Abstract
The filamentous fungus Talaromyces cellulolyticus (formerly Acremonium cellulolyticus) is currently being intensively studied as a promising industrial producer of a number of secreted cellulolytic enzymes. In this study, the T. cellulolyticus gene lacA, which encodes a protein orthologous to the fungal extracellular β-galactosidases of family 35, was identified. The substitution of the lacA upstream region with a constitutive promoter demonstrated that the product of this gene is effectively secreted and possesses β-galactosidase activity. The optimal pH and temperature values for the hydrolysis of o-nitrophenyl-β-D-galactopyranoside by this enzyme were determined to be pH 4.5–5.5 and 50 °C, respectively. The negligible production of β-galactosidase activity by strains expressing lacA under native regulation raises the possibility of using lacA as a reporter gene. To test this hypothesis, the native promoter of lacA was replaced with the strong inducible promoter of the T. cellulolyticus cellobiohydrolase I gene. The cultivation of the resulting strain in various media showed that the β-galactosidase activity depends on cultivation conditions similar to the cellobiohydrolase activity. Thus, the suitability of lacA as a reporter for evaluating promoters with a wide range of expression profiles was demonstrated.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The filamentous fungus Talaromyces cellulolyticus Y-94, formerly known as Acremonium cellulolyticus, is currently being intensively studied as an industrial microorganism capable of efficiently processing lignocellulose substrates into monosugars [1]. T. cellulolyticus Y-94 (CBS 136886, FERM BP-5826) was isolated in 1982 from soil in Japan as an organism that secretes large amounts of cellulolytic enzymes [2]. Subsequently, random mutagenesis was used to derive the strains TN and S6-25 from Y-94 as mutants that exhibit high cellulase production [3]. Significant success in the study of this fungus has been achieved in recent years: effective methods for the transformation of protoplasts and starch-inducible homologous expression systems have been developed [4, 5]. The draft genome sequence of T. cellulolyticus Y-94 has been determined and deposited in the GenBank database [6]. Further investigation of T. cellulolyticus and the development of new producers based on this fungus require the design of genetic tools for this organism. In particular, the presence of a convenient reporter gene would facilitate the study of regulation of gene expression and the search for promoters with the required pattern of regulation and strength. One of the commonly used reporter genes for genetic engineering research is the gene encoding β-galactosidase (β-1,4-galactosidase, E.C. 3.2.1.23). The advantage of this gene is the easy detection of the corresponding activity in solid and liquid media as well as the ability to use colored substrates for direct visualization of the results. T. cellulolyticus does not produce significant intracellular or extracellular β-galactosidase activity when cultivated in media supplemented with cellulose, which are used for cellulase production [7, 8]. Therefore, the use of the β-galactosidase-encoding gene as a reporter for this organism is possible, since the natively produced activity will not interfere with the activity produced by the reporter gene.
Extracellular β-galactosidase is produced by many fungi as a part of enzymatic complexes for the breakdown of hemicelluloses. The role of this enzyme is the cleavage of terminal β-D-galactose residues from polysaccharides [9]. This enzyme has been well studied in the industrially important genera Aspergillus, Penicillium and Hypocrea (Trichoderma). Some fungi have been shown to produce several isozymes or possess multiple genes encoding β-galactosidase [10,11,12,13,14,15,16,17,18,19,20].
The objective of this work was to clarify whether T. cellulolyticus harbors a gene encoding a functional β-galactosidase and whether this gene can be used as a reporter in further studies.
Materials and Methods
Strains, Materials and Media
The T. cellulolyticus strains Y-94, S6-25 and F09 (uracil-deficient mutant of S6-25) were kindly provided by the Ajinomoto (Kawasaki, Tokyo, Japan) research team. The strain E. coli DH5α was used for the maintenance and amplification of plasmids.
Restriction endonucleases, DNA polymerases, and the plasmid pUC57 were purchased from Thermo Fisher Scientific Inc. (USA). The In-fusion HD EcoDry Cloning Kit was purchased from Takara Bio (USA, Cat# 639690). Gibson Assembly® Master Mix was purchased from New England Biolabs Inc. (USA). The primers for polymerase chain reaction (PCR) were produced by Evrogen (Russia). Phusion High-Fidelity DNA Polymerase (F530) was used for the amplification of chromosomal fragments of T. cellulolyticus for cloning, and Taq DNA polymerase was used for other applications. X-Gal and ortho-nitrophenyl-β-galactoside (ONPG) were purchased from Thermo Fisher Scientific Inc. (USA). PD medium, PD agar (PDA), corn steep liquor (Csl), microcrystalline cellulose (Avicel), methylumbelliferyl β-D-cellobioside (MUC) and 4-methylumbelliferone were purchased from Sigma–Aldrich (Sigma–Aldrich, Missouri, USA), and yeast extract (0251/0-MG-L) was purchased from Biospringer (Maisons-Alfort, France).
The MM medium was described previously [4]. It contained 10 g/L glucose, 0.54 g/L NH4Cl, 1.4 g/L KH2PO4, 0.5 g/L KCl, 0.05 g/L MgSO4·7H2O, 0.01 g/L ZnSO4·7H2O, 0.01 g/L MnSO4·5H2O and 0.01 g/L CuSO4·5H2O; pH 6.5. For the preparation of MMAg (solid MM medium), 2% agar was added to MM. The AC-C medium was prepared by modifying the medium described previously [3]. It contained 40 g/L Avicel (microcrystalline cellulose), 5 ml/L Csl, 24 g/L KH2PO4, 5 g/L (NH4)2SO4, 1.2 g/L MgSO4·7H2O, 4 g/L Urea, 0.01 g/L ZnSO4·7H2O, MnSO4·5H2O 0.01 g/L; CuSO·5H2O 0.01 g/L; Tween 80 1 g/L; pH 4.0. AC-G and AC-GC media were prepared as AC-C, but instead of 40 g/L Avicel they contained 20 g/L glucose or 20 g/L glucose and 2 g/L Avicel, respectively. Strains Y-94 and S6-25 were maintained on PDA plates, and F09 was maintained on PDA plates supplemented with 1 g/L uracil [4] at 30 °C. After transformation, the protoplasts were spread on MMSAg plates (MMAg supplemented with 1 M sucrose). For the detection of β-galactosidase activity, the transformants were transferred to MMAg plates supplemented with 0.004% X-Gal.
To study the influence of various carbon sources on β-galactosidase synthesis, the strains were cultivated on plates with MMAg supplemented with these carbon sources instead of glucose.
