Abstract
This study revealed the prognostic significance of long non-coding RNA (lncRNA) CCDC144NL-AS1 in NSCLC patients and discussed the effect and mechanism of proliferation, migration, and invasion of non-small cell lung cancer (NSCLC) cells. 128 pairs of NSCLC tissues and paracancerous tissues were collected, and qRT-PCR was used to detect the differential expression of lncRNA CCDC144NL-AS1 in all tissues and cells lines. Kaplan–Meier analysis and Cox proportional hazards model analysis were used to estimate the prognostic value of lncRNA CCDC144NL-AS1. CCK-8 and Transwell assays confirmed the effect of lncRNA CCDC144NL-AS1 on the proliferation, migration, and invasion of NSCLC. Bioinformatics was used to predict the microRNAs that lncRNA CCDC144NL-AS1 might bind to miR-490-3p. The regulation of lncRNA CCDC144NL-AS1 on miR-490-3p was verified by luciferase activity assay with wide type or mutation. The expression of lncRNA CCDC144NL-AS1 was enhanced in both NSCLC tissues and cell lines. Patients with overexpression of lncRNA CCDC144NL-AS1 have a poor prognosis, and lncRNA CCDC144NL-AS1 is an independent prognostic factor for NSCLC. Increased the relative expression level of lncRNA CCDC144NL-AS1 can promote the proliferation, migration, and invasion of NSCLC cells. LncRNA CCDC144NL-AS1 might target miR-490-3p. LncRNA CCDC144NL-AS1 can be used as an oncogene of NSCLC to predict patient prognosis and promote tumor proliferation, migration, and invasion by targeting miR-490-3p.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lung cancer is one of the malignancies with the highest morbidity and mortality in the world [1]. Lung cancer can be divided into small cell lung cancer and non-small cell lung cancer (NSCLC) according to the different clinicopathological types [2, 3]. NSCLC is the main type of lung cancer. Due to the lack of early clinical manifestations, tumor cells usually have lymphatic metastasis and distant metastasis, with poor prognosis and low five-year survival rate when patients show obvious clinical symptoms [4, 5]. Many genetic alterations represent feasible molecular therapeutic targets for the treatment of NSCLC, which acts as applicable predictor biomarkers using as personalized medicine [6]. These novel targeted therapies, including checkpoint immunotherapy, have shown promising results in the treatment of advanced-stage NSCLC [7, 8]. Therefore, it is very important to search for more effective prognostic markers for NSCLC to improve the survival rate of patients with NSCLC.
Long non-coding RNAs (lncRNAs) are over 200 bp in length, which are different from miRNAs around 20 bp in length. lncRNAs were once considered as dark matter in RNA because of the limited protein-coding ability. However, it was later found that lncRNAs have the role of regulating transcription and gene expression levels after transcription [9,10,11]. The competitive endogenous RNA mechanism is the recognized regulation mechanism of lncRNAs. This mechanism suggests that lncRNAs can competitively bind to miRNAs and exert inhibitory effects on target genes through negative regulation of miRNAs, thus, participating in the regulation of various signaling pathways and biological functions [12].
A large number of studies have confirmed that lncRNAs are involved in tumor cell proliferation, migration, invasion, angiogenesis, and other pathological processes and affect the prognosis of tumor patients [13,14,15]. Abnormal expression of lncRNAs is associated with the occurrence and development of a variety of tumors. Recent studies have found that lncRNA CCDC144NL-AS1 plays an important regulatory role in the development of gastric cancer, osteosarcoma, and colorectal cancer, but its role in NSCLC remains unclear [16,17,18]. In this research, we aimed to study the expression level of CCDC144NL-AS1 in NSCLC, reveal the biological effect and prognostic significance of CCDC144NL-AS1 on NSCLC, and further explore the mechanism of lncRNA CCDC144NL-AS1-miR-490-3p in the occurrence and development of NSCLC.
