Abstract
Lung adenocarcinoma (LUAD) patients exhibit poor prognosis, primarily due to metastasis. Emerging studies have demonstrated that long noncoding RNAs (lncRNAs) play critical roles in cancer progression and metastasis besides their physiological function. Here, we investigated the potential role of lncRNA MAF BZIP Transcription Factor G Antisense RNA 1 (MAFG-AS1) in LUAD metastasis by analyzing its expression in The Cancer Genome Atlas (TCGA) LUAD database, and its function in LUAD using in vitro and in vivo experiments. We performed bioinformatics analysis, western blotting, dual-luciferase reporter gene assay, RNA immunoprecipitation (RIP), and rescue assays to reveal the molecular mechanisms underlying MAFG-AS1 function. We observed augmented expression of MAFG-AS1 in LUAD tissues compared with normal adjacent tissues, and its association with poor prognosis. Furthermore, MAFG-AS1 overexpression promoted LUAD cell migration, proliferation, invasion, and epithelial mesenchymal transition (EMT). Besides, MAFG-AS1 also targeted miR-3196 directly by acting as an endogenous sponge, thereby rescuing the inhibition of SOX12, a target of miR-3196. Thus, the rescue assays demonstrated that MAFG-AS1 promotes cell migration, invasion, and EMT by modulating the miR-3196/SOX12 pathway. In conclusion, our findings suggest that MAFG-AS1/miR-3196/SOX12 axis regulates LUAD progression and is a potential therapeutic target for LUAD.
Similar content being viewed by others

Avoid common mistakes on your manuscript.
Introduction
Lung adenocarcinoma (LUAD) is the most predominant subtype of lung cancer, accounting for 40–50% of all lung cancer cases, and has the highest fatality rate [1, 2]. Despite remarkable progress in early diagnosis, targeted therapy, immunotherapy, and surgical interventions for the treatment of lung cancer, LUAD patients still have a low 5‐year survival rate [3]. At present, invasion and early metastasis of LUAD are primarily responsible for unsatisfactory prognosis, with the 5-year survival rate of 17% [4, 5]. Therefore, early diagnosis and effective treatment of LUAD necessitates further investigations for identifying novel therapeutic targets.
LncRNAs are RNA molecules of over 200 nucleotides in length and do not encode for proteins [6]. They participate in a variety of biological processes such as the regulation of gene expression, mRNA stabilization, facilitating interactions among proteins or between proteins and RNAs, and direct or indirect molecular orientation via epigenetic modifications [7]. Various studies have demonstrated the involvement of several lncRNAs in the regulation of lung cancer pathogenesis and progression. Certain lncRNAs modulate the progression of lung cancer by acting as competing endogenous RNAs (ceRNAs) [8, 9]. For instance, lncRNA MNX1-AS1 expedites lung cancer progression via the miR-527/BRF2 pathway [10]. LncRNA LCAT1 serves as a ceRNA to regulate the function of RAC1 by sponging miR-4715-5p in lung cancer [11]. Likewise, lncRNA SNHG14 sponges miR-613 to facilitate tumor formation in LUAD [9]. MAFG-AS1 (MAFG-DT) is a lncRNA that plays significant roles in the progression of various cancers, such as bladder cancer [12], breast cancer [13], and colorectal cancer [14]; however, its role in LUAD, especially in context of cell invasion and migration, remains to be delineated.
LncRNA expression analysis using The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) revealed substantial overexpression of MAFG-AS1 in LUAD. Moreover, the expression of lncRNA MAFG-AS1 was closely related to prognosis of LUAD patients as significant difference was observed in the overall survival (OS), and a certain trend in disease-free survival (DFS) among patients with differential MAFG-AS1 expression. Hence, MAFG-AS1 was selected for further investigations in the present study. In this study, we aimed to investigate the effects of MAFG-AS1 on LUAD progression and delineate the potential underlying mechanisms. We discovered that LUAD patients overexpressing MAFG-AS1 exhibit poor survival rates. In vitro, lncRNA gain- and loss-of-function experiments uncovered that MAFG-AS1 acted as ceRNA to promote cell migration, proliferation, EMT, and invasion and regulated miR-3196/SOX12 axis. In addition, LUAD tumor growth was suppressed by MAFG-AS1 knockdown in vivo. In conclusion, our findings revealed that by acting as a ceRNA, MAFG-AS1 influences LUAD carcinogenesis and thus, is a potential therapeutic target for LUAD.
Materials and Methods
Bioinformatics Analysis
Datasets from TCGA database (https://portal.gdc.cancer.gov/) and Genotype-Tissue Expression (GTEx) were used for studying differential lncRNA expression in LUAD. The list of differentially expressed lncRNAs (P value < 0.05, |log2FC|> 1) was retrieved with the help of R software. The results were corroborated with GEPIA (http://gepia.cancer-pku.cn/). StarBase (http://starbase.sysu.edu.cn/) and miRDB (http://mirdb.org) were used to predict the lncRNA-miRNA interactions and the miRNA target genes, respectively.
