Abstract
Hepatocellular carcinoma (HCC) has a high dead rate partly due to late diagnosis. This study aimed to investigate the prognostic value of miR-937 in HCC and its role in the HCC progression. HCC tissue and adjacent non-cancerous tissues (NCT) (n = 125) were detected about the expression level of miR-937 via real-time quantitative PCR. The relationship between miR-937 expression and each important clinical characteristic was evaluated. And the prognostic significance of miR-937 was assessed by Kaplan–Meier curve and Cox regression analysis. CCK-8 and Transwell assays were conducted to observe the effects of miR-937 on HCC cell proliferation, migration, and invasion. The miR-937 expression was upregulated in HCC tissues, as well as in HCC cell lines. The upregulation of miR-937 showed a significant association with lymph node metastasis and TNM stage. Upregulation of miR-937 predicted poor prognosis of HCC patients. Overexpression of miR-937 promoted HCC cell ability of proliferation, migration, and invasiveness, while knockdown of miR-937 inhibited these cellular behaviors. miR-937 expression was upregulated in HCC and may serve as a promising prognostic factor and treated target for HCC patients. miR-937 might exert a promoter role in HCC through accelerated tumor cell proliferation, migration, and invasion.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hepatocellular carcinoma (HCC) is high-incidence primary liver cancer and ranked in the third leading cause of cancer death worldwide [1]. Among the high-incidence regions, China accounts for nearly half of all cases estimated to occur [2]. The outcome of patients with HCC at very early or early stages has improved due to advances in diagnosis and multidisciplinary therapy [3]. However, most HCC patients cannot benefit from such a possibility because of being diagnosed at an advanced stage leading to limited therapeutic options [4]. The overall survival (OS) rate of HCC remained unsatisfactory. Therefore, sensitive and invasive prognosis assessment for HCC need to be further explored.
MicroRNAs (miRNAs) are typically 20 to 23 nucleotides in length and classified as non-coding RNA [5]. Although short and non-coding, miRNAs can regulate post-transcriptional silencing of downstream target genes and play important role in a range of biological processes [6]. The level of complementarity between miRNA and messenger RNA (mRNA) can either inhibit the translation of the target mRNAs or degrade them [7]. Aberrant miRNAs expression is associated with various diseases, including cancers [8, 9]. Specifically, miRNAs show exclusive expression due to the type, stage, and metastasis of cancer, brought the potential of diagnostic and prognostic non-invasive biomarkers including hepatocellular carcinoma. For instance, miRNA-221 was accessed and verified to be an important prognostic marker for HCC [10]. Serum miRNA-27a and miRNA-18b expression levels were able to serve as potential predictive biomarkers of chronic hepatitis, post-chronic hepatitis cirrhosis, and HCC [11]. Recently, miR-1203 can predict a poor prognosis in HCC patients and has the potential to be a therapeutic target for HCC [12]. MicroRNA-937 (miRNA-937) level has been reported to be increased in breast cancer tissues and colon cancer tissues compared with adjacent normal ones, respectively, and may act as prognostic biomarkers for breast cancer and colon cancer. Shi et al. have shown that miR-937 was with an increased expression in HCC patients [13]. However, the prognostic value of miRNA-937 in HCC has no report.
In the current study, we aimed to investigate the expression level of miR-937 and its prognostic significance in HCC. The effect of miR-937 on HCC cellular function was involved too.
Materials and Methods
Sample Collection
A total of 125 tumor tissue samples and corresponding adjacent non-cancerous tissues (NCT) were obtained from HCC patients during surgery. The tumor stage was determined according to the 7th edition of TNM. Samples were collected at The Second Affiliated Hospital of Kunming Medical University between 2011 and 2014. All acquired slices of the tumor and adjacent tissue were verified pathologically to confirm. After surgical resection, the collected samples were immediately placed in liquid nitrogen and stored until analysis. Significant clinicopathological data were collected from each participant. Patients’ key characteristics are presented in Table 1.