Fungal Transformation
The transformation of T. cellulolyticus was performed as described previously [4] with minor modifications. Strain F09 was cultivated in 20 ml of PD medium supplemented with 1 g/L uracil at 30 °C in a rotary shaker at 200 rpm. The mycelium was collected, washed, suspended in hyperosmotic buffer (10 mM KH2PO4 and 0.8 M NaCl (pH 6.0 adjusted with NaOH) containing 10 mg/ml yatalase (Takara Bio, Ohtsu, Japan) and then gently agitated at 30 °C for approximately 2 h (the formation of protoplasts was monitored by microscopy). After hydrolysis, the suspension was passed through a sterile glass filter with a pore size of 140–160 µm, and protoplasts were collected from the resulting suspension by centrifugation at 750 g for 15 min. The protoplasts were washed twice with solution A (1.2 M sorbitol, 10 mM Tris–HCl pH 7.5, 10 mM CaCl2) and then suspended in this solution at a final concentration of 5 × 107 protoplasts/ml. For transformation, 200 µl of protoplast suspension was mixed with 50 µl of solution B (50% PEG4000, 10 mM Tris–HCl pH 7.5, 10 mM CaCl2) and 10 µg of DNA diluted in a volume not exceeding 20 µl. The mixture was incubated at room temperature for 30 min; then, 1 ml of solution B was added and incubated under the same conditions for 20 min. Finally, the mixture was gently mixed with 8.5 ml of solution A and centrifuged at 750 g for 15 min, and the supernatant was removed. The pellet was mixed with 200 µl of solution A and spread on MMSAg plates. After cultivation for 5–7 days at 30 °C, the colonies that appeared were transferred to MMAg plates.
Bioinformatics Analysis
To identify the proteins of T. cellulolyticus orthologous to the known proteins, a searching for reciprocal best hits was performed [21]. The search was performed in the T. cellulolyticus Y-94 protein database (taxid: 1472165) using the “blastp” algorithm of BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). To identify β-galactosidase, the whole amino acid sequence of β-galactosidase precursors of Aspergillus niger, Aspergillus oryzae and Penicillium sp. (GenBank accession numbers AAA32696.1, 4IUG_A and CAF32457.1, respectively) were used as queries. The search revealed several proteins with significant similarity to the query proteins and with various lengths of homologous sequences. All three query proteins had the highest similarity (62, 64 and 67% identical amino acids, respectively, with 100% coverage of query and E-value 0.0) with the hypothetical protein TCE0_017f03666 (accession number GAM35377.1). The reverse search using the sequence of TCE0_017f03666 as a query against the A. niger, A. oryzae and Penicillium sp. databases yielded the best hits with AAA32696.1, 4IUG_A and CAF32457, respectively. To identify the hypothetical translation elongation factor 1-alpha and hypothetical gamma-actin, the sequences of A. oryzae KBN616 translation elongation factor 1-alpha (GenBank BAA76296) and Penicillium chrysogenum gamma-actin (UniProtKB/Swiss-Prot: Q9URS0.1), respectively, were used as queries.
To design oligonucleotide primers for PCR amplification of the fragments of T. cellulolyticus DNA, the amino acid sequences of the identified proteins were used as a query to search the genome database. Since the nucleotide sequence of T. cellulolyticus is not fully published, the search was also performed in the database of Talaromyces pinophilus 1–95 (taxid:128442). This species is closely related to T. cellulolyticus; moreover, based on genome sequence analysis, some researchers believe that these two organisms belong to the same species [22].
The alignments were performed using the Clustal Omega tool (https://www.ebi.ac.uk/Tools/msa/clustalo/).
Plasmids and Strains Construction
The oligonucleotide primers used in this study are listed in Table 1.
The design of the promoter replacement experiment was based on the “Split-Marker” recombination method previously developed for Saccharomyces cerevisiae and then adapted for various fungi [23,24,25]. The order of plasmid construction and the design of the experiment for the substitution of the native promoter of lacA (PlacA) with the promoter of tef1 (Ptef1) are shown in Fig. 1.
First, the plasmid pTcpyrF-Ptef1, containing the pyrF gene of T. cellulolyticus, encoding orotate phosphoribosyltransferase, and the promoter of the tef1 gene of T. cellulolyticus, encoding putative translation elongation factor 1-alpha, was constructed. For this purpose, the DNA fragment containing the pyrF coding region and the promoter and terminator of this gene was produced by PCR using the primers pUC57-pyrF-51 and TcpyrF-Ptef1 and chromosomal DNA of T. cellulolyticus S6-25 as the template. The DNA fragment containing 0.80 kb of the tef1 upstream region and start codon of this gene was amplified by PCR using the primers Ptef1-51 and Ptef1-31-pUC57 and chromosomal DNA of T. cellulolyticus S6-25 as the template. The resulting fragments were cloned into the plasmid pUC57 digested with Eco321 using the In-Fusion® HD EcoDry Cloning Kit.
Then, the plasmid pUlacA-pyrF-Ptef1 was assembled. For this purpose, the 3-kb DNA region located 0.8 kb upstream of the lacA coding region was amplified by PCR using the primers p57Nr-bgal and bgal-pyrF and chromosomal DNA of T. cellulolyticus S6-25 as the template. The linear DNA fragment containing pyrF, Ptef1 and the pUC57 backbone was produced by PCR using pTcpyrF-Ptef1 as the template and the primers pUC57-Bsp68I_L and TcpyrF-511. The resulting DNA fragments were joined using Gibson Assembly® Master Mix.
The plasmid ppyrF-Ptef1-lacA was assembled as follows. The 3.3-kb DNA fragment of the coding region of lacA from the start codon was amplified by PCR using the primers Ptef-TclacA and lacA-pUC57-432 and chromosomal DNA of T. cellulolyticus S6-25 as the template. The linear DNA fragment containing pyrF, Ptef1 and the pUC57 backbone was produced by PCR using pTcpyrF-Ptef1 as the template and the primers Ptef1-31 and pUC57-432. The resulting DNA fragments were joined using Gibson Assembly® Master Mix.
The plasmid ppyrF-PlacA-lacA was assembled as follows. The DNA fragment containing 0.85 kb of the lacA upstream region was amplified by PCR using the primers TcpyrF-PlacA and PlacA-TClacA and chromosomal DNA of T. cellulolyticus S6-25 as the template. The second linear DNA fragment was produced by PCR using ppyrF-Ptef1-lacA as the template and the primers TcpyrF-32 and TclacA-56. The resulting DNA fragments were joined using Gibson Assembly® Master Mix.