Samples
The cancer and cancer-adjacent samples were obtained from patients with LUAD who visited our hospital between January 2015 and January 2019. The tumors were surgically resected before blanket inclusion of patients who received chemoradiotherapy and stored in liquid nitrogen (− 196 °C) until further experiments. All patients enrolled in the study provided written informed consent.
Culture and Transfection of Cells
The LUAD-derived (H1373, A549, HCC827, and PC-9 cells) as well as normal lung epithelial 16HBE cells were procured from the Cell Resource of CAMS (Beijing, P. R. China). RPMI-1640 medium supplemented with 10% FBS (Gibco, Thermo Fisher Scientific Corp., MA, USA) was used for culturing cells in 5% CO2 at 37 °C.
The small interfering (si-MAFG-AS1) and an overexpression plasmid (MAFG-AS1 OE) for MAFG-AS1 were purchased from Shanghai GenePharma Co., Ltd, Shanghai, P. R. China. The miR-3196 mimics and negative control (miR-NC) were procured from RiboBio Co., Ltd., Guangzhou, P. R. China. Cells (5 × 104 cells/well) were seeded into 12-well cell culture plates. After 24 h, the cells were transfected with 50 nM siRNA using Lipofectamine 3000 in accordance with manufacturer’s protocol (Invitrogen, Thermo Fisher Scientific Corp., MA, USA).
Quantitative Real‐Time Polymerase Chain Reaction (qRT-PCR)
In accordance manufacturer’s protocol, TRIzol® reagent (Invitrogen, Thermo Fisher Scientific Corp., MA, USA) was used to isolate total RNA from tissues and cells. The cDNA synthesis was performed using a reverse transcription kit (Takara Bio Inc., Liaoning, P. R. China) as per the manufacturer’s protocol. The qRT-PCR assays were performed using a SYBR-Green Master Mix kit (Takara Bio Inc.) on ABI 7000 real-time PCR system (Applied Biosystems, Invitrogen, Thermo Fisher Scientific Corp., MA, USA). GAPDH was used for evaluating the relative expression of MAFG-AS1 and SOX12, while U6 was used to evaluate the expression of miR-3196, and 2−ΔΔCt method was used to calculate the relative lncRNA and mRNA expression. The primer sequences are enlisted in Table 1.
Fluorescence In Situ Hybridization (FISH)
To assess the subcellular localization of MAFG-AS1 in LUAD cells, we performed RNA FISH using the Ribo™ Fluorescent in Situ Hybridization Kit (RiboBio Co., Ltd., P. R. China). The MAFG-AS1 probe was designed and synthesized by RiboBio Co., Ltd. and labeled with FAM fluorescent dye. DAPI (Beyotime Biotechnology., Beijing, P. R. China) was used to stain the nuclei. RNA FISH was performed on air-dried cells according to the manufacturer's instructions. Fluorescence was detected using a fluorescence microscope (Olympus Corporation, Tokyo, Japan). The experiment was repeated at least three times.
RNA Immunoprecipitation (RIP)
To study the interplay between MAFG-AS1 and miR-3196, RIP assays were performed using a Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore (Shanghai) Trading Co., Ltd., Shanghai, P. R. China) in compliance with the manufacturer's protocol. Cells were lysed using a lysis buffer containing protease inhibitor cocktail and RNase inhibitor. The magnetic beads were preincubated with antibodies against Ago2 (Abcam, Cambridge, UK) or rabbit IgG (Merck Millipore., MA, USA) for 1 h at room temperature, followed by immunoprecipitation of cell lysates with the magnetic beads at 4 °C overnight. Finally, RNA was eluted from the beads, purified, reverse-transcribed using a cDNA Synthesis Kit (TaKaRa Bio Inc), and qRT-PCR were performed.
Western Blotting
Total proteins were extracted from cells using RIPA lysis buffer and quantitated using the bicinchoninic acid assay. Subsequently, protein samples were resolved on SDS-PAGE mini gels, transferred onto PVDF membranes, and probed with primary antibodies (anti-SOX12; 1:1000 dilution and anti-GAPDH; 1:2000 dilution, both Abcam), and EMT Antibody Sampler Kit (Cell Signaling Technology, Inc., MA, USA; 1:1000 dilution) and secondary antibodies. The immunoreactive signals were visualized using an ECL substrate (Merck Millipore., MA, USA) and the NIH-ImageJ software was used for densitometry analysis, with GAPDH used as an endogenous control.
Cell Counting Kit-8 (CCK-8), EdU, and Colony Formation Assays
We utilized the CCK-8 kit (MyBioSource, Inc., CA, USA; Catalog no. MBS176412) for studying the proliferation of HCC827 and PC-9 cells 48 h after transfection. Briefly, cells (4 × 103 cells/0.1 mL medium) were added to each well of a 96-well cell culture plate post-transfection. The cells were incubated at 37 °C and absorbance was measured at 450 nm every 24 h for a total of 72 h. The CCK-8 solution was added into each well 2 h before the measurement of absorbance.