Written informed consent was provided from each participant. The study was approved by the Ethics Committee of the Second Affiliated Hospital of Kunming Medical University.
Cell Lines Culture and Transfection
The human HCC cell lines PLC/PRF/5, SNU-449, and SK-HEP-1 were purchased from ATCC; HuH-7 and normal liver cell line THLE-2 were obtained from Cell Bank of Chinese Academy of Sciences (Shanghai, China). PLC/PRF/5 and SNU-449 cells were cultured in RPMI 1640 (Thermo Fisher Scientific, IL, USA) containing 10% fetal bovine serum (FBS) (Thermo Fisher Scientific, IL, USA). HuH-7 and SK-HEP-1cells were cultured in DMEM (Thermo Fisher Scientific, IL, USA) containing 10% FBS. The culture was carried out at 37 °C in a humidified chamber with 5% CO2.
The mimic negative control (NC, sense 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense 5′-ACGUGACACGUUCGGAGAATT-3′), miR-937 mimic (sense 5′-AUCCGCGCUCUGACUCUCUGCC-3′ and antisense 5′-GGCAGAGAGUCAGAGCGCGGAUUU-3′), inhibitor NC (5′-CAGUACUUUUGUGUAGUACAA-3′), and miR-937 inhibitor (sense 5′-GGCAGAGAGUCAGAGCGCGGAU-3′) were acquired from GenePharma (Shanghai, China). Lipofectamine 3000 reagents (Invitrogen; USA) were applied for cell transfection according to the manufacturer’s instructions.
RNA Extraction and qRT-PCR
Total RNA was purified from cultured cells or tissue samples using the TRIzol reagent (Invitrogen; USA). RNA concentration was ascertained by NanoDrop 2000c (Thermo Scientific). Then, total RNA was reverse-transcribed to be cDNA using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems; Thermo Fisher Scientific, IL, USA). MiR-937 was determined along with U6 (reference gene) by RT-qPCR equipped with TaqMan® miRNA assays following the manufacturer’s protocol. The primers used were as follows: miR-937, forward: 5′-GCCGAGATCCGCGCTCTGAC-3′ and reverse: 5′-CTCAACTGGTGTCGTGGA-3′; U6, forward: 5′-CGCTTCGGCAGCACATATACTA-3′ and reverse: 5′-CGCTTCACGAATTTGCGTGTCA-3′. The expression levels of miR-937 normalized to U6 genes were analyzed using the \(2^{{ - \Delta \Delta C_{{\text{t}}} }}\) method.
Cell Proliferation Assay
The cell proliferation assay was performed using a CCK-8 cell counting kit (Dojindo Laboratories, Kumamoto, Japan). PLC/PRF/5 (3 × 103 cells per well) and SNU-449 (2 × 103 cells per well) cells were seeded into 96-well plates. Cells were incubated for 0, 24, 48, and 72 h at 37 °C with a 5% CO2 in air atmosphere. CCK-8 reagent was added to each well at every time point, and cells were incubated for a further 2 h at 37 °C. Then, the optical density (OD) value was measured using a microplate reader (Bio-Tek Company, Winooski, VT, USA) under 450 nm wavelength.
Transwell Migration and Invasion Assays
Transwell chambers (Millipore, Burlington, MA, USA) were utilized to assess HCC cell migratory and invasive abilities. For migration assays, the upper chambers were filled with transfected HCC cells (1 × 105 cells) in the serum-free culture medium and the lower chamber with a complete culture medium containing 10% FBS. For invasion assay, the upper chamber was pre-coated with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) instead, and the other process was similar to migration assays. After 24 h of incubation, migrated or invaded cells on the filter inserts were fixed, stained, and counted in five random fields under a microscope.
Prediction of miR-937 Target Gene
miR-937 target genes were predicted in silico using TargetScan 7.2 (http://www.targetscan.org/vert_72/) and miRWalk (http://mirwalk.umm.uni-heidelberg.de/).