The plasmid ppyrF-Pcbh1-lacA was assembled as follows. The DNA fragment containing 1.35 kb of the cbh1 upstream region was amplified by PCR using the primers TcpyrF-Pcbh1 and Pcbh1-TclacA and chromosomal DNA of T. cellulolyticus S6-25 as the template. The second linear DNA fragment was produced by PCR using ppyrF-Ptef1-lacA as the template and the primers TcpyrF-32 and TclacA-56. The resulting DNA fragments were joined using Gibson Assembly® Master Mix.
Strain F09 pyrF+ Ptef1-lacA was constructed as follows. Two linear DNA fragments were produced with PCR (Fig. 1). The first fragment, which consisted of the lacA upstream region and the 5′ part of pyrF, was produced using pUlacA-pyrF-Ptef1 as the template and the primers p57Nr-bgal and pyrF-in-R. The second fragment was produced using ppyrF-Ptef1-lacA as the template and the primers pyrF-in-L and lacA-pUC57-432 and comprised the 3′ region of pyrF, Ptef1 and the upstream part of lacA. The resulting DNA fragments were mixed and used for transformation of the F09 strain (uracil-deficient mutant of S6-25). The resulting strain was designated F09 pyrF+ Ptef1-lacA.
The strains F09 pyrF+ and F09 pyrF+ Pcbh1-lacA were constructed in a manner similar to the construction of F09 pyrF+ Ptef1-lacA, but the plasmids ppyrF-PlacA-lacA and ppyrF-Pcbh1-lacA were used instead of the plasmid ppyrF-Ptef1-lacA.
The integration of the promoters upstream of lacA was confirmed by PCR analysis using a primer that annealed upstream of the fragment that participates in homologous recombination during integration and a primer that annealed on the coding region of lacA (Fig. 2); final confirmation was done by sequence analysis.
Verification of the integration of Ptef1 upstream of lacA by PCR. a Scheme for annealing of the primers. Pr.3, the primer SbfI-bga; Pr.2, the primer Tcbg-33. The estimated fragment size for the wild-type allele was 4281 bp and that for the region with integrated Ptef1 was 6614 bp. b Electrophoresis of the PCR products. 1–4, the transformants F09 pyrF+ Ptef1-lacA; 5, F09; 6, molecular weight marker
Determination of Enzymatic Activity
The strains were cultivated at 30 °C in 20 ml of liquid media in 750-ml flasks on a rotary shaker at 200 rpm for 72 h. The cultivation procedure in liquid medium was previously described [5]. After cultivation, samples were collected and centrifuged at 15,000 g for 10 min. The supernatant was either immediately used for enzymatic assays or frozen and stored at −20 °C. The growth of the strains was assessed by the yield of wet biomass when the strains were cultivated in PD or AC-C media; when cultivation was carried out in media supplemented with Avicel, biomass formation was not assessed, since it is impossible to separate fungal mycelium from a suspension of cellulose.
Determination of β-galactosidase activity was performed according to a previously described method [15] with minor modifications. The reaction was carried out in 800 µl of 50 mM Na acetate buffer (pH 4.6) containing 5 mM ONPG and the sample (usually 10 µl). The reaction mixture was incubated at 37 °C for 15 min, and the reaction was stopped by the addition of 400 µl of 1 M Na2CO3. The optical density of the reaction mixture was then measured at 420 nm. One unit of activity was defined as the amount of enzyme that liberated one µmol of o-nitrophenol per minute. The molar coefficient of the absorbance of o-nitrophenol was assumed to equal 4.5.
The effects of different pH values on β-galactosidase activity were estimated using 50 mM sodium acetate buffer solutions with pH values of 4.0–5.5 and in 50 mM potassium phosphate solutions with pH values of 6.0–7.5.
Cellobiohydrolase activity was measured by estimating the rate of hydrolysis of 4-methylumbelliferyl β-D-cellobioside (MUC) [26]. The reaction was carried out by mixing 75 μl of 0.75 M sodium acetate, pH 4.8, 25 μl of 1.8 mM MUC dissolved in the same buffer and 50 μl of culture supernatant and the following incubation at 50 °C for 10 min. Then, the reaction was quenched with 150 μl of 1 M Na2CO3, and the fluorescence of the sample was measured using a microplate reader (Tecan Infinite M200, Tecan Group Ltd, Switzerland) with excitation at 365 nm and emission at 445 nm. The amount of produced 4-methylumbelliferon was determined from a calibration curve plotted using 4-methylumbelliferone standards. One unit of activity was defined as the amount of enzyme that liberated one µmole of 4-methylumbelliferyl per minute.
Reverse Transcription-Quantitative Polymerase Chain Reaction (RT–qPCR)
Total RNA was isolated according to the protocol described in [27]. Reverse transcription was carried out using a RevertAid H Minus First Strand cDNA Synthesis Kit, K1631 (Thermo Fisher Scientific) and oligo(dT)18 primers. SYBR Green-based qPCR analysis was performed on a CFX96 Touch Deep Well Real-Time PCR Detection System (Bio–Rad, Hercules, USA) using a qPCRmix-HS SYBR kit (PK147L, Evrogen, Moscow, Russia). As a reference gene, the gene encoding the protein annotated as “actin” (GenBank: GAM40862.1) was used. Serial dilutions of reverse transcription mixtures were used to plot the standard curves to estimate PCR efficiency.
Protein Concentration and Gel Electrophoresis
The culture broth obtained after the cultivation of T. cellulolyticus strains in PD medium for 72 h was used for the preparation of concentrated and partially purified β-galactosidase. All manipulations were performed at 4 °C.
The enzyme was concentrated using several steps. First, ammonium sulfate was added to 3.5 ml of culture broth at up to 20% saturation, which allowed the removal of polysaccharides and other impurities. The sample was then desalted and concentrated using an Amicon® Ultra4 Centrifugal Filter Unit. At this stage, the sample was concentrated tenfold. In the next step, ammonium sulfate was added until complete saturation, and the precipitate was dissolved in 500 μl of Tris–HCl buffer (pH 7.5) and concentrated using a Microcon® 10 Centrifugal Filter Unit. Finally, the protein concentration was increased approximately 20-fold.
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) was performed according to the standard method [28] in 10% PAAG. The proteins on the gel were stained with Coomassie Blue R-250.
For the visualization of the protein possessing β-galactosidase activity, electrophoresis was performed without a denaturation step. PAAG (8%) was used, and the samples were neither boiled nor treated with mercaptoethanol; the process was performed at 4 °C. After electrophoresis, the SDS in the gel was removed by washing with 100 mM sodium citrate buffer (pH 5.0). Subsequently, 1% X-Gal solution in sodium citrate buffer (pH 5.0) was carefully spread over the gel surface, and the gel was incubated for 2 h at 30 °C (until a blue bend appeared).