Materials and Methods
Patients and Tissues Collection
A total of 128 patients with NSCLC diagnosed in People's Hospital of Rizhao from February 2012 to December 2015 were selected, and the NSCLC tissues and corresponding adjacent normal tissues were surgically removed. Clinical characteristics of the patients were also collected. Inclusion criteria for patients: (1) NSCLC diagnosed by histopathological examination; (2) all patients had not received radiotherapy or chemotherapy before surgery; (3) according to the lung cancer TNM staging of its clinical pathological information classification, including I/II and III/IV. Tissue specimens were immediately frozen and preserved in liquid nitrogen. This study was approved by the Ethics Committee of People's Hospital of Rizhao, and informed consent was signed with all patients. To investigate the effect of CCDC144NL-AS1 on prognosis in patients with NSCLC, all enrolled patients were followed up for 5 years.
Cell Culture and Transient Transfection
Human normal lung cell lines BEAS-2B and NSCLC cell lines H1299, NCI-H650, A549, and HCC827 were purchased from the Institute of Cell Biology, Chinese Academy of Sciences, Shanghai (Shanghai, China). All cell lines were cultured in RPMI-1640 medium containing 10% fetal bovine serum (FBS; Gibco, NY, USA) at 37 °C and 5% CO2 saturated humidity. The medium was changed once every other day, and the cells in the logarithmic growth phase were selected for the experiment.
For cell transfection, si-CCDC144NL-AS1 was applied for interfered RNAs designed for CCDC144NL-AS1, and si-NC was siRNA negative control. si-CCDC144NL-AS1 or si-NC purchased Genepharm. Co. (Shanghai, China) was transfected into H1299 and A549 cells. The miR-490-3p mimics, inhibitor negative control (inhibitor NC), or miR-490-3p inhibitor was constructed by Ribobio (Guangzhou, China). Transfection was performed according to the instructions of the Lipofectamine 2000 kit (Invitrogen, Thermo Fisher Scientific, Inc., Waltham, MA, USA), and cells in each group were collected for subsequent experiments.
RNA Isolation and qRT-PCR for RNA Expression Level
Total RNA was extracted from cell lines and tissues using TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA), and the total RNA of lncRNA CCDC144NL-AS1 or miR-490-3p was reverse-transcribed into cDNA according to the instructions of TaqMan miRNA reverse transcription kit (Thermo Fisher Scientific) or the Superscript III transcriptase (Invitrogen, Grand Island, NY), respectively. Subsequently, SYBR PrimeScript RT-PCR kit (TaKaRa, Dalian, China) was used for qRT-PCR assay, and the experiment was completed on an ABI 7500 PCR System. GAPDH was used as the internal reference gene for lncRNA CCDC144NL-AS1, and U6 was used for miR-490-3p. The results were analyzed by 2−ΔΔCt method.
Nuclear and Cytoplasmic Separation Experiment
The nuclear and cytoplasmic separation experiment was performed following the protocols of a PARIS kit (Ambion, Austin, USA). Nuclear and cytoplasmic RNAs were separated from H1299 and A549 cells (1 × 107), and then the expression of lncRNA CCDC144NL-AS1 in nuclear and cytoplasm was detected using qRT-PCR assay using GAPDH (cytoplasmic control) and U6 (nuclear control) for standardization.
Cell Proliferation Assay
Cell-Counting Kit‑8 (CCK-8) assays were conducted to detect the ability of cell proliferation. H1299 and A549 cells in each group were spread into 96-well plates. 10 μL CCK-8 solution was added to each well for 24, 48, and 72 h, respectively. After incubation for 2 h, an absorbance value at 450 nm was detected by the SpectraMax Plus 384 Microplate Reader (Molecular Devices, LLC). Each group is provided with 3 duplicate holes.