The EdU assay was performed using the BeyoClick™ EdU Cell Proliferation Kit (Beyotime Biotechnology., Beijing, P. R. China) in accordance with the manufacturer’s protocol. Cells (1 × 104 cells/well) were seeded onto 96-well cell culture plates and incubated overnight in 5% CO2 at 37 °C. Subsequently, the cells were fixed in 4% paraformaldehyde for 30 min and permeabilized for 10 min using 0.5% Triton X-100. The cells were imaged using a fluorescent microscope (Leica, Wetzlar, Germany). The assay was performed at least three times.
For colony formation assays, the cells (700 cells/well) were seeded into 6-well cell culture plates in 10% FBS-containing medium, which was replaced every 4 days and discarded 14 days later. Subsequently, the cells were fixed using methanol, the colonies were stained using Giemsa, and the images were captured. In every assay, each well was replicated and assessed in triplicate, and the experiments were independently repeated three times. The number of colonies was evaluated with the help of NIH-ImageJ software.
Wound Healing Assay
Cell migration was evaluated using the wound-healing assays. Briefly, 5 × 104 cells were plated in 6-well cell culture plates and cultured overnight in high-glucose DMEM containing 10% FBS in 5% CO2 at 37 °C. Subsequently, the cells were then cultured in high-glucose DMEM without FBS. After 24 h serum starvation, the wounds were created by scratching the cell monolayers with a 200 µL sterile pipette tip. The cells were washed to remove floating cells and incubated under serum-starved conditions. The images of the wound were capture immediately after scratching, and at specified time points. The wound distance was measured using NIH-ImageJ software to calculate the migration rate.
Transwell Assay
The assess the migratory potential of LUAD cells, 2 × 105 cells were seeded on the upper chamber of Matrigel-coated Transwell membrane filter inserts (BD Biosciences, NJ, USA) containing 500 μL basal medium. The bottom chamber was filled with medium (200 μL) containing 10% FBS as a chemoattractant. After 24 h incubation, the invaded cells on the lower membrane surface were fixed with paraformaldehyde for 15 min and then stained with a 0.1% crystal violet solution for 15 min. Finally, images were captures using an inverted microscope and the cells invaded to the lower membrane were counted using the NIH-ImageJ software.
Dual-Luciferase Reporter Gene Assay
The lncRNA MAFG-AS1 or SOX12 3′-UTR with the predicted or mutant target site for miR-3196 were synthesized and cloned into pGL3 reporter vectors (Promega Corporation, WI, USA). Subsequently, miR-3196 mimics or miR-NC and pGL3-MAFG-AS1-wild-type (Wt), pGL3-SOX12-Wt, pGL3-MAFG-AS1-mutant (Mut), or pGL3-SOX12-Mut constructs were transfected into cells. The cells were lysed after 48 h to perform the dual-luciferase reporter assays using the Dual-Luciferase Reporter Assay System (Promega (Beijing) Biotech Co., Ltd., Beijing, P. R. China) as per the manufacturer’s instructions. The experiment was repeated at least three times.
Xenograft Mouse Model
Eighteen BALB/c male nude mice (aged 6 weeks) were purchased from Beijing Vital River Co., Ltd. (Beijing, P. R. China) and randomly distributed into six groups (n = 3 per group). The mice were housed under standard conditions (24 ± 2 °C, 50 ± 10% relative humidity, 12 h light/dark cycles) with unlimited access to standard rodent feed (Beijing Keao Xieli Feed Co., Ltd., Beijing, P. R. China) and water. Hygienic conditions were maintained by weekly cage changes. Animal health and behavior were monitored daily, and body weight was assessed weekly over the course of the study. Animal experiments were carried out in accordance with the National Institute of Health’s Guide for the Care and Use of Laboratory Animals, with the approval of the Animal Research Committee of our hospital.
The right flank of mice was subcutaneously injected with HCC827 cells (5 × 106 cells) transfected with MAFG-AS1 shRNA or shRNA NC (n = 3 per group, three replicates). All 18 nude mice eventually developed tumors. The tumor dimensions were measured at an interval of 7 days using Vernier caliper and the tumor volumes were calculated as follows: volume = (length × width2)/2. All animals were euthanized after 4 weeks with pentobarbital overdose (> 120 mg/kg body weight) administered via intraperitoneal injection. Death was verified by loss of spontaneous breathing. Xenograft tumor tissues were resected for subsequent analyses.
Statistical Analysis
The data were statistically analyzed using IBM SPSS Statistics 20.0 (IBM Corporation, NY, USA) and GraphPad Prism 8.0 (GraphPad Software Inc., CA, USA). Data collected from at least three separate experiments are presented as mean ± standard deviation. Intergroup comparisons were analyzed using Student’s t-test or one-way analysis of variance. The association between MAFG-AS1 and SOX12 expression was analyzed using Spearman’s correlation analysis. Statistical significance was set at P < 0.05.