Statistical Analysis
To assess the effect of genetic modifications or media composition on the level of enzyme production, the enzymatic activity was measured in at least three biological replicates (parallel cultures or repeated experiments). To determine the optimal temperature and pH, the measurement of the β-galactosidase activity was carried out in three analytical replicates for each temperature and pH value. RT–qPCR analysis was performed for four independently grown cultures for each strain. Statistical analysis was performed using Student’s t test with p value = 0.05.
Results
Search for Genes Orthologous to the Known Fungal β-galactosidase Genes in the Genome of T. cellulolyticus
The whole amino acid sequences of the β-galactosidase precursors of A. niger, A. oryzae and Penicillium sp. were used as a query for search against the T. cellulolyticus Y-94 protein database. On the basis of their molecular structure, these enzymes are assigned to glycosyl hydrolase family 35 (GHF-35) and are well studied [10,11,12,13,14,15,16,17,18]. The search revealed the hypothetical protein TCE0_017f03666 (accession number GAM35377.1) as a reciprocal best hit. This protein had 62, 64 and 67% identical amino acids, respectively, with the query proteins and 100% coverage of queries.
TCE0_017f03666 consists of 1010 amino acid residues, which is similar to the size of precursors of the β-galactosidases of A. niger, A. oryzae and Penicillium sp. [11,12,13,14,15,16,17,18]. Multiple sequence alignment with these proteins showed that for the region of TCE0_017f03666 from Leu42 to Ser395, which corresponds to the catalytic domains of these proteins [14, 16, 18], the similarity level is higher than for the whole protein. For example, with the Penicillium sp. domain, it was 78% identical and had 86% similar residues. Moreover, in the positions that are conserved for β-galactosidases [18], TCE0_017f03666 contains the same amino acid residues. For example, Tyr98, Asn142, and Glu144 at the positions responsible for the binding of β-galactose, and Glu202 and Glu299 at the positions responsible for hydrolysis of the glycoside bond (Fig. 3). These results indicate that TCE0_017f03666 is highly likely to possess β-galactosidase activity. By analogy with the gene encoding β-galactosidase in A. niger [11], it was designated lacA.
Multiple sequence alignment of the first 418 residues of TCE0_017f03666 with the corresponding regions of β-galactosidases of Penicillium sp (Psp), A. oryzae (Ao) and A. niger (Anig). The conserved amino acids, conserved replacement, and semiconserved replacement are marked with asterisks, colon, and point, respectively. The conserved amino acids of the studied enzymes, which form hydrogen bonds with β-galactose and hydrolyze the glycoside bond, are shown in boxes. The gray box marks the site recognized by the Kex2 peptidase. The pre-pro-sequences are underlined. The signal peptides predicted by the “SignalP-5.0” service are in italics
Targeting extracellular proteins into the secretory pathway occurs due to the presence of a signal sequence (also called pre-sequence) at their N-terminus. This sequence is usually cleaved off while the protein is translocated through the membrane of the endoplasmic reticulum. The typical length of signal peptides is 15–30 amino acids [29], and they have a conserved structure: a positively charged N-terminal region, followed by a central hydrophobic region and a neutral but polar C-region [30, 31]. In some proteins, the signal peptide is followed by another removable region, called a pro-sequence. It is believed that pro-sequences are required for the correct folding of their associated proteins [32, 33]. Cleavage of the pro-sequence is carried out by Kex2-like peptidase, and cleavage occurs immediately after the sequence “KR” or “RR” [34,35,36,37]. The precursors of β-galactosidases of A. niger, A. oryzae and Penicillium sp., used as a query to search the T. cellulolyticus database, are of this type of protein and contain per-pro-sequences that are 40 amino acids in length. These regions end with the amino acid residue “KR”, which is consistent with the consensus sequence (Fig. 3). The alignment demonstrated that the corresponding region of TCE0_017f03666 consisted of 42 amino acid residues, with identity levels to A. niger, A. oryzae and Penicillium sp. per-pro-sequences approximately 25% (Fig. 3). Despite this low identity, the online tool for prediction of signal peptides “SignalP-5.0” (https://services.healthtech.dtu.dk/service.php?SignalP-5.0) predicted with more than 0.5 probability that the first 20 amino acids of TCE0_017f03666 are a signal peptide (for comparison, for the N-ends of the sequences of A. niger, A. oryzae and Penicillium sp. this probability is higher than 0.9). The sequence of TCE0_017f03666, which corresponded to the pro-regions of, ended in residues “QR” that differed from the consensus sequence (Fig. 3). However, the pro-peptide of Trichoderma harzianum α-1,3-glucanase ends with the sequence “ER”, and this enzyme is correctly processed in Aspergillus oryzae [38]. Thus, it is highly likely that the first 42 residues of TCE0_017f03666 function as a pre-pro-region.
Substitution of the Native lacA Promoter with the Promoter of tef1 Led to the Appearance of Significant Extracellular β-galactosidase Activity
To verify whether lacA actually encodes a functional extracellular enzyme, the upstream region of this gene was replaced with a strong constitutive promoter. For this purpose, the promoter of the gene encoding translation elongation factor 1-alpha (tef1) was chosen. This promoter is often used for gene overexpression. To the best of our knowledge, T. cellulolyticus tef1 has not been identified experimentally. For this reason, the upstream region of a T. cellulolyticus gene encoding a protein GAM39229 orthologous to A. oryzae translation elongation factor 1-alpha (BAA76296) was used. The resulting strain was designated F09 pyrF+ Ptef1-lacA. As a control, the strain F09 pyrF+ was constructed. This strain contains the pyrF gene upstream of the native lacA promoter region.
The growth of the parental strain and several resulting transformants, F09 pyrF+ Ptef1-lacA and F09 pyrF+, was compared on MMAg plates supplemented with X-Gal. Contrary to the parental strain F09 and transformants F09 pyrF+, the transformants in which the promoters were replaced formed blue colonies surrounded by a blue halo, which indicated active β-galactosidase secretion (Fig. 4).
The β-galactosidase activity assay in transformants grown in PD medium showed similar results: the strains produced the same amount of biomass, but the supernatant of the F09 pyrF+ culture medium possessed negligible β-galactosidase activity (0.010 ± 0.004), whereas the supernatant of F09 pyrF+ Ptef1-lacA possessed an activity of approximately 1 U/ml (1.09 ± 0.14). RT–qPCR analysis confirmed that the replacement of the lacA promoter led to an increase in lacA transcription by at least 28,000-fold (details of analysis are given in Online Resource 1).