Luciferase Reporter Assays
Luciferase reporter containing the human wild-type 3-UTR of SEC14L2 (SEC14L2-wt) or a mutant reporter (SEC14L2-mut) was constructed by GenePharma (Shanghai, China). For the reporter assays, PLC/PRF/5 and SNU449 cells were cultured in 24-well plates and transfected with both the SEC14L2-wt/SEC14L2-mut plasmids and the miR-937 mimic/Mimic NC or miR-937 inhibitor/inhibitor NC, according to the Lipofectamine 3000 transfection system protocol. The cells were harvested 72 h after transfection. Firefly and Renilla luciferase activities were measured at a Dual-Luciferase Reporter Assay System (Promega, USA) and were normalized to the Renilla luciferase activity.
Statistical Analysis
All experiments were repeated at least three times. Data analysis was performed by the SPSS 20.0 software (SPSS, Chicago, IL, USA) and GraphPad 5.0 software (GraphPad Software, Inc., La Jolla, CA, USA). Comparison of the data sets between HCC and non-cancer tissues was analyzed by t test. The differences of multiple groups were performed by one-way ANOVA. Correlation between HCC patients’ clinical data and expression levels of miR-937 was analyzed by Chi-squared test. The survival curve was plotted via the K–M method and compared using a log-rank test. Differences were considered to be statistically significant when P < 0.05.
Results
miR-937 Presented an Elevated Expression in Both HCC Tissues and Cell Lines
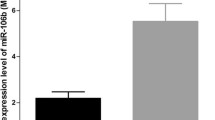
miR-937 expression levels in 125 pairs of HCC tissues and NCT were quantified by qRT-PCR. The detection results showed the relative expression of miR-937 was significantly higher in HCC tissues than that in NCT (P < 0.001, Fig. 1A). Then, the expression of miR-937 in HCC cell lines was also determined. Consistent with the result of HCC tissues, higher miR-937 expression levels were observed in four HCC cell lines compared to the normal liver cell line (P < 0.001, Fig. 1B).
miR-937 expression is upregulated in HCC via qRT-PCR. A The relative expression of miR-937 was significantly upregulated in HCC tissues compared with NCT. Differences were tested using a 2-tailed Student’s t test. B The expression of miR-937 was significantly higher in HCC cells than that in normal liver cells. Differences were evaluated by one-way ANOVA followed by Tukey’s post hoc test. **P < 0.01, ***P < 0.001
The Correlation Between miR-937 Expression and Key Clinical Parameters of HCC Patients
The enrolled HCC patients were classified as high expression group and low group based on the median expression level of miR-937 (2.30). To access the relationship between miR-937 expression and HCC progression, Chi-Squared tests were conducted. As shown in Table 1, a high level of miR-937 was closely associated with positive lymph node metastasis (LNM, P < 0.01) and advanced TNM stage (P < 0.01), but not with other parameters (P > 0.05).
Increased Expression of miR-937 Associated with Poor Prognosis of Patients with HCC
The Kaplan–Meier survival analysis with log-rank test was performed to predict the prognostic value of miR-937 in HCC patients. As shown in Fig. 2, patients with high levels of miR-937 had evidently shorter mean overall survival time (41.3 months, 95% CI 44.8–55.7) compared with those with lower miR-937 expression (50.3 months, 95% CI 36.0–46.5) (log-rank P = 0.021). Notably, the multivariate Cox regression analysis implied that miR-937 expression was an independent factor for predicting the 5-year overall survival of HCC patients (HR 3.237, 95% CI 1.385–7.567; P = 0.007, shown in Table 2). These results indicated that miR-937 might be a promising biomarker for predicting the prognosis of HCC patients.