Cell Migration and Invasion Assays
Cell migration assay: 200 μL serum-free medium cell suspension contains transfection cell (5 × 104 cells/well) that was inoculated in Transwell chamber, and under the condition of 37 °C and 5% CO2 cultivate 24 h, takes out the chamber, wipes gently with cotton swabs upper cells after removal of nutrient solution, PBS washing, adds 4% paraformaldehyde-fixed 30 min, 0.1% crystal violet staining for 10 min, PBS washing, microscope, and photographed counting. Cell invasion assay: RPMI-1640 medium was added at a ratio of 1:5 to dilute Matrigel and then spread in the upper chamber of the Transwell chamber. After drying at room temperature, the cell invasion assay was carried out according to the steps. Finally, the number of cells stained with crystal violet under the microscope was the number of invaded cells.
Luciferase Reporter Assay
The 3'UTR luciferase expression vector WT-CCDC144NL-AS1 and MUT-CCDC144NL-AS1 were constructed to target wild-type and mutated CCDC144NL-AS1 genes. H1299 cells in the logarithmic growth phase were seeded into 24-well plates (5 × 104 cells per well). Cells in WT-CCDC144NL-AS1 or MUT-CCDC144NL-AS13 groups were transfected with miR-490-3p mimic, miR-NC, or miR-490-3p inhibitor using Lipofectamine 2000 when 80% of the cells were grown and fused. Dual-luciferase reporter assay was performed as described in the Dual-luciferase reporter assay kit (Promega). The results were statistically analyzed by the ratio of firefly luciferase activity to sea kidney luciferase activity. The experiment was repeated 3 times.
Statistical Analyses
Statistical analysis was performed using SPSS 20.00 (SPSS Inc., Chicago, IL, USA) or GraphPad 5.0 (GraphPad Software, Inc., La Jolla, CA, USA). Measurement data were expressed as mean ± standard deviation, a 2-tailed student’s t test was performed for comparison between two groups, and one-way or two-way analysis of variance (ANOVA) was used for comparison between multiple groups. Spearman correlation analyses were used to test the correlation between CCDC144NL-AS1 and miR-490-3p expression. P < 0.05 indicated statistically significant difference.
Results
Expression of lncRNA CCDC144NL-AS1 in NSCLC Tissues and Cell Lines
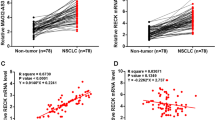
The expression of lncRNA CCDC144NL-AS1 in NSCLC tissues and cell lines was evaluated by qRT-PCR. Compared with human normal lung tissues, CCDC144NL-AS1 was all highly expressed in NSCLC tissues (P < 0. 001, Fig. 1a). Furthermore, the expression of lncRNA CCDC144NL-AS1 was compared in the tissues of patients with or without lymph node metastasis (LNM). The data revealed that the expression levels of lncRNA CCDC144NL-AS1 were higher in the lymph node metastasis-positive tissues than that in negative lymph node metastasis tissues (P < 0.001, Fig. 1b). Meanwhile, the expression level of lncRNA CCDC144NL-AS1 was significantly increased in four lung cancer cell lines (H1299, NCI-H650, A549, and HCC827), compared with human normal lung cell lines BEAS-2B. It is worth noting that the expression level of CCDC144NL-AS1 was the highest in H1299 cell line and was followed by A549 cell line (P < 0. 05, Fig. 1c). Besides, lncRNA CCDC144NL-AS1 was expressed in the nuclear and cytoplasm of H1299 and A549 cells, whereas the expression levels in the cytoplasm were higher than that in nuclear (P < 0.001, Fig. 1d), which suggesting that the lncRNA CCDC144NL-AS1 acted a major role in the cytoplasm of NSCLC.