Based on significant homology of TCE0_017f03666 to β-galactosidases of GHF-35 and appearance of clearly detectable β-galactosidase activity after substitution of the native T. cellulolyticus lacA promoter, hereinafter we refer TCE0_017f03666 as β-galactosidase, realizing that the determination of substrate specificity of this enzyme is required to confirm name assignment.
Study of the Properties of the β-galactosidase of T. cellulolyticus
Despite the fact that β-galactosidase activity was easily detected in the culture broth of strain F09 pyrF+ Ptef1-lacA, no protein bands were observed by SDS–PAGE analysis (Fig. 5a, line 2). For this reason, the culture broth of the strains F09 pyrF+ Ptef1-lacA after 72 h of cultivation in PD medium was concentrated 20-fold and purified by precipitation with ammonium sulfate. The resulting protein preparations were also subjected to SDS–PAGE. After these manipulations, several bands became visible, and these included a band with a molecular weight between 95 kDa and 130 kDa, which corresponded to the calculated molecular weight of β-galactosidase (approximately 110 kDa, Fig. 5a, line 3).
a SDS–PAGE of T. cellulolyticus culture broth. 1, molecular weight marker; 2, culture broth of strain F09 pyrF+ Ptef1-lacA; 3, concentrated and partially purified culture broth of strain F09 pyrF+ Ptef1-lacA. b Visualization of β-galactosidase activity. 1, molecular weight marker; 2, concentrated proteins from F09 pyrF+ Ptef1-lacA
To ensure that the band with a molecular weight of approximately 110 kDa was indeed β-galactosidase, electrophoresis was performed under conditions that prevent protein denaturation. After treatment with X-Gal solution, a specific blue zone appeared in the region corresponding to approximately 110 kDa (Fig. 5b).
To determine the conditions suitable for detection of the activity of T. cellulolyticus β-galactosidase, the optimal temperature and pH for the β-galactosidase produced by strain F09 pyrF+ Ptef1-lacA were determined. The optimal temperature was approximately 50 °C (Fig. 6a), and the optimal pH was approximately 4.5–5.5 (Fig. 6b). These values are similar to other known fungal β-galactosidases with an acidic pH optimum (Table 2).
Study of the Applicability of lacA as a Reporter in T. cellulolyticus Cells Grown on Various Carbon Sources
It is obvious that the use of lacA as a reporter gene is possible only in the absence of significant synthesis of other enzymes with β-galactosidase activity. As mentioned above, for some fungi, the production of several enzymes with β-galactosidase activity encoded by different genes has been demonstrated. In this work, a BLAST search of the protein database of T. cellulolyticus also revealed several proteins with significant similarity to known fungal β-galactosidases. Thus, to verify in experiments with which carbon sources lacA can be used as a reporter, a test of F09 on X-Gal-containing plates supplemented with various carbon sources was performed. Strains Y-94 and S6-25, which are parental strains of F09, were also tested. The formation of a weak blue color (indicating weak β-galactosidase activity) was observed for all three strains in the presence of 1% xylan from oat spelts, 0.1% D-galactose, L-arabinose, D-xylose or lactose (Table 3 and Online Resource 2). The color obtained with L-arabinose and D-xylose was more intense than that obtained with other sugars. This finding was consistent with the data showing that xylose and arabinose, as well as xylan, induce the synthesis of hemicellulose-degrading enzymes in some Aspergillus and Penicillium species [42,43,44,45,46,47,48]. When sugars were used at a concentration of 1%, formation of the blue color was not observed. Importantly, the formation of a blue color during the growth of Y-94, S6-25 and F09 on all tested media was much slower than that for strain F09 pyrF+ Ptef1-lacA, in which lacA is expressed under a constitutive promoter (Fig. 4). Moreover, even after five days of cultivation, blue halos did not appear around colonies of Y-94, S6-25 and F09. These results indicated a very low level of β-galactosidase activity produced under the tested conditions. Thus, lacA can be applied as a reporter gene in T. cellulolyticus strains grown on tested carbon sources.
Study the Applicability of lacA as a Reporter for Evaluating Strong Promoters
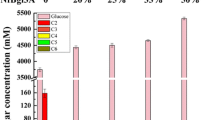
To determine whether LacA can be secreted at a level sufficient for evaluating promoters of highly expressed genes, the promoter of lacA was replaced with the strong promoter of T. cellulolyticus cbh1, encoding cellobiohydrolase I (Cel7A or Cbh1) [5, 49]. This enzyme is intensively produced when T. cellulolyticus is cultivated in cellulose-containing media without glucose, which inhibits its synthesis [7, 50, 51]. Strain F09 pyrF+ Pcbh1-lacA was cultivated in medium supplemented with either glucose, a mixture of glucose and cellulose, or only cellulose (AC-G, AC-GC and AC-C, respectively). Then, the β-galactosidase and cellobiohydrolase activities were studied in the supernatants. In general, the dependence of the β-galactosidase activity on the medium composition was similar to that of the cellobiohydrolase activity: the activity was minimal in the case of AC-G and maximal in the case of AC-C (Fig. 7). The ratio of changes in β-galactosidase and cellobiohydrolase activities depending on the carbon source was not exactly the same: the increase in activity in AC-GC compared to that in AC-G was higher for β-galactosidase, whereas the increase in AC-C compared to AC-GC was higher for cellobiohydrolase (Fig. 7). The most likely reason for this discrepancy is that several enzymes with hydrolytic activities contributed to overall (measured by MUC hydrolysis) cellobiohydrolase activity, but some limitation of β-galactosidase secretion in AC-C also cannot be excluded. However, this difference does not seem to be very significant. Therefore, LacA is secreted at near the same level as Cbh1, and this secretion ability of LacA is enough to use lacA as a reporter gene to study regulatory elements for high expression levels.
Effect of medium composition on β-galactosidase and cellulase activity produced by F09 pyrF+ Pcbh1-lacA. AC-G, AC-GC and AC-C, media supplemented with glucose, a mixture of glucose and cellulose, or with cellulose alone, respectively. The activity during cultivation in AC-C medium was taken to be 100%. The error bars show the ± 95% confidence intervals
Discussion
Despite the emergence of powerful tools such as RT–qPCRRT-qPCR, RNA array and RNA sequencing, reporter genes remain a convenient tool for studying the regulation of gene expression. The use of reporter genes remains the best choice in the case of studying the regulation of a small number of genes under different conditions because easy detection allows studying gene expression under a wide range of conditions with low labor costs. The disadvantage of this method is the necessity to assemble transcriptional or translational fusion cassettes prior to carrying out experiments.