Upregulation of miR-937 Promotes HCC Cell Proliferation, Migration, and Invasion In Vitro
To explore the functional role of miR-937 on HCC cell behavior, HCC cells with overexpression or knockdown of miR-937 were constructed using miR-937 mimics or inhibitors. The qRT-PCR results verified transfection efficiency in PLC/PRF/5 and SNU-449 cells (P < 0.001, Fig. 3A). CCK-8 assay was used to judge the effects of miR-937 on cell proliferation. The OD values in PLC/PRF/5 and SNU-449 cells increased significantly after the upregulation of miR-937 expression but reduced significantly after the knockdown of miR-937 (P < 0.01, Fig. 3B). These results revealed that miR-937 promotes cell proliferation in HCC cells. In the transwell assay, the numbers of migration and invasion cells increased significantly after the overexpression of miR-937 in PLC/PRF/5 and SNU-449 cells but decreased significantly after the knockdown of miR-937 (P < 0.001, Fig. 4A and B). These results suggested that miR-937 accelerated HCC cell migration and invasion.
Overexpression of miR-937 accelerated cell proliferation, while knockdown of miR-937 inhibited cell proliferation, compared to untreated cells. Difference analysis was conducted using one-way ANOVA followed by Tukey’s post hoc test. A Transfection efficiency of miR-937 mimic and inhibitor was supported using qRT-PCR. B CCK-8 assays were performed 0, 24, 48, and 72 h after transfection to determine the proliferation of PLC/PRF/5 and SNU-449 cells. *P < 0.05, **P < 0.01, ***P < 0.001
miR-937 promoted HCC cell invasion and migration. A The migration assays demonstrated overexpression of miR-937 pushed HCC cell migration, while downregulation of miR-937 repressed the migration. B The invasion assays showed upregulation of miR-937 promoted HCC cell migration, while downregulation of miR-937 inhibited the invasion. Differences were analyzed via one-way ANOVA followed by Tukey’s post hoc test. ***P < 0.001
In Silico Prediction of the Target Genes of miR-937
To identify the downstream target genes of miR-937, target gene prediction was conducted using TargetScan 7.2 and miRWalk. And we found SEC14L2, which has been reported to show a reduced expression in hepatocellular carcinoma and inhibits cancer cells [14], may be the downstream gene of miR-937 (Fig. 5A). In HCC cells, overexpression of miR-937 can reduce the relative expression of SEC14L2 mRNA, while knockdown of miR-937 can increase the relative expression of SEC14L2 mRNA (Fig. 5B). Further, the Luciferase reporter assay showed that miR-937 mimic can reduce the luciferase activity of SEC14L2-wt but can influence the value of SEC14L2-mut, while miR-937 inhibitor can significantly increase the luciferase activity value of SEC14L2-wt but can influence the value of SEC14L2-mut, both in PLC/PRF/5 and SNU-449 cells (Fig. 5C and D). Combining these results, miR-937 may exert its role in HCC cells partly at least by regulating the expression of SEC14L2.
SEC14L2 may be a target gene of miR-937 in HCC. A The binding sites between SEC14L2 and miR-937 by TargetScan 7.2. B RT-qPCR of SEC14L2 mRNA in mimic/inhibitor miR937-transfected PLC/PRF/5 and SNU-449 cells, in comparison to cells transfected with respective negative controls. C and D PLC/PRF/5 and SNU-449 cells were co-transfected with SEC14L2-mut or SEC14L2-wt, together with mimic NC, miR-937 mimic, inhibitor NC, or miR-937 inhibitor as indicated. Firefly luciferase activity was measured and normalized to Renilla luciferase activity. Differences were analyzed via two-way ANOVA by Sidak's multiple comparisons test. ***P < 0.001
Discussion
It is well recognized that miRNAs can play roles of oncogenes or tumor suppressor genes in many cancers and affect the progression of different human cancers [15, 16]. Accumulating evidence has linked miRNAs with poor prognosis after curative HCC resection [17, 18]. This suggests the key roles for miRNAs in promoting HCC progression [19]. miRNAs may directly aggravate HCC progression by dysregulating their biogenesis and expression [20]. Thus, the interaction between miRNAs and HCC cells is a key factor in HCC progression.