Results
MAFG-AS1 Expression is Upregulated in LUAD Tissues
Based on TCGA dataset analysis, lncRNAs overexpressed in LUAD and closely related to the prognosis of LUAD patients were selected for further investigations. The assessment of a series of samples, including 483 LUAD tissues and 347 cancer-adjacent or normal lung tissues from the TCGA or GTEx databases, respectively indicated that MAFG-AS1 was remarkably overexpressed in LUAD tissues compared to the cancer-adjacent or normal lung tissues (Fig. 1a). This finding is consistent with that of GEPIA (http://gepia.cancer-pku.cn/) database. Furthermore, the Kaplan Meier survival analyses revealed that MAFG-AS1 overexpression is strongly associated with the OS of LUAD patients (Fig. 1b). Meanwhile, the DFS of LUAD was also affected to a certain extent (Fig. 1c). To confirm these findings, we measured MAFG-AS1 expression in LUAD tissues (n = 47) obtained from our hospital and found that MAFG-AS1 expression was remarkably higher in LUAD tissues than in the adjacent normal tissues (Fig. 1d). The patient characteristics are summarized in Table 2. Moreover, the relative expression levels of MAFG-AS1 in LUAD cell lines (PC-9, H1373, HCC827, and A549) were remarkably higher than those in the normal human lung epithelial cell line (16HBE) (Fig. 1e). Among these LUAD cell lines, the expression level of MAFG-AS1 was the highest in HCC827 cells and the lowest in PC-9 cells. Consequently, PC-9 and HCC827 cells were selected for further studies.
MAFG-AS1 is upregulated in LUAD tissues. a TCGA dataset analysis of MAFG-AS1 expression in LUAD tissues (n = 483) and adjacent normal tissues (n = 347). b Kaplan–Meier analysis of the overall survival in LUAD patients (n = 477, P = 0.014) according to TCGA datasets. c Kaplan–Meier analysis of disease-free survival in LUAD patients (n = 477, P = 0.074) according to TCGA datasets. d qRT-PCR analysis of MAFG-AS1 expression in LUAD tissues (n = 47) and adjacent normal tissues. e qRT-PCR analysis of MAFG-AS1 expression in LUAD cell lines and normal human lung epithelial cell line (16HBE). *P < 0.05 and **P < 0.01
MAFG-AS1 is Primarily Located in the Cytoplasm of LUAD Cells
To elucidate the potential mechanism by which MAFG-AS1 functions in LUAD cells, the subcellular localization of MAFG-AS1 was investigated by performing RNA FISH and subcellular fractionation assays. The results of RNA FISH (Fig. 2a) and subcellular fractionation assays (Fig. 2b) showed that MAFG-AS1 was predominantly located in the cytoplasm of PC-9 and HCC827 cell lines.
MAFG-AS1 locates in the cytoplasm. a FISH assays were used to show the subcellular localization of MAFG-AS1 in HCC827 and PC-9 cells (scale bar = 10 μm). b A subcellular fractionation assay indicated that MAFG-AS1 was mostly located in the cytoplasm, and U6 and GAPDH acted as endogenous controls for the nucleus and cytoplasm
MAFG-AS1 Boosts Proliferation of LUAD Cells
To investigate the effect of MAFG-AS1 on LUAD cell proliferation, the PC-9 cells and HCC827 cells were transfected with MAFG-AS1 overexpressing vectors (MAFG-AS1 OE) and MAFG-AS1 siRNA (si-MAFG-AS1), respectively (Fig. 3a). CCK8 and EdU assays showed that silencing MAFG-AS1 expression using specific siRNA inhibited while overexpressing MAFG-AS1 using MAFG-AS1 OE constructs augmented the proliferation of LUAD cells (Fig. 3b, c). Furthermore, the results of colony formation assay indicated that MAFG-AS1 overexpression remarkably promoted colony forming ability of LUAD cells, while silencing MAFG-AS1 remarkably inhibited this characteristic (Fig. 3d). Overall, our findings provide evidence that MAFG-AS1 promotes the proliferation and colony formation abilities of LUAD cells.