Relative lncRNA CCDC144NL-AS1 expression in NSCLC tissues and cell lines. A The qRT-PCR analysis detected the expression level of lncRNA CCDC144NL-AS1 in 128 pairs of NSCLC tissues and the corresponding normal tissues with three independent repeats. B The expression of lncRNA CCDC144NL-AS1 was compared in lymph node metastasis-positive or negative tissues with three independent repeats. C qRT-PCR analysis of lncRNA CCDC144NL-AS1 in NSCLC cell lines and human normal lung cell lines. The expression was measured with three independent repeats. D Detection of lncRNA CCDC144NL-AS1 expression in H1229 and A549 cells by nuclear and cytoplasmic separation experiments with three repeats. ***P < 0.001, **P < 0.01, *P < 0.05
Correlation Between Expression of lncRNA CCDC144NL-AS1 and Clinical Data of Patients with NSCLC
First, 128 NSCLC patients were divided into high expression group (n = 66) and low expression group (n = 62) according to the average expression level of CCDC144NL-AS1. Then, the relationship between the lnc CCDC144NL-AS1 expression with clinical characteristics in NSCLC was explored. The results are supplied in Table 1. It can be the inference that lymph node metastasis (P = 0.003) and TNM stage (P = 0.001) were significant in correlation with the expression level of CCDC144NL-AS1, the difference was statistically significant. However, there was no significant difference in other clinical features, including age, gender, tumor size, smoking status, and cell differentiation (P > 0.05).
Upregulation of lncRNA CCDC144NL-AS1 is Associated with Poor Prognosis in NSCLC Patients
The Kaplan–Meier curve is plotted according to the relationship between the expression level of CCDC144NL-AS1 and the 5-year survival rate of NSCLC patients. As shown in Fig. 2, the high lncRNA CCDC144NL-AS1 expression group showed a lower 5-year survival rate compared to those in the low expression group (P < 0.05). Multivariate analyses were introduced to study independent prognostic factors associated with overall survival. In Table 2, the results revealed that lncRNA CCDC144NL-AS1 (HR = 2.577, P = 0.003), lymph node metastasis (HR = 1.920, P = 0.036), and TNM stage (HR = 1.960, P = 0.030) were the prognostic factors of NSCLC.
The Effect of lncRNA CCDC144NL-AS1 on Cell Proliferation, Migration and Invasion
Two NSCLC cell lines (H1299 and A549) with the lowest expression of lncRNA CCDC144NL-AS1 were selected. For cell proliferation, the results of RT-qPCR showed that the relative expression of si-CCDC144NL-AS1 decreased significantly compared with that of control and si-NC (P < 0.001, Fig. 3a, b). Then, the CCK-8 experiment showed that knockdown of lncRNA CCDC144NL-AS1, OD value decreased, which represented the down-regulation of lncRNA CCDC144NL-AS1expression weakened cell proliferation ability (P < 0.001, Fig. 3c, d). For migration and invasion, transwell assays were first conduct, the number of cell migration and invasion was significantly reduced when lncRNA CCDC144NL-AS1 expression was knocked down, compared with that of untreated cells and cells transfected si-NC (P < 0.001, Fig. 3e–h).
Effects of lncRNA CCDC144NL-AS1 expression levels on cell proliferation, migration, and invasion abilities in H1299 and A549 cells. A and B The expression level of lncRNA CCDC144NL-AS1 was verified by qRT-PCR after transient transfection with si-NC/si-lncRNA CCDC144NL-AS1. The data were analyzed using one-way ANOVA followed Turkey’s post hoc test with 5 times experiments. C and D The CCK-8 assay was performed to examine cell proliferation with 3 independent repeats. E and F The migration abilities of H1299 and A549 cells were assessed with Transwell assay with 5 repeats. G and H The invasive abilities of H1299 and A549 cells were assessed with Transwell assay with 5 repeats. ***P < 0.001, **P < 0.01
LncRNA CCDC144NL-AS1 Targets miR-490-3p
Bioinformatics analysis showed that lncRNA CCDC144NL-AS1 had an adsorption-binding site with miR-490-3p (Fig. 4a). Luciferase reporter gene assay results showed that the luciferase activity of WT-CCDC144NL-AS1 of the miR-490-3p mimic group was significantly decreased compared with control, mimic NC or inhibitor NC group after transfection of WT-CCDC144NL-AS1 in the H1299 cell line, and the luciferase activity in miR-490-3p inhibitor group was significantly increased (P < 0.001). However, no effect was found on luciferase activity in MUT-CCDC144NL-AS1-transfected cells (P > 0.05, Fig. 4b). The expression level of miR-490-3p in NSCLC tissues was also analyzed, the results showed that the expression level of miR-490-3p in NSCLC tissues was significantly higher than that in normal tissues (P < 0.001, Fig. 4c). In addition, Fig. 4d reveals that the expression of lncRNA CCDC144NL in NSCLC tissues was significantly negatively correlated with the expression of miR-490-3p (P < 0.0001). Then H1299 cells were transfected with si-NC, si- CCDC144NL. The results of Fig. 4e showed that si-CCDC144NL can promote the relative expression of miR-490-3p (P < 0.001).
