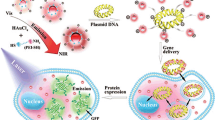
Abstract
The efficient carrier design for transferring therapeutic genes into target cells as well as tracking the delivered agents has attracted lots of attention in the field of DNA-based therapeutics. Here, we demonstrate this concept by a fast and facilitated method using BSA gold nanocluster (BSA AuNcs) conjugated with chimeric peptide with ability of DNA binding/packaging, endosome disruption and cell nuclear localization. An extensive characterization of photoluminescence properties, electrophoresis mobility and size distribution of the nanocarrier demonstrating the stable complexes composed of plasmid DNA, chimeric peptide and BSA AuNcs were successfully formed through electrostatic interactions. In the hybrid complexes, chimeric peptide could effectively decrease the cytotoxicity of AuNcs as well as enhance internalization of plasmid harboring firefly luciferase gene into HEK 293 T. The designed nanocarrier could be a promising vector in gene delivery systems for improved theranostics applications.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
DNA-based pharmaceuticals have attracted a lot of attention in the field of medicine and biotechnology due to the potentials for treating diseases such as inherited disorders, various cancers and viral infections [1]. To achieve successful DNA-based drugs, development of proper gene delivery and tracking vectors is essential. Several gene transfer barriers need to be overcome for development of such carriers [2]. Various viral and none-viral carriers have been designed to overcome the barriers [3]. Viral carriers, although highly efficient, but their broad use is affected by the limited size of the genetic material and safety concerns [4]. Hence, non-viral carriers including cationic lipids, polymers and peptides have been developed for specific delivery of DNA-based drugs [5]. One of the promising non-viral carriers is multifunctional peptide capable of DNA binding/packaging, endosomal disrupting and crossing the nuclear envelope through its specialized motifs such as histone H1 (H), nuclear localization signal of simian virus 40 (N) and histidine-rich motif H5WYG (H) [6]. Such biomimetic peptides have been previously designed in our lab and are abbreviated as HNH [7]. It has been indicated that peptide-based carriers can be rationally engineered for efficient intracellular gene delivery [8, 9]. Moreover, high transfection efficiency was achieved through genetically functionalization with anti-HER2-ScFV for targeted gene delivery into HER2-positive breast cancer cells and labeling with iRGD sequence [10]. Moreover, a novel peptideticles including two main peptides, MiRGD and HNH were designed to identify the tumor vasculature zip code, penetrate in tumor tissue and target the cancer microenvironment for gene delivery [9].
Besides transfection efficiency of the carrier, the ability of tracking the delivered cargo is another challenge of gene therapy. Optical imaging based on fluorescence probes due to its excellent accuracy, high sensitivity and the non-destructive nature provides a comprehensive tool for investigating many aspects of the gene delivery process such as the location, degradation and release of the genetic material in target cell [11, 12]. Metal nanoclusters (NCs) as new classes of luminescent materials consist of a small number of atoms surrounded by stabilizing ligands such as DNA, peptide and protein. The nanomaterials have exhibited excellent photostability, high biocompatibility and good aqueous solubility which resulted in remarkable biomedical applications for long-term imaging and accurate tracking of biomolecular. In recent decades, many fluorescent nanoclusters, especially nanoclusters of Au and Ag, have been widely used in biotracking [13,14,15].
In this study, we used the fluorescent BSA AuNcs and HNH chimeric peptide for preparation of peptide–cluster hybrid nanocomplexes as a novel gene carrier. In the hybrid complexes, chimeric peptide could effectively decrease the cytotoxicity of AuNcs as well as promote condensation, endosome escape and nuclear internalization of the plasmid harboring firefly luciferase gene into HEK 293 T. In addition, the AuNcs with two different green and red emissions fluorescence and excellent photostability provided a great potential to track the transfection behavior.