In the current study, it is demonstrated that miR-937 was significantly upregulated in HCC tissues and cell lines compared with matching NCT and normal liver cell lines, respectively. And, this upregulation was closely related to the survival time of HCC patients, that is to say, HCC patients with high-expressed miR-937 would behave a shorter survival time than those with low-expressed miR-937. Furthermore, miR-937 significantly affected HCC progression on the proliferation, migration, and invasion, shown as upregulation of miR-937 promoting the progression, while knockdown of miR-937 reversing the auxo-action. Taken together, miR-937 may promote HCC progression and has the potential to be a prognostic biomarker for HCC.
HCC is known as a kind of malignant neoplasm with high mortality [21]. Even application of sorafenib and regorafenib in HCC, the overall survival outcome of HCC remains unsatisfactory with the poor median survival in advanced HCC [22]. Thus, it is of great value to explore the novel prognosis markers for HCC to predict overall survival. In recent years, microRNAs are involved in a variety of human cancers, including liver cancer, might be candidate biomarkers for the prognosis and the therapeutic target of HCC [23,24,25].
In this study, expression of miR-937 in HCC tissues and cell lines was compared with those in NCT and liver normal cell line by qRT-PCR and the results revealed that the expression of miR-937 was upregulated in both HCC tissues and cell lines. There is also a rising trend of miR-937 expression in colon cancer from a recent study by Liu et al. and the upregulated expression leads to promotion in cancer proliferation, migration, and invasion, and miR-937 may be a prognostic factor and potential target for colon cancer therapy [26]. So we further evaluated the prognosis value of miR-937 for HCC patients and it showed that miR-937 may act as a promising prognostic biomarker for HCC via the results of KM curve and multivariate Cox regression analysis between clinical characteristics and 5-year follow-up information. Consistently, miR-937 has also shown the ability of prognosis in breast cancer [27]. Besides, the effects of aberrant expression of miR-937 on the HCC process using CCK-8 assay and transwell assay, as it turned out, overexpression of miR-937 push HCC cells proliferation, migration, and invasion, while inhibition of miR-937 lead to an inhibitory effect. Similar consequences of abnormal miR-937 expression were found in lung cancer. As is reported, miR-937 expression was increased in lung cancer and contributed to lung cancer cell proliferation [28].
To gain insight into the biological role of miR937, it is integral to identify its target gene, since miRNAs negatively regulate their target genes and then make fine-scale adjustments to protein outputs [29]. By computational methods, TargetScan 7.2 and miRWalk represent SEC14L2 maybe a downstream gene of miR-937 with bind sites at the 3′-UTR. The in silico-predicted target SEC14L2 was verified by cloning 3′UTR of target genes into the 3′-UTR region of a reporter vector, followed by co-transfection with miR-937 mimic and miR-937 inhibitor. In other studies, SEC14L2 has been identified to be a master regulator gene in HCC and represents a decreased expression in HCC cells [14, 30]. In addition, SEC14L2 has been reported to be a target gene of miR-1307-5p in HCC [31]. This study indicated that SEC14L2 was one of the miR-937 targets. miR-937 may exert its role in HCC cells partly at least by regulating SEC14L2. However, further study is still needed to clarify the detailed mechanism.
In summary, miR-937 is upregulated in HCC and promotes the progression of HCC cells such as proliferation, migration, and invasion. Besides, the high expression of miR-937 is closely related to the shorter overall survival of HCC patients. Thus, miR-937 may serve as a tumor-treated target and has the potential to be a novel prognostic marker for HCC.
References
Forner, A., Reig, M., & Bruix, J. (2018). Hepatocellular carcinoma. Lancet (London, England)., 391(10127), 1301–1314.
Hartke, J., Johnson, M., & Ghabril, M. (2017). The diagnosis and treatment of hepatocellular carcinoma. Seminars in Diagnostic Pathology, 34(2), 153–159.