miR-490-3p acted as the target of lncRNA CCDC144NL-AS1. A The binding sites between lncRNA CCDC144NL-AS1 and miR-490-3p. B The interaction between miR-490-3p and lncRNA CCDC144NL-AS1 was confirmed in H1299 cells co-transfected with WT-LINC00460 or MUT-LINC00460 by luciferase reporter assay with 3 independent repeats. C The qRT-PCR analysis detected the expression level of miR-490-3p in 128 pairs of NSCLC tissues and the corresponding normal tissues. D correlation analysis between lncRNA CCDC144NL-AS1 and miR-490-3p in 128 tumor tissues. r = − 0.7170, P < 0.0001. E qRT-PCR was used to determine the expression of miR-490-3p with 5 repeats, and the groups were divided into control, NC, si-NC, and si- CCDC144NL-AS1. ***P < 0.001
Discussion
NSCLC has a high incidence in China, which seriously threatens people's life and health [19,20,21]. It has been found that the progress of NSCLC involves a series of changes at the molecular level, so the study at the molecular level has become a new focus of tumor research [22, 23]. Targeted therapies and immunotherapy are developing rapidly, whereas the overall survival rate of NSCLC patients remains dissatisfactory [24]. LncRNA is an important part of tumor biology and can regulate the progression of tumors through a variety of mechanisms, and are also closely related to tumor metastasis, invasion, and patient prognosis [25, 26]. More and more new lncRNAs have been found to play an important role in NSCLC. For example, a previous study demonstrated that lnc-ROR acts as an oncogene in NSCLC and is associated with poor prognosis in patients with NSCLC [27]. Recent studies have shown that the expression of lnc-PINT was decreased in NSCLC and also proved that lnc-PINT plays a vital biological role in NSCLC by sponging miR-543 and inducing PTEN [28].
In this study, lncRNA CCDC144NL-AS1 has been shown to be a possible prognostic marker for NSCLC and has important significance in the development of NSCLC. First, the expression of lncRNA CCDC144NL-AS1 showed an upregulation tendency in both NSCLC tissues and cell lines, and the expression levels were higher in lymph node metastasis-positive tissues. Besides, lncRNA CCDC144NL-AS1 was closely associated with poor prognosis in patients with NSCLC. Based on the above results, we speculate that lncRNA CCDC144NL-AS1 may be involved in the development process of NSCLC and maybe an oncogene of NSCLC. Then, cellular function assays were carried out. Overexpression of lncRNA CCDC144NL-AS1 demonstrated to enhance the ability of proliferation, migration, and invasion in NSCLC cells. These conclusions were consistent with the results of lncRNA CCDC144NL-AS1 in gastric cancer. In a study on gastric cancer, it was found that overexpression lncRNA CCDC144NL-AS1 was an independent predictor of poor prognosis, and lncRNA CCDC144NL-AS1 could promote the tumor cellular functions, illustrating the carcinogenic role of lncRNA CCDC144NL-AS1/miR-143-3p/MAP3K7 axis in gastric cancer [16]. In addition, lncRNA CCDC144NL-AS1 not only shows great research value in tumors but also plays an important impact in diseases. For example, in endometriosis, lncRNA CCDC144NL-AS1 may be involved in the pathogenesis of endometriosis and reduces the migration and invasion of endometrial stromal cells, providing a new idea for the treatment of endometriosis [29]. Consequently, lncRNA CCDC144NL-AS1 is a potential biomarker that should be attended to both in tumors and diseases.