Materials and Methods
Overexpression of the H1-NLS-H5WYG Fusion Peptide in E. coli
The fusion peptide was overexpressed in the E. coli strain C41 (DE3) pLysS using the pET-21a vector. A transformed single-colony was inoculated into 10 ml LB with 100 μg/ml kanamycin and grown overnight at 37 °C in a rotary shaker (200 RPM). The overnight culture (1 ml) was added with 300 ml of 2-XY medium and incubated at 37 °C until OD600 reached 1.0. Then isopropyl-β-d-thiogalactopyranoside was added to a final concentration of 0.4 mM to induce peptide expression. Cells were harvested at 4 h post-induction by centrifugation at 5000 RPM for 10 min. The cell pellet was resuspended by pipetting on ice in 3 ml lysis buffer (500 mM NaCl, 20 mM Tris-Base, 8 M urea, and 5 mM imidazole; pH 11). Cell lysis was continued by sonication on ice bath for 10 min in 20 s bursts. Insoluble materials were removed by centrifugation at 12,000 RPM for 30 min and the supernatant was filtered through a 0.45 μm syringe filter.
Purification/Refolding of H1-NLS-H5WYG Fusion Peptide
Immobilized Metal Affinity Chromatography (IMAC) purification was used to purify the His-tagged fusion peptide. The supernatant was loaded on to 2 ml Ni-NTA-Agarose column equilibrated with lysis buffer for 1 h. Then the column was washed by five washing buffers (500 mM NaCl, 20 mM Tris-Base; pH 8) with increasing imidazole (5 mM, 10 mM, 20 mM, 30 mM and 40 mM imidazole) and decreasing urea concentration (6.0 M, 4.0 M, 2.0 M and 0 M urea). Increasing imidazole decreases non-specific protein binding and gradual removal of urea refolds chimeric peptide. The bound peptide was eluted by 3 ml elution buffer containing 20 mM Tris-Base, 500 mM NaCl, and 300 mM imidazole. The purity of the peptide was analyzed by 15% sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SDS-PAGE) and dialyzed in phosphate-buffered saline (PBS) buffer for 48 h using a dialysis tubing (Cut off 3 kDa).
Synthesis of BSA AuNcs and Characterization
2 ml of HAuCl4 (20 mM) was added to 2 ml of BSA solution (50 mg/ml) under vigorous stirring at 800 RPM. After 10 min, 500 μl of 1 M sodium hydroxide (NaOH) was added and heated at 37 °C for 4 h.
Characterization of BSA AuNcs is carried out using different methods, which include Ultraviolet–visible spectroscopy (Biowave II), Fluorescence emission spectroscopy (Cytation™ 3 Cell Imaging Multi-Mode Reader), Fourier transform infrared spectroscopy (Thermo Scientific Nicolet IR100), Transmission electron microscope (Philips CM30), and Dynamic light scattering (Zetasizer Nano, Malvern).
Conjugation of HNH, pDNA, and BSA AuNcs
First, 20 ng/μl of pDNA harboring firefly luciferase gene was added to chimeric peptide at different ratios (1.5 to 31; Table 1). Then, 10 μg/μl of BSA AuNcs solution was mixed to the prepared pDNA/HNH complexes (Table 1). BSA AuNcs (10 μg/μl) was also added to different concentrations of purified peptide (50–1000 ng/μl).
The characterization of the complexes is carried out using different methods including Electrophoretic mobility shift assay (1% Agarose Gel), Fluorescence emission spectroscopy (Cytation™ 3 Cell Imaging Multi-Mode Reader), Transmission electron microscope (Philips CM30), and Dynamic light scattering (Zetasizer Nano, Malvern).
Cytotoxicity Assay
MTT assay was used to detect the cytotoxicity of BSA AuNcs, HNH peptide and the prepared complexes. 1 × 104 HEK 293 T cells were seeded per well of 96-well plates in DMEM/F12 medium containing l-glutamine (4 mM), penicillin/streptomycin (50 mg/ml), and 10% FBS. Different concentrations of chimeric peptide (100–1000 ng/μl), a series of diluted BSA AuNcs (6, 12, 25, 50 μg/μl), and the prepared complexes (Table 1) were added to the cells. After 24 h incubation, the medium was replaced once again with a DMEM medium plus % 10 v/v MTT reagent (5 mg/ml). After 4 h incubation, the medium/MTT was removed and 100 µl of DMSO was added to dissolve the formazan crystals and the plates were read using cytation™ 3 Cell Imaging Multi-Mode Reade at 570 nm.