Grandhi, M. S., Kim, A. K., Ronnekleiv-Kelly, S. M., Kamel, I. R., Ghasebeh, M. A., & Pawlik, T. M. (2016). Hepatocellular carcinoma: From diagnosis to treatment. Surgical Oncology, 25(2), 74–85.
Ren, Z., Ma, X., Duan, Z., & Chen, X. (2020). Diagnosis, therapy, and prognosis for hepatocellular carcinoma. Analytical Cellular Pathology (Amsterdam)., 2020, 8157406.
Lu, T. X., & Rothenberg, M. E. (2018). MicroRNA. The Journal of Allergy and Clinical Immunology, 141(4), 1202–1207.
Mohr, A. M., & Mott, J. L. (2015). Overview of microRNA biology. Seminars in Liver Disease, 35(1), 3–11.
Ying, S. Y., Chang, D. C., & Lin, S. L. (2008). The microRNA (miRNA): Overview of the RNA genes that modulate gene function. Molecular Biotechnology, 38(3), 257–268.
Li, X., Zeng, Z., Wang, J., Wu, Y., Chen, W., Zheng, L., et al. (2020). MicroRNA-9 and breast cancer. Biomedicine & Pharmacotherapy [Biomedecine & Pharmacotherapie], 122, 109687.
Lopez-Pedrera, C., Barbarroja, N., Patiño-Trives, A. M., Luque-Tévar, M., Torres-Granados, C., Aguirre-Zamorano, M. A., et al. (2020). Role of microRNAs in the development of cardiovascular disease in systemic autoimmune disorders. International Journal of Molecular Sciences. https://doi.org/10.3390/ijms21062012
Chen, F., Li, X. F., Fu, D. S., Huang, J. G., & Yang, S. E. (2017). Clinical potential of miRNA-221 as a novel prognostic biomarker for hepatocellular carcinoma. Cancer Biomarkers: Section A of Disease Markers, 18(2), 209–214.
Rashad, N. M., El-Shal, A. S., Shalaby, S. M., & Mohamed, S. Y. (2018). Serum miRNA-27a and miRNA-18b as potential predictive biomarkers of hepatitis C virus-associated hepatocellular carcinoma. Molecular and Cellular Biochemistry, 447(1–2), 125–136.
Shi, J., Li, X., Hu, Y., Zhang, F., Lv, X., Zhang, X., et al. (2020). MiR-1203 is involved in hepatocellular carcinoma metastases and indicates a poor prognosis. Neoplasma, 67(2), 267–276.
Shi, B., Zhang, X., Chao, L., Zheng, Y., Tan, Y., Wang, L., et al. (2018). Comprehensive analysis of key genes, microRNAs and long non-coding RNAs in hepatocellular carcinoma. FEBS Open Bio, 8(9), 1424–1436.
Li, Z., Lou, Y., Tian, G., Wu, J., Lu, A., Chen, J., et al. (2019). Discovering master regulators in hepatocellular carcinoma: One novel MR, SEC14L2 inhibits cancer cells. Aging, 11(24), 12375–12411.
Carvalho de Oliveira, J., Molinari Roberto, G., Baroni, M., Bezerra Salomão, K., Alejandra Pezuk, J., & Sol Brassesco, M. (2018). MiRNA dysregulation in childhood hematological cancer. International Journal of Molecular Sciences. https://doi.org/10.3390/ijms19092688
Harrandah, A. M., Mora, R. A., & Chan, E. K. L. (2018). Emerging microRNAs in cancer diagnosis, progression, and immune surveillance. Cancer Letters, 438, 126–132.
Qadir, M. I., & Rizvi, S. Z. (2017). miRNA in hepatocellular carcinoma: pathogenesis and therapeutic approaches. Critical Reviews in Eukaryotic Gene Expression., 27(4), 355–361.
Nagy, Á., Lánczky, A., Menyhárt, O., & Győrffy, B. (2018). Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Scientific Reports, 8(1), 9227.
Li, S., Yao, J., Xie, M., Liu, Y., & Zheng, M. (2018). Exosomal miRNAs in hepatocellular carcinoma development and clinical responses. Journal of Hematology & Oncology., 11(1), 54.
