province (Grant No. 2019JZZY020608).
Author information
Authors and Affiliations
Contributions
AXL, XL, and JW conceived and designed research. XL, JW, XLH, and MQL conducted experiments. XYS and CH analyzed data. JW and CH wrote the manuscript. All authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, J., Li, X., Sun, X. et al. Establishment and Application of a Multiplex PCR Assay for Detection of Sclerotium rolfsii, Lasiodiplodia theobromae, and Fusarium oxysporum in Peanut. Mol Biotechnol 65, 1369–1377 (2023). https://doi.org/10.1007/s12033-022-00647-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-022-00647-1