MAFG-AS1 boosts cell proliferation. a Transfection efficiency of MAFG-AS1 siRNA and MAFG-AS1 overexpression vector. b CCK-8 and c EdU were used to determine the effect of MAFG-AS1 on cell proliferation. d Colony formation assay was used to determine the effect of MAFG-AS1 on cell colon. Data represent the mean ± SD of three independent experiments. **P < 0.01 and ***P < 0.001
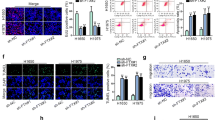
MAFG-AS1 Promotes the Migration, Invasion, and EMT of LUAD Cells
To understand the effect of MAFG-AS1 on the migration and invasion of LUAD cells, wound healing (Fig. 4a) and Transwell migration (Fig. 4b) assays were performed. The results revealed that in contrast to si-NC, si-MAFG-AS1 distinctively inhibited the migratory and invasive potential of HCC827 cells, while MAFG-AS1 overexpression remarkably improved the motility of PC-9 cells relative to control (Fig. 4a, b). EMT is a well-established core phenotypic indicator of cancer invasion and metastasis. Hence, we tested the impact of varying MAFG-AS1 expression on primary EMT markers expressed in LUAD cells. The results showed that silencing MAFG-AS1 expression augmented the expression of E-cadherin in HCC827 cells (Fig. 4c), whereas the expression of N-cadherin and Snail1 were downregulated. In contrast, opposite trends were observed in PC-9 cells overexpressing MAFG-AS1. Overall, these findings indicate that MAFG-AS1 plays an anti-metastatic role in LUAD carcinogenesis.
MAFG-AS1 promotes cell migration, invasion and EMT. a Wound healing assay was used to determine the effect of MAFG-AS1 on cell migration. b Transwell assay was used to examine cell invasion. c Western blot analysis was utilized to explore the effect of MAFG-AS1 on EMT in two LUAD cell lines. Data represent the mean ± SD of three independent experiments. **P < 0.01
MAFG-AS1 Acts as a ceRNA of miR-3196
In the cytoplasm, several lncRNAs act as csRNA and sponge certain miRNAs [15, 16]. Hence, we attempted to identify possible target miRNAs of MAFG-AS1 by exploring the ceRNA regulatory network. Accordingly, putative miRNA binding sites were predicted using the starBase (http://starbase.sysu.edu.cn) (Table S1) and miRDB (http://mirdb.org) (Table S2) databases, of which six were identified to be common in both (Fig. 5a). Furthermore, the RNA pull-down assay results demonstrated that MAFG-AS1 was more likely to pull down miR-3196 (Fig. 5b). As a result, miR-3196 was selected for further analysis. miRNAs which are predominantly distributed in the cytoplasm, could be a component of the RNA-induced silencing complex (RISC). Ago2 is required for miRNA-mediated gene silencing. In this study, we analyzed whether MAFG-AS1 and miR-3196 are present in the same RISC using an RNA-binding protein immunoprecipitation (RIP) assay. The results revealed that MAFG-AS1 and miR-3196 were enriched in miR-containing Ago2 complexes compared with the IgG control (Fig. 5c). To further ascertain whether miR-3196 is a direct target of MAFG-AS1, two reporter plasmids coupled with full-length MAFG-AS1 3′-UTR harboring wild-type (Wt) or mutant (Mut) miR-3196 binding sites were constructed (Fig. 5d). Dual-luciferase assays in 293 T cells demonstrated that miR-3196 inhibited the luciferase activity in the MAFG-AS1 Wt + miR-3196 mimic transfected cells, but not in the MAFG-AS1 Mut + miR-3196 mimic transfected cells compared to scramble control transfected cells (Fig. 5e). These findings provide evidence that MAFG-AS1 functions as a ceRNA for miR-3196.
MAFG-AS1 acts as a ceRNA of miR-3196. a Venn diagram showed 6 mutual miRNAs (miR-3180-3p, miR-3180, miR-6816-5p, miR-3196, miR-24-3p, and miR-744-5p) interacting with MAFG-AS1 that were found after searching starBase and miRDB databases. b RNA pull-down assays detected the interaction of MAFG-AS1 with the selected 6 miRNAs. c Bioinformatics predicted and mutated miR-3196 binding sites of MAFG-AS1. d Dual-Luciferase reporter assay. e RIP assays exhibited the interaction between MAFG-AS1 and miR-3196. Data represent the mean ± SD of three independent experiments. **P < 0.01 and ***P < 0.001
MiR-3196 is Indispensable for MAFG-AS1 -Mediated Migration, Invasion, and EMT of LUAD Cells
To determine whether MAFG-AS1-induced LUAD cell metastasis and EMT were mediated by miR-3196, Transwell migration and western blotting assays were performed. Accordingly, MAFG-AS1 overexpression vectors were used to increase the expression of MAFG-AS1 in the HCC827 cells (MAFG-AS1 OE) (Fig. 6a), and miR-3196 mimics were used to upregulate miR-3196 expression in HCC827 and PC-9 cells (Fig. 6b). The results of Transwell migration assays showed miR-3196 mimics inhibited the migration and invasion of LUAD cells (Supplementary Fig. 1). Furthermore, the miR-3196 mimics reversed MAFG-AS1-triggered elevation of cell migration and invasion in HCC827 and PC-9 cells (Fig. 6c). Our immunoblotting studies revealed that overexpression of MAFG-AS1 attenuated the expression of E-cadherin, but increased N-cadherin expression in both the LUAD cell lines, which could be reversed by miR-3196 mimics (Fig. 6d). Thus, our results validated our hypothesis, and provided evidence that MAFG-AS1 can directly target miR-3196 to regulate LUAD cell metastasis and EMT.