Li, D., Zhang, J., & Li, J. (2020). Role of miRNA sponges in hepatocellular carcinoma. Clinica Chimica Acta, 500, 10–9.
Clark, T., Maximin, S., Meier, J., Pokharel, S., & Bhargava, P. (2015). Hepatocellular carcinoma: Review of epidemiology, screening, imaging diagnosis, response assessment, and treatment. Current Problems in Diagnostic Radiology, 44(6), 479–486.
Nishikawa, H., Arimoto, A., Wakasa, T., Kita, R., Kimura, T., & Osaki, Y. (2013). Comparison of clinical characteristics and survival after surgery in patients with non-B and non-C hepatocellular carcinoma and hepatitis virus-related hepatocellular carcinoma. Journal of Cancer, 4(6), 502–513.
Tietze, L., & Kessler, S. M. (2020). The good, the bad, the question-H19 in hepatocellular carcinoma. Cancers. https://doi.org/10.3390/cancers12051261
Zhang, T. Q., Su, Q. Q., Huang, X. Y., Yao, J. G., Wang, C., Xia, Q., et al. (2018). Micro RNA-4651 serves as a potential biomarker for prognosis when selecting hepatocellular carcinoma patients for postoperative adjuvant transarterial chemoembolization therapy. Hepatology Communications, 2(10), 1259–1273.
D’Anzeo, M., Faloppi, L., Scartozzi, M., Giampieri, R., Bianconi, M., Del Prete, M., et al. (2014). The role of micro-RNAs in hepatocellular carcinoma: From molecular biology to treatment. Molecules (Basel, Switzerland)., 19(5), 6393–6406.
Liu, H., Ma, L., Wang, L., & Yang, Y. (2019). MicroRNA-937 is overexpressed and predicts poor prognosis in patients with colon cancer. Diagnostic pathology., 14(1), 136.
Li, D., Zhong, J., Zhang, G., Lin, L., & Liu, Z. (2019). Oncogenic role and prognostic value of microRNA-937-3p in patients with breast cancer. OncoTargets and Therapy, 12, 11045–11056.
Zhang, L., Zeng, D., Chen, Y., Li, N., Lv, Y., Li, Y., et al. (2016). miR-937 contributes to the lung cancer cell proliferation by targeting INPP4B. Life Sciences, 155, 110–115.
Fujiwara, T., & Yada, T. (2013). miRNA-target prediction based on transcriptional regulation. BMC Genomics, 14(Suppl 2), S3.
Zhou, D., Dong, L., Yang, L., Ma, Q., Liu, F., Li, Y., et al. (2020). Identification and analysis of circRNA-miRNA-mRNA regulatory network in hepatocellular carcinoma. IET Systems Biology, 14(6), 391–398.
Eun, J. W., Seo, C. W., Baek, G. O., Yoon, M. G., Ahn, H. R., Son, J. A., et al. (2020). Circulating exosomal microRNA-1307-5p as a predictor for metastasis in patients with hepatocellular carcinoma. Cancers. https://doi.org/10.3390/cancers12123819
Acknowledgements
We deeply thanks to the help from Jinyun Li and Yan Wang (Department of Pathology, The Second Affiliated Hospital of Kunming Medical University) for the tissue pathological confirmation.
Author information
Authors and Affiliations
Contributions
All authors initiated and designed the work, analyzed the data, and wrote and revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Approval
The study was approved by the Ethics Committee of Affiliated Hospital of the Second Affiliated Hospital of Kunming Medical University.
Consent to Participate
Written informed consent was provided from each participant.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, Z., Zhang, Y. Upregulation of MicroRNA-937 Predicts a Poor Prognosis and Promotes Hepatocellular Carcinoma Cell Proliferation, Migration, and Invasion. Mol Biotechnol 64, 33–41 (2022). https://doi.org/10.1007/s12033-021-00388-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-021-00388-7