To further study the mechanism of lncRNA CCDC144NL-AS1 in NSCLC, the bioinformatics method and luciferase activity assays were applied. The results displayed that miR-490-3p might be the target gene of lncRNA CCDC144NL-AS1, and the luciferase activity of WT- CCDC144NL-AS1 was significantly reduced when miR-490-3p mimic was introduced. In view of these, we proposed that lncRNA CCDC144NL-AS1 combined with miR-490-3p was involved in the regulation of tumor biological function in NSCLC.
miR-490-3p, which is located on chromosome 7q33, has previously been suggested to act as a suppressor or oncogene for a variety of tumors [30, 31]. Recent studies have demonstrated that miR-490-3p was down-regulated in lung cancer and suppresses tumor progression [16, 32, 33]. In the present study, we also confirmed that miR-490-3p expression was down-regulated in NSCLC tissues and markedly negative correlation between lncRNA CCDC144NL-AS1 and miR-490-3p. Furthermore, the relative expression of miR-490-3p was correspondently increased when the expression of lncRNA CCDC144NL-AS1 was decreased in the H1299 cell line. These results indicate that miR-490-3p can be used as a target of lncRNA CCDC144NL-AS1, and the two interact to affect tumor progression. The reciprocal regulation of lncRNA CCDC144NL-AS1 and miR-490-3p has also been studied in osteosarcoma. Juliang He et al. demonstrated that CCDC144NL-AS1 exerts a cancer-promoting effect in osteosarcoma, and mainly by functioning as a sponge for miR-490-3p and increasing HMGA2 expression [17].
Although we try our best to explore the role of lncRNA CCDC144NL-AS1 in the occurrence and development of NSCLC, there are still some unsatisfactory aspects. The sample size is limited due to we can only collect volunteer patients in our hospital and have not been able to verify our results in animals. In future research, we will collect samples from a variety of ways, expand the sample size, carry out in vivo experiments, and explore the mechanism of abnormally expressed lincRNA in NSCLC.
Taken together, this study proposed that lncRNA CCDC144NL-AS1 was up-regulated in NSCLC tissues and cell lines, and overexpression of lncRNA CCDC144NL-AS1 was closely associated with poor prognosis in NSCLC patients. Down-regulation of lncRNA CCDC144NL-AS1 expression can inhibit the proliferation, migration, and invasion of lung cancer cells, and the mechanism may be related to the targeting of miR-490-3p. These results suggested that lncRNA CCDC144NL-AS1 can be used as a prognostic marker of lung cancer, which provides a basis for future molecular research on the treatment of NSCLC.
References
Romaszko, A. M., & Doboszyńska, A. (2018). Multiple primary lung cancer: A literature review. Advances in Clinical and Experimental Medicine, 27, 725–730.
Herbst, R. S., Morgensztern, D., & Boshoff, C. (2018). The biology and management of non-small cell lung cancer. Nature, 553, 446–454.
Jonna, S., & Subramaniam, D. S. (2019). Molecular diagnostics and targeted therapies in non-small cell lung cancer (NSCLC): An update. Discovery Medicine, 27, 167–170.
Broderick, S. R. (2020). Adjuvant and neoadjuvant immunotherapy in non-small cell lung cancer. Thoracic Surgery Clinics, 30, 215–220.
Valentino, F., Borra, G., Allione, P., & Rossi, L. (2018). Emerging targets in advanced non-small-cell lung cancer. Future Oncology, 14, 61–72.
Camidge, D. R., Doebele, R. C., & Kerr, K. M. (2019). Comparing and contrasting predictive biomarkers for immunotherapy and targeted therapy of NSCLC. Nature Reviews. Clinical Oncology, 16, 341–355.