MiR-3196 is indispensable for migration, invasion and EMT of LUAD cells facilitated by MAFG-AS1. a Transfection efficiency of MAFG-AS1 overexpression vector in HCC827 cells. b Transfection efficiency of miR-3196 mimics in HCC827 and HC-9 cells. c Transwell assay was used to examine cell invasion and migration. d Western blot analysis was utilized to explore the effect of MAFG-AS1 and miR-3196 on EMT in two LUAD cell lines. Data represent the mean ± SD of three independent experiments. **P < 0.01 and ***P < 0.001
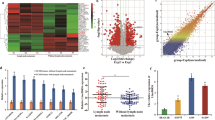
SOX12 is a Target of miR-3196
To determine the target gene of miR-3196, bioinformatics analysis was performed using miRDB, which predicted 167 target genes (Table S3). Furthermore, the correlation between batches of lncRNAs and mRNAs was assessed according to the LUAD tissue-associated data obtained from the TCGA database; 473 mRNAs were found to be positively correlated with MAFG-AS1 expression (Table S4). Of the MAFG-AS1 positively correlated mRNAs, only PPFIA3 and SOX12 that bound miR-3196, (Fig. 7a) were positively correlated with MAFG-AS1 in LUAD as shown in Fig. 7b and Table S3 (Target Score > 70) and Table S4 (Spearman's Correlation > 0.3). When miR-3196 was overexpressed in HCC827 cells, the SOX12 mRNA level, but not that of PPFIA3 was found to be decreased (Fig. 7c). Consequently, SOX12 was selected for further experiments. To ascertain whether SOX12 is a direct target of miR-3196, we constructed two reporter plasmids coupled with full-length SOX12 3′-UTR harboring Wt or Mut miR-3196 binding sites (Fig. 7d). Dual-luciferase reporter assays in 293 T cells showed that miR-3196 weakened the luciferase activity of the SOX12 Wt group compared to that of the SOX12 Mut group (Fig. 7e). Furthermore, qRT-PCR results showed increased SOX12 expression in 47 LUAD tissues than in cancer-adjacent tissues (Fig. 7f). Similar to TCGA database, SOX12 and MAFG-AS1 were also positively correlated in the 47 LUAD tissues (Fig. 7g). Additionally, our western blotting assays revealed that miR-3196 mimics distinctively hindered the expression of SOX12 protein, while MAFG-AS1 overexpression increased SOX12 protein expression (Fig. 7h and i). Moreover, miR-3196 mimics abrogated MAFG-AS1 overexpression-induced augmented SOX12 protein levels to a limited degree. Taken together, these data indicate that SOX12 is a target of miR-3196 and is inhibited by miR-3196.
SOX12 is a target of miR-3196 and inhibited by miR-3196. a Venn diagram showed 2 mutual mRNAs (PPFIA3 and SOX12) binding with miR-3196. b Spearman’s correlation analysis indicated the relationship between SOX12 and MAFG-AS1 based on TCGA datasets. c RT-PCR showed that miR-3196 inhibited the expression of SOX12. d Bioinformatics predicted and mutated miR-3196 binding sites of SOX12. e Dual-Luciferase reporter assay. f qRT-PCR analysis of SOX12 expression in LUAD tissues (n = 47) and adjacent normal tissues. g Spearman's correlation analysis indicated the relationship between SOX12 and MAFG-AS1 based on 47 LUAD samples. h Western blot assay was utilized to explore the effect of miR-3196 on the protein expression of SOX12. i Western blot assay was utilized to explore the effect of miR-3196 and MAFG-AS1 on the protein expression of SOX12. Data represent the mean ± SD of three independent experiments. **P < 0.01 and ***P < 0.001
MAFG-AS1 Downregulation Inhibits LUAD Tumor Growth In Vivo
To determine the role of MAFG-AS1 in the tumorigenicity of LUAD cells, in vivo assays were performed. The results revealed that MAFG-AS1 knockdown weakened the tumor‐forming ability of HCC827 cells (Fig. 8a). Furthermore, compared to the sh-NC group, the sh-MAFG-AS1 group harbored tumors of smaller volume and weight (Fig. 8b, c). Subsequent analysis of MAFG-AS1 expression showed that the sh-MAFG-AS1 group exhibited remarkably reduced expression of MAFG-AS1 in the xenograft tumors compared to the sh-NC group (Fig. 8d). Overall, these findings suggest that MAFG-AS1 downregulation abrogates LUAD tumor growth in vivo.