Cellular Bioimaging
Fluorescence microscopy technique was used for tracking the fluorescence of nanocomplexes into the living cells. 4 × 104 HEK 293 T cells were cultured in 10 cm petri dish for 24 h. After 2 h serum starvation, the prepared pDNA/HNH/BSA AuNcs complexes were added to the cells and incubated for 6 h. Then cells were washed three times with PBS and finally, fluorescence imaging was done at different wavelengths ((ex:469 nm; em:525 nm) and (ex:585 nm; em:647 nm)) by olympus ix81 fluorescence microscope.
Transfection Assay
HEK 293 T cells were plated in 10 cm petri dish and grown at 37 °C for 48 h. The cells were 70% confluent at the time of transfection. Before transfection, the cells were serum starved for 2 h in serum free medium then the prepared pDNA/HNH/BSA AuNcs complexes were added to the cells. After 6 h, the media of the cells were removed and the DMEM medium supplemented with 10% FBS was added and incubated for 24 h. Then the transfected cells were lysed with 50 μl CCLR (Cell Culture Lysis Reagent) buffer. The luciferase activity was recorded (RLU/s) using luciferase substrate (2 mM Luciferin, 4 mM ATP and 100 4 mM MgSO4) and luminometer (Berthold detection systems, GmbH).
Results and Discussion
BSA AuNcs Characterization
The UV–Vis spectra of BSA AuNcs (Fig. 1a) revealed an absorption shoulder in the wavelength range of 300–400 nm resulted from the multiple absorptions of the AuNcs. Similar to previous reports, we observed two distinct emission peaks at 450 and 650 nm, arising from BSA scaffold and AuNcs, respectively (Fig. 1b) [16]. The FTIR spectrum of BSA and BSA AuNcs is illustrated in Fig. 1c. However, the characteristic peaks of O–H/N–H stretching (band 3320 cm−1) and bending vibrations of amide bands (1560, 1338, 1230 cm−1) observed in both spectrum, but significant change in intensity of the peaks observed after cluster formation, indicating a different intra- or intermolecular bonding in BSA AuNcs than BSA [17]. DLS and TEM micrograph demonstrated average size of AuNcs around 10 nm (Fig. 1d).
a UV–Vis absorption spectra of the AuNcs prepared through BSA-mediated synthesis (green line) and BSA and HAuCl4 (red and blue lines). b Emission spectra of the BSA AuNcs. Inset shows: Photographs of the AuNcs with excitation at 365 nm. c FTIR spectrum of both BSA and the BSA AuNcs and d size distribution of BSA AuNcs determined by DLS and Inset shows: TEM image of BSA AuNcs
Peptide Expression and Preparation of Nanocomplexes
The purified peptide appeared as single bands in 15% polyacrylamide gel (Fig. 2a). Peptide motifs containing positively charged amino acids are capable of binding to the negatively charged pDNA and BSA AuNcs. The electrophoretic mobility shift assay demonstrated that the dual and ternary stable complexes including pDNA/HNH, BSA AuNcs/HNH and pDNA/HNH/BSA AuNcs were successfully formed through electrostatic interactions (Fig. 2b–e). Prepared ratios for both complexes are indicated in Table 1. The ratios are calculated based on positively chargeable amino acids of chimeric peptide to negatively charged amino acids of BSA protein and phosphate groups of plasmid as provided in S1.
a Coomassie blue stained purified peptide in 15% SDS-PAGE (MW 10 kDa). b Agarose-based gel retardation of pDNA/HNH complexes followed by ethidium bromide staining. c Gel retardation of the BSA AuNcs/HNH complexes under 360 nm UV. d Gel retardation of pDNA/HNH/BSA AuNcs complexes under 360 nm UV (without emission filter) and e HNH/pDNA/BSA AuNcs complexes under 360 nm UV with 520/740 emission filters
Figure 2b, c demonstrates that the