Proto, C., Ferrara, R., Signorelli, D., Lo Russo, G., Galli, G., Imbimbo, M., Prelaj, A., Zilembo, N., Ganzinelli, M., Pallavicini, L. M., De Simone, I., Colombo, M. P., Sica, A., Torri, V., & Garassino, M. C. (2019). Choosing wisely first line immunotherapy in non-small cell lung cancer (NSCLC): What to add and what to leave out. Cancer Treatment Reviews, 75, 39–51.
Ruiz-Cordero, R., & Devine, W. P. (2020). Targeted therapy and checkpoint immunotherapy in lung cancer. Surgical Pathology Clinics, 13, 17–33.
Jarroux, J., Morillon, A., & Pinskaya, M. (2017). History, discovery, and classification of lncRNAs. Advances in Experimental Medicine and Biology, 1008, 1–46.
Isin, M., & Dalay, N. (2015). LncRNAs and neoplasia. Clinica Chimica Acta, 444, 280–288.
Wu, T., & Du, Y. (2017). LncRNAs: From basic research to medical application. International Journal of Biological Sciences, 13, 295–307.
Gupta, C., Su, J., Zhan, M., Stass, S. A., & Jiang, F. (2019). Sputum long non-coding RNA biomarkers for diagnosis of lung cancer. Cancer Biomarkers, 26, 219–227.
Gao, S., Zhao, Z. Y., Wu, R., Zhang, Y., & Zhang, Z. Y. (2018). Prognostic value of long noncoding RNAs in gastric cancer: A meta-analysis. Oncotargets and Therapy, 11, 4877–4891.
Quan, J., Pan, X., Zhao, L., Li, Z., Dai, K., Yan, F., Liu, S., Ma, H., & Lai, Y. (2018). LncRNA as a diagnostic and prognostic biomarker in bladder cancer: A systematic review and meta-analysis. Onco Targets and Therapy, 11, 6415–6424.
He, Y., Meng, X. M., Huang, C., Wu, B. M., Zhang, L., Lv, X. W., & Li, J. (2014). Long noncoding RNAs: Novel insights into hepatocelluar carcinoma. Cancer Letters, 344, 20–27.
Fan, H., Ge, Y., Ma, X., Li, Z., Shi, L., Lin, L., Xiao, J., Chen, W., Ni, P., Yang, L., & Xu, Z. (2020). Long non-coding RNA CCDC144NL-AS1 sponges miR-143-3p and regulates MAP3K7 by acting as a competing endogenous RNA in gastric cancer. Cell Death & Disease, 11, 521.
He, J., Guan, J., Liao, S., Wu, Z., Liu, B., Mo, H., & Yuan, Z. (2021). Long noncoding RNA CCDC144NL-AS1 promotes the oncogenicity of osteosarcoma by acting as a molecular sponge for microRNA-490-3p and thereby increasing HMGA2 expression. Onco Targets and Therapy, 14, 1–13.
Liu, H., Gu, X., Wang, G., Huang, Y., Ju, S., Huang, J., Ju, S., Huang, J., & Wang, X. (2019). Copy number variations primed lncRNAs deregulation contribute to poor prognosis in colorectal cancer. Aging (Albany NY), 11, 6089–6108.
Li, B., Zhang, F., & Li, H. (2020). miR-1225-5p inhibits non-small cell lung cancer cell proliferation, migration and invasion, and may be a prognostic biomarker. Experimental and Therapeutic Medicine, 20, 172.
Naylor, E. C., Desani, J. K., & Chung, P. K. (2016). Targeted therapy and immunotherapy for lung cancer. Surgical Oncology Clinics of North America, 25, 601–609.
Pennell, N. A., Arcila, M. E., Gandara, D. R., & West, H. (2019). Biomarker testing for patients with advanced non-small cell lung cancer: Real-world issues and tough choices. American Society of Clinical Oncology Educational Book, 39, 531–542.