Discussion
The vital role played by lncRNAs in the physiological process as well various cancers has been confirmed. For instance, HORAS5 promotes survival of castration-resistant prostate cancer by activating the androgen receptor transcriptional program [17]. SOX2OT promotes the proliferation of pancreatic cancer cells by binding to FUS [18]. Aberrant expression of lncRNA SNHG15 correlates with liver metastasis and poor survival in colorectal cancer [19]. CASC9 promotes LIN7A expression via miR-758-3p to increase the malignancy of ovarian cancer [20]. Accordingly, our lncRNA expression study based on TCGA analysis revealed that lncRNA MAFG-AS1 was remarkably overexpressed in LUAD and demonstrated a close association with LUAD prognosis. Hence, it was selected for further experiments to evaluate whether it affects cancer-related processes such as metastasis, invasion, and EMT in LUAD. Our functional assays confirmed that MAFG-AS1 acts as an oncogenic lncRNA that promotes the proliferation, metastasis, and EMT of LUAD cells.
Several lncRNAs act as ceRNA and are involved in the development of various cancers [11, 21]. Our results of FISH and subcellular fractionation assays showed that MAFG-AS1 is primarily located in the cytoplasm. Thus, we used starBase, miRDB, and performed pull-down assays to screen for miRs that could bind with MAFG-AS1. Our results revealed that miR-3196 bound to MAFG-AS1 and regulated the promotion of MAFG-AS1-mediated LUAD cell metastasis and EMT. miRNAs can silence mRNAs by binding to their 3'-UTR. Furthermore, we confirmed that SOX12 was the downstream target of miR-3196 and its expression levels were remarkably higher in LUAD. Moreover, MAFG-AS1 and miR-3196 showed positive and negative regulation of SOX12 expression, respectively. Li et al. have confirmed that SOX12 functions as an oncogenic molecule during the development of human lung cancer [22], which is consistent with the conclusions presented in the present study.
This study had several limitations. First, we were unable to ascertain the clinical relevance of MAFG-AS1 in our study. Thus, a larger sample size is required to conclusively determine the clinical significance of MAFG-AS1. Second, the relationship between MAFG-AS1 and EGFR mutation status in lung adenocarcinoma was not analyzed, which is important since EGFR mutations play key roles in lung cancer; this should be explored in the future studies to validate our findings.
In conclusion, our study revealed a ceRNA network in LUAD comprising MAFG-AS1/miR-3196/SOX12 axis and highlighted that MAFG-AS1 is a promising target for LUAD treatment and prognosis.
Data Availability
Datasets used and analyzed during this study are available from the corresponding author upon reasonable request.
Abbreviations
- LUAD:
-
Lung adenocarcinoma
- TCGA:
-
The cancer genome atlas
- lncRNAs:
-
Long noncoding RNA
- EMT:
-
Epithelial-mesenchymal transition
- FBS:
-
Fetal bovine serum
- qRT-PCR:
-
Quantitative real-time polymerase chain reaction
- EdU:
-
5-Ethynyl-2′-deoxyuridine
- FISH:
-
Fluorescent in situ hybridization
- RIP:
-
RNA immunoprecipitation
- ceRNAs:
-
Competing endogenous RNAs
References
Kim, Y., Kim, H., Bang, S., Jee, S., & Jang, K. (2021). MicroRNA-130b functions as an oncogene and is a predictive marker of poor prognosis in lung adenocarcinoma. Laboratory Investigation, 101, 155–164.
Mullen, D. J., Yan, C., Kang, D. S., et al. (2020). TENET 2.0: Identification of key transcriptional regulators and enhancers in lung adenocarcinoma. PLoS Genetics, 16, e1009023.
Cao, X., Fang, X., Malik, W. S., et al. (2020). TRB3 interacts with ERK and JNK and contributes to the proliferation, apoptosis, and migration of lung adenocarcinoma cells. Journal of Cellular Physiology, 235, 538–547.
Zhan, J., Wang, P., Li, S., et al. (2019). HOXB13 networking with ABCG1/EZH2/Slug mediates metastasis and confers resistance to cisplatin in lung adenocarcinoma patients. Theranostics, 9, 2084–2099.
Tsoyi, K., Osorio, J. C., Chu, S. G., et al. (2019). Lung adenocarcinoma syndecan-2 potentiates cell invasiveness. American Journal of Respiratory Cell and Molecular Biology, 60, 659–666.
Pan, Q., Zhao, Z., Liao, Y., et al. (2019). Identification of an interferon-stimulated long noncoding RNA (LncRNA ISR) involved in regulation of influenza A virus replication. International Journal of Molecular Sciences. https://doi.org/10.3390/ijms20205118
Wang, X., Yang, J., Guo, G., et al. (2019). Novel lncRNA-IUR suppresses Bcr-Abl-induced tumorigenesis through regulation of STAT5-CD71 pathway. Molecular Cancer, 18, 84.
Guo, H., Feng, Y., Yu, H., Xie, Y., Luo, F., & Wang, Y. (2020). A novel lncRNA, loc107985872, promotes lung adenocarcinoma progression via the notch1 signaling pathway with exposure to traffic-originated PM25 organic extract. Environmental Pollution, 266, 115307.