The choice of a reporter gene depends on the purpose of the planned experiment. There are many reviews devoted to this topic, for example [52,53,54]. Here, we briefly describe several types of experiments and the types of reporter genes suitable for them to demonstrate the type of experiments in which lacA of T. cellulolyticus can be used. The isolation of regulatory genes and selection regulatory mutants have been achieved by cloning under the control of the promoter of interest housekeeping genes, genes responsible for the utilization of specific nutrients, or genes conferring antibiotic resistance [55, 56]. This approach is also suitable for designing promoter analysis systems aimed at determining the minimal length of promoters [57]; reporters changing the color or morphology of organisms [58] are also very convenient for such purposes. The main advantage of these approaches is that they do not require any additional experimental procedures, such as measuring enzymatic activity or fluorescence. The genes encoding enzymes with activity that can be detected on a plate test, such as β-galactosidase or β-glucuronidase, are also suitable for this purpose. When it is necessary to compare several promoters or to distinguish several levels of transcription provided by one promoter, the most convenient approach is to use fluorescent proteins or enzymes with easily detectable activity. In the case of studies of unicellular organisms, the protein encoded by the reporter gene can be localized intracellularly, since its production can be normalized by the number of cells [59,60,61], or even a graph of the distribution of production by cells can be plotted [62]. Since filamentous fungi are multicellular organisms that often form dense pellets when cultured in liquid media, the quantification of intracellular products is relatively laborious. For this reason, it is preferable that the product of the reporter gene would be secreted into the culture medium.
The results obtained in this work demonstrate that T. cellulolyticus lacA encodes a functional extracellular enzyme possessing β-galactosidase activity. The negligible production of β-galactosidase activity by strains expressing lacA under its native regulation allows using lacA as a reporter gene for comparison of promoters of this organism and for studying regulation of gene expression during cultivation with the commonly used carbon sources. The secretory ability of LacA was found to be sufficient for the evaluation of very strong promoters, such as the promoter of cbh1. The sensitivity of the assay was sufficient to reliably measure levels that differed by at least three orders of magnitude.
The technique of replacement of the lacA promoter used in this work to study lacA properties can also be applied for studying promoters of interest using lacA as a reporter. This approach requires fewer cloning steps than the assembly of expression cassettes containing reporter genes under the control of the investigated promoter. It also does not require the search for integration sites for expression cassettes and decreases the risk of undesirable genome modification due to the choice of an improper integration site. Additionally, when using this method, simultaneous integration of several copies of the cassette expressing the reporter gene cannot occur, which eliminates the need to check the obtained strains for the presence of multiple cassette integration.
References
Fujii, T., Hoshino, T., Inoue, H., & Yano, S. (2014). Taxonomic revision of the cellulose-degrading fungus Acremonium cellulolyticus nomen nudum to Talaromyces based on phylogenetic analysis. FEMS Microbiology Letters, 351, 32–41.
Yamanobe, T., Mitsiushi, Y., & Takasaki, Y. (1987). Isolation of a cellulolytic enzyme producing microorganism, culture conditions and some properties of the enzymes. Agricultural and Biological Chemistry, 51, 65–74.
Fang, X., Yano, S., Inoue, H., & Sawayama, S. (2009). Strain improvement of Acremonium cellulolyticus for cellulase production by mutation. Journal of Bioscience and Bioengineering, 107, 256–261.
Fujii, T., Iwata, K., Murakami, K., Yano, S., & Sawayama, S. (2012). Isolation of uracil auxotrophs of the fungus Acremonium cellulolyticus and the development of a transformation system with the pyrF gene. Bioscience, Biotechnology, and Biochemistry, 76, 245–249.
Inoue, H., Fujii, T., Yoshimi, M., Taylor, L. E., 2nd., Decker, S. R., Kishishita, S., Nakabayashi, M., & Ishikawa, K. (2013). Construction of a starch-inducible homologous expression system to produce cellulolytic enzymes from Acremonium cellulolyticus. Journal of Industrial Microbiology and Biotechnology, 40, 823–830.
Fujii, T., Koike, H., Sawayama, S., Yano, S., & Inoue, H. (2015). Draft genome sequence of Talaromyces cellulolyticus Strain Y-94, a source of lignocellulosic biomass-degrading enzymes. Genome Announcements, 3, e00014–e00015. https://doi.org/10.1128/genomeA.00014-15
Inoue, H., Decker, S. R., Taylor, L. E., 2nd., Yano, S., & Sawayama, S. (2014). Identification and characterization of core cellulolytic enzymes from Talaromyces cellulolyticus (formerly Acremonium cellulolyticus) critical for hydrolysis of lignocellulosic biomass. Biotechnology for Biofuels, 7, 151.
Ptitsyn, L. R., Yampolskaya, T. A., & Kutukova, E. A. (2017). Identification of core cellulolytic enzymes from Talaromyces cellulolyticus strain S6–25. FEBS J. 284, P.1.3–026.
van den Brink, J., & de Vries, R. P. (2011). Fungal enzyme sets for plant polysaccharide degradation. Applied Microbiology and Biotechnology, 91, 1477–1492.
Widmer, F., & Leuba, J. L. (1979). β-Galactosidase from Aspergillus niger. Separation and characterization of three multiple forms. European Journal of Biochemistry, 100, 559–567.
Kumar, V., Ramakrishnan, S., Teeri, T. T., Knowles, J. K., & Hartley, B. S. (1992). Saccharomyces cerevisiae cells secreting an Aspergillus niger beta-galactosidase grow on whey permeate. Biotechnology (N. Y), 10, 82–85.
Niu, D., Tian, X., Mchunu, N., Jia, C., Singh, S., Liu, X., Prior, B., & Lu, F. (2017). Biochemical characterization of three Aspergillus niger β-galactosidases. Electronic Journal of Biotechnology., 27, 37–43.
Rico-Díaz, A., Vizoso Vázquez, Á., Cerdán, M. E., Becerra, M., & Sanz-Aparicio, J. (2014). Crystallization and preliminary X-ray diffraction data of β-galactosidase from Aspergillus niger. Acta Crystallogr F Struct Biol Commun., 70, 1529–1531. https://doi.org/10.1107/S2053230X14019815
Rico-Díaz, A., Ramírez-Escudero, M., Vizoso-Vázquez, Á., Cerdán, M. E., Becerra, M., & Sanz-Aparicio, J. (2017). Structural features of Aspergillus niger β-galactosidase define its activity against glycoside linkages. FEBS Journal, 284, 1815–1829. https://doi.org/10.1111/febs.14083
Park, Y., Santi, M., & Pastore, G. (1979). Production and characterization of β-galactosidase from Aspergillus oryzae. Journal of Food Science, 44, 100–103.