Chen, S., Shi, F., Zhang, W., Zhou, Y., & Huang, J. (2019). miR-744-5p inhibits non-small cell lung cancer proliferation and invasion by directly targeting PAX2. Technology in Cancer Research & Treatment, 18, 1533033819876913.
Yang, Z. Q., Wu, C. A., & Cheng, Y. X. (2018). Prognostic value of microRNA-133a expression and its clinicopathologic significance in non-small cell lung cancer: A comprehensive study based on meta-analysis and the TCGA database. Oncology Research and Treatment, 41, 762–768.
Osmani, L., Askin, F., Gabrielson, E., & Li, Q. K. (2018). Current WHO guidelines and the critical role of immunohistochemical markers in the subclassification of non-small cell lung carcinoma (NSCLC): Moving from targeted therapy to immunotherapy. Seminars in Cancer Biology, 52, 103–109.
Li, Y., Jiang, T., Zhou, W., Li, J., Li, X., Wang, Q., Jin, X., Yin, J., Chen, L., Zhang, Y., Xu, J., & Li, X. (2020). Pan-cancer characterization of immune-related lncRNAs identifies potential oncogenic biomarkers. Nature Communications, 11, 1000.
Sun, W., Zu, Y., Fu, X., & Deng, Y. (2017). Knockdown of lncRNA-XIST enhances the chemosensitivity of NSCLC cells via suppression of autophagy. Oncology Reports, 38, 3347–3354.
Qu, C. H., Sun, Q. Y., Zhang, F. M., & Jia, Y. M. (2017). Long non-coding RNA ROR is a novel prognosis factor associated with non-small-cell lung cancer progression. European Review for Medical and Pharmacological Sciences, 21, 4087–4091.
Wang, S., Jiang, W., Zhang, X., Lu, Z., Geng, Q., Wang, W., Li, N., & Cai, X. (2020). LINC-PINT alleviates lung cancer progression via sponging miR-543 and inducing PTEN. Cancer Medicine, 9, 1999–2009.
Zhang, C., Wu, W., Zhu, H., Yu, X., Zhang, Y., Ye, X., Cheng, H., Ma, R., Cui, H., Luo, J., Guan, J., & Chang, X. (2019). Knockdown of long noncoding RNA CCDC144NL-AS1 attenuates migration and invasion phenotypes in endometrial stromal cells from endometriosis†. Biology of Reproduction, 100, 939–949.
Liu, X., He, B., Xu, T., Pan, Y., Hu, X., Chen, X., & Wang, S. (2018). MiR-490-3p functions as a tumor suppressor by inhibiting oncogene VDAC1 expression in colorectal cancer. Journal of Cancer, 9, 1218–1230.
Tian, J., Xu, Y. Y., Li, L., & Hao, Q. (2017). MiR-490-3p sensitizes ovarian cancer cells to cisplatin by directly targeting ABCC2. American Journal of Translational Research, 9, 1127–1138.
Lu, H., Yang, D., Zhang, L., Lu, S., Ye, J., Li, M., & Hu, W. (2019). Linc-pint inhibits early stage pancreatic ductal adenocarcinoma growth through TGF-β pathway activation. Oncology Letters, 17, 4633–4639.
Xu, Y., Wang, H., Li, F., Heindl, L. M., He, X., Yu, J., Yang, J., Ge, S., Ruan, J., Jia, R., & Fan, X. (2019). Long non-coding RNA LINC-PINT suppresses cell proliferation and migration of melanoma via recruiting EZH2. Frontories in Cell and Developmental Biology, 7, 350.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author reports no conflict of interest in this work.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhang, L., Chi, B., Chai, J. et al. LncRNA CCDC144NL-AS1 Serves as a Prognosis Biomarker for Non-small Cell Lung Cancer and Promotes Cellular Function by Targeting miR-490-3p. Mol Biotechnol 63, 933–940 (2021). https://doi.org/10.1007/s12033-021-00351-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-021-00351-6