Xu, Z. N., Wang, Z. X., Xu, L., et al. (2019). Long noncoding RNA SNHG14 exerts oncogenic functions in lung adenocarcinoma through acting as a sponge to miR-613. European Review for Medical and Pharmacological Sciences, 23, 10810–10817.
Liu, H., Han, L., Liu, Z., & Gao, N. (2019). Long noncoding RNA MNX1-AS1 contributes to lung cancer progression through the miR-527/BRF2 pathway. Journal of Cellular Physiology, 234, 13843–13850.
Yang, J., Qiu, Q., Qian, X., et al. (2019). Long noncoding RNA LCAT1 functions as a ceRNA to regulate RAC1 function by sponging miR-4715-5p in lung cancer. Molecular Cancer, 18, 171.
Xiao, M., Liu, J., Xiang, L., et al. (2020). MAFG-AS1 promotes tumor progression via regulation of the HuR/PTBP1 axis in bladder urothelial carcinoma. Clinical and Translational Medicine, 10, e241.
Feng, J., Wen, T., Li, Z., et al. (2020). Cross-talk between the ER pathway and the lncRNA MAFG-AS1/miR-339-5p/ CDK2 axis promotes progression of ER+ breast cancer and confers tamoxifen resistance. Aging, 12, 20658–20683.
Ruan, Z., Deng, H., Liang, M., et al. (2020). Downregulation of long non-coding RNA MAFG-AS1 represses tumorigenesis of colorectal cancer cells through the microRNA-149-3p-dependent inhibition of HOXB8. Cancer Cell International, 20, 511.
Wang, Q. Y., Peng, L., Chen, Y., et al. (2020). Characterization of super-enhancer-associated functional lncRNAs acting as ceRNAs in ESCC. Molecular Oncology, 14, 2203–2230.
Chu, J., Tao, L., Yao, T., et al. (2021). Circular RNA circRUNX1 promotes papillary thyroid cancer progression and metastasis by sponging MiR-296-3p and regulating DDHD2 expression. Cell Death & Disease, 12, 112.
Parolia, A., Venalainen, E., Xue, H., et al. (2019). The long noncoding RNA HORAS5 mediates castration-resistant prostate cancer survival by activating the androgen receptor transcriptional program. Molecular Oncology, 13, 1121–1136.
Zhu, S., Zhao, D., Li, C., et al. (2020). BMI1 is directly regulated by androgen receptor to promote castration-resistance in prostate cancer. Oncogene, 39, 17–29.
Huang, L., Lin, H., Kang, L., et al. (2019). Aberrant expression of long noncoding RNA SNHG15 correlates with liver metastasis and poor survival in colorectal cancer. Journal of Cellular Physiology, 234, 7032–7039.
Hu, X., Li, Y., Kong, D., Hu, L., Liu, D., & Wu, J. (2019). Long noncoding RNA CASC9 promotes LIN7A expression via miR-758-3p to facilitate the malignancy of ovarian cancer. Journal of Cellular Physiology, 234, 10800–10808.
Wang, Y., Zeng, X., Wang, N., et al. (2018). Long noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335-5p and miR-1972 in osteosarcoma. Molecular Cancer, 17, 89.
Wang, L., Hu, F., Shen, S., et al. (2017). Knockdown of SOX12 expression inhibits the proliferation and metastasis of lung cancer cells. American Journal of Translational Research, 9, 4003–4014.
Acknowledgements
Not applicable.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
QW performed the experiments, generated data, and data analysis and interpretation of data. JJ made substantial contributions to the conception and design of the present study. All authors contribute to the drafting and revision of the manuscript. All authors read, revised and approved the manuscript and agreed to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interest or personal relationships that could influence the results reported in this manuscript.
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the Ethics Committee of Quzhou People’s Hospital Affiliated to Wenzhou Medical University and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Animal studies were carried out in accordance with the National Institute of Health’s Guide for the Care and Use of Laboratory Animals, with the approval of the Animal Research Committee of Quzhou People’s Hospital Affiliated to Wenzhou Medical University.
Informed Consent
All patients provided written informed consent.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
12033_2022_455_MOESM1_ESM.tif
Supplementary Figure 1. miR-3196 mimics inhibits cell migration and invasion of LUAD cells. (A) Wound healing assay was used to determine the effect of miR-3196 mimics on cell migration. (B) Transwell assay was used to examine cell invasion. *P<0.05 and **P<0.01 (TIF 26154 kb)
12033_2022_455_MOESM5_ESM.xlsx
Table S4. The mRNAs are found to be positively correlated with the expression of lncRNA MAFG-AS1 (Spearman's Correlation>0.3) in the TCGA database (XLSX 1263 kb)
Rights and permissions
About this article
Cite this article
Wu, Q., Jiang, J. LncRNA MAFG-AS1 Promotes Lung Adenocarcinoma Cell Migration and Invasion by Targeting miR-3196 and Regulating SOX12 Expression. Mol Biotechnol 64, 970–983 (2022). https://doi.org/10.1007/s12033-022-00455-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-022-00455-7