Maksimainen, M. M., Lampio, A., Mertanen, M., Turunen, O., & Rouvinen, J. (2013). The crystal structure of acidic β-galactosidase from Aspergillus oryzae. International Journal of Biological Macromolecules, 60, 109–115. https://doi.org/10.1016/j.ijbiomac.2013.05.003
Nikolaev, I. V., Epishin, S. M., Zakharova, E. S., Kotenko, S. V., & Vinetskiĭ Iu. P. (1992). [Molecular cloning of the gene for secreted beta-galactosidase of the filamentous fungus Penicillium canescens]. [Article in Russian] Mol Biol (Mosk). 26, 869–875.
Rojas, A. L., Nagem, R. A., Neustroev, K. N., Arand, M., Adamska, M., Eneyskaya, E. V., Kulminskaya, A. A., Garratt, R. C., Golubev, A. M., & Polikarpov, I. (2004). Crystal structures of β-galactosidase from Penicillium sp. and its complex with galactose. Journal of Molecular Biology, 343, 1281–1292. https://doi.org/10.1016/j.jmb.2004.09.012
Adalberto, P. R., Massabni, A. C., Goulart, A. J., Contiero, J., Carmona, E. C., Cardello, L., & Monti, R. (2006). Production of β-galactosidase by Trichoderma reesei FTKO-39 in wheat bran: Partial purification of two isozymes. Applied Biochemistry and Biotechnology, 133, 163–170.
Maksimainen, M., Hakulinen, N., Kallio, J. M., Timoharju, T., Turunen, O., & Rouvinen, J. (2011). Crystal structures of Trichoderma reesei β-galactosidase reveal conformational changes in the active site. Journal of Structural Biology, 174, 156–163. https://doi.org/10.1016/j.jsb.2010.11.024
Tatusov, R. L., Koonin, E. V., & Lipman, D. J. (1997). A genomic perspective on protein families. Science, 278, 631–637. https://doi.org/10.1126/science.278.5338.631
Yilmaz, N., Visagie, C. M., Houbraken, J., Frisvad, J. C., & Samson, R. A. (2014). Polyphasic taxonomy of the genus Talaromyces. Studies in Mycology, 78, 175–341. https://doi.org/10.1016/j.simyco.2014.08.001
Fairhead, C., Llorente, B., Denis, F., Soler, M., & Dujon, B. (1996). New vectors for combinatorial deletions in yeast chromosomes and for gap-repair cloning using ‘split-marker’ recombination. Yeast, 12, 1439–1457.
Catlett, N. L., Lee, B., Yoder, O. C., & Turgeon, B. G. (2003). Split-marker recombination for efficient targeted deletion of fungal genes. Fungal Genetics Reports. https://doi.org/10.4148/1941-4765.1150
Nielsen, M. L., Albertsen, L., Lettier, G., Nielsen, J. B., & Mortensen, U. H. (2006). Efficient PCR-based gene targeting with a recyclable marker for Aspergillus nidulans. Fungal Genetics and Biology, 43, 54–64. https://doi.org/10.1016/j.fgb.2005.09.005
Chernoglazov, V. M., Jafarova, A. N., & Klyosov, A. A. (1989). Continuous photometric determination of endo-1,4-beta-D-glucanase (cellulase) activity using 4-methylumbelliferyl-beta-D-cellobioside as a substrate. Analytical Biochemistry, 179, 186–189. https://doi.org/10.1016/0003-2697(89)90222-4
Collart, M. A., & Oliviero, S. (2001). Preparation of yeast RNA. Current Protocols in Molecular Biology. https://doi.org/10.1002/0471142727.mb1312s23
Smith, B. J. (1984). SDS polyacrylamide gel electrophoresis of proteins. Methods in Molecular Biology (Clifton, NJ), 1, 41–55. https://doi.org/10.1385/0-89603-062-8:41
Lum, G., & Min, X. J. (2011). FunSecKB: the Fungal Secretome KnowledgeBase. Database, 2011, bar001. https://doi.org/10.1093/database/bar001
von Heijne, G. (1985). Signal sequences. The limits of variation. Journal of Molecular Biology, 184, 99–105. https://doi.org/10.1016/0022-2836(85)90046-4
Nielsen, H., Engelbrecht, J., Brunak, S., & von Heijne, G. (1997). Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein engineering, 10, 1–6. https://doi.org/10.1093/protein/10.1.1
Baker, D., Shiau, A. K., & Agard, D. A. (1993). The role of pro regions in protein folding. Current Opinion in Cell Biology, 5, 966–970. https://doi.org/10.1016/0955-0674(93)90078-5
Eder, J., & Fersht, A. R. (1995). Pro-sequence-assisted protein folding. Molecular microbiology, 16, 609–614. https://doi.org/10.1111/j.1365-2958.1995.tb02423.x
Mizuno, K., Nakamura, T., Ohshima, T., Tanaka, S., & Matsuo, H. (1989). Characterization of KEX2-encoded endopeptidase from yeast Saccharomyces cerevisiae. Biochemical and Biophysical Research Communications, 159, 305–311.
Brenner, C., & Fuller, R. S. (1992). Structural and enzymatic characterization of a purified prohormone-processing enzyme: Secreted, soluble Kex2 protease. Proceedings of the National Academy of Sciences of the United States of America., 89, 922–926.
Bevan, A., Brenner, C., & Fuller, R. S. (1998). Quantitative assessment of enzyme specificity in vivo: P2 recognition by Kex2 protease defined in a genetic system. Proceedings of the National Academy of Sciences of the United States of America, 95, 10384–10389.
Jalving, R., van de Vondervoort, P. J., Visser, J., & Schaap, P. J. (2000). Characterization of the kexin-like maturase of Aspergillus niger. Applied and Environment Microbiology, 66, 363–368.
Fuglsang, C. C., Berka, R. M., Wahleithner, J. A., Kauppinen, S., Shuster, J. R., Rasmussen, G., Halkier, T., Dalboge, H., & Henrissat, B. (2000). Biochemical analysis of recombinant fungal mutanases. A new family of alpha1,3-glucanases with novel carbohydrate-binding domains. Journal of Biological Chemistry, 275, 2009–2018.
O’Connell, S., & Walsh, G. (2008). Application relevant studies of fungal beta-galactosidases with potential application in the alleviation of lactose intolerance. Applied Biochemistry and Biotechnology, 149, 129–138.
Cardoso, B. B., Silvério, S. C., Abrunhosa, L., Teixeira, J. A., & Rodrigues, L. R. (2017). β-galactosidase from Aspergillus lacticoffeatus: A promising biocatalyst for the synthesis of novel prebiotics. International Journal of Food Microbiology, 257, 67–74.
Cruz, R., Cruz, V., Belote, J. N., Khenayfes, M. D., Dorta, C., & Oliveira, L. H. (1999). Properties of a new fungal β-galactosidase with potential application in the dairy industry. Revista de Microbiologia, 30, 265–271.
v. d. Veen, P., Flipphi, M.J., Voragen, A.G. and Visser, J. (1993). Induction of extracellular arabinases on monomeric substrates in Aspergillus niger. Archives of Microbiology, 159, 66–71.
Fernández-Espinar, M., Piñaga, F., Graaff, L., Visser, J., Ramón, D., & Vallés, S. (2004). Purification, characterization and regulation of the synthesis of an Aspergillus nidulans acidic xylanase. Applied Microbiology and Biotechnology., 42, 555–562.
Kumar, S., & Ramón, D. (1996). Purification and regulation of the synthesis of a β-xylosidase from Aspergillus nidulans. FEMS Microbiology Letters., 135, 287–293.
de Vries, R. P., Visser, J., & de Graaff, L. H. (1999). CreA modulates the XlnR-induced expression on xylose of Aspergillus niger genes involved in xylan degradation. Research in Microbiology, 150, 281–285.
Rizzatti, A. C., Freitas, F., Bertolini, M., Peixoto-Nogueira, S. C., Terenzi, H., Jorge, J., & Polizeli, M. D. (2007). Regulation of xylanase in Aspergillus phoenicis: A physiological and molecular approach. Journal of Industrial Microbiology & Biotechnology., 35, 237–244.
Nikolaev, I. V., & Vinetski, Y. P. (1998). L-Arabinose induces synthesis of secreted beta-galactosidase in the filamentous fungus Penicillium canescens. Biochemistry (Moscow), 63, 1294–1298.
Vavilova, E. A., & Vinetskiĭ, Iu. P. (2003). Induction of endo-1,4-beta-xylanase and beta-galactosidase in the original and recombinant strains of the fungus Penicillium canescens]. [Article in Russian. Prikladnaia Biokhimiia i Mikrobiologiia, 39, 167–172.
Midoh, N., Sumida, N., Okakura, K., Murakami, T., & Yamanobe, T. (2009). New promoter for expressing protein products. Japanese patent JP4257759
Hideno, A., Inoue, H., Fujiim, T., Yano, S., Tsukahara, K., Murakami, K., Yunokawa, H., & Sawayama, S. (2013). High-coverage gene expression profiling analysis of the cellulase-producing fungus Acremonium cellulolyticus cultured using different carbon sources. Applied Microbiology and Biotechnology, 97, 5483–5492. https://doi.org/10.1007/s00253-013-4689-0
Fujii, T., Inoue, H., & Ishikawa, K. (2013). Enhancing cellulase and hemicellulase production by genetic modification of the carbon catabolite repressor gene, creA, in Acremonium cellulolyticus. AMB Express, 3, 73. https://doi.org/10.1186/2191-0855-3-73
Pardy, K. (1994). Reporter enzymes for the study of promoter activity. Molecular Biotechnology, 2, 23–27. https://doi.org/10.1007/BF02789287
Kain, S. R., & Ganguly, S. (2001). Overview of genetic reporter systems. Current Protocols in Molecular Biology, 9(Unit9), 6. https://doi.org/10.1002/0471142727.mb0906s36
Ghim, C. M., Lee, S. K., Takayama, S., & Mitchell, R. J. (2010). The art of reporter proteins in science: Past, present and future applications. BMB Reports, 43, 451–460. https://doi.org/10.5483/bmbrep.2010.43.7.451
Saloheimo, A., Aro, N., Ilmén, M., & Penttilä, M. (2000). Isolation of the ace1 gene encoding a Cys(2)-His(2) transcription factor involved in regulation of activity of the cellulase promoter cbh1 of Trichoderma reesei. The Journal of biological chemistry, 275, 5817–5825. https://doi.org/10.1074/jbc.275.8.5817
Chulkin, A. M., Vavilova, E. A., & Benevolenskiĭ, S. V. (2011). The mutational analysis of carbon catabolite repression in filamentous fungus Penicillium canescens. Molekuliarnaia Biologiia, 45, 871–878. https://doi.org/10.1134/S0026893311050049
Honda, Y., Tanigawa, E., Tsukihara, T., Nguyen, D. X., Kawabe, H., Sakatoku, N., Watari, J., Sato, H., Yano, S., Tachiki, T., Irie, T., Watanabe, T., & Watanabe, T. (2019). Stable and transient transformation, and a promoter assay in the selective lignin-degrading fungus Ceriporiopsis subvermispora. AMB Express, 9, 92. https://doi.org/10.1186/s13568-019-0818-1
Song, H. Y., Choi, D., Han, D. M., Kim, D. H., & Kim, J. M. (2018). A Novel Rapid Fungal Promoter Analysis System Using the Phosphopantetheinyl Transferase Gene, npgA, in Aspergillus nidulans. Mycobiology, 46, 429–439. https://doi.org/10.1080/12298093.2018.1548806
Miller, J. H. (1972). Experiments in molecular genetics. ColdSpring Harbor Laboratory Press.
Bitter, G. A., Chang, K. K., & Egan, K. M. (1991). A multi-component upstream activation sequence of the Saccharomyces cerevisiae glyceraldehyde-3-phosphate dehydrogenase gene promoter. Molecular & General Genetics: MGG, 231, 22–32. https://doi.org/10.1007/BF00293817
Flagfeldt, D. B., Siewers, V., Huang, L., & Nielsen, J. (2009). Characterization of chromosomal integration sites for heterologous gene expression in Saccharomyces cerevisiae. Yeast, 26, 545–551. https://doi.org/10.1002/yea.1705
Nevoigt, E., Kohnke, J., Fischer, C. R., Alper, H., Stahl, U., & Stephanopoulos, G. (2006). Engineering of promoter replacement cassettes for fine-tuning of gene expression in Saccharomyces cerevisiae. Applied and Environmental Microbiology, 72, 5266–5273. https://doi.org/10.1128/AEM.00530-06
Acknowledgements
We would like to thank Dr. Natalia P. Zakataeva (Ajinomoto-Genetika Research Institute, 1st Dorozhny proezd, 1-1, Moscow 117545, Russia) for critical reading of the manuscript and for fruitful discussions.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Orleneva, A.P., Serebrianyi, V.A., Kutukova, E.A. et al. Identification of the Talaromyces cellulolyticus Gene Encoding an Extracellular Enzyme with β-galactosidase Activity and Testing it as a Reporter for Gene Expression Assays. Mol Biotechnol 64, 637–649 (2022). https://doi.org/10.1007/s12033-022-00453-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-022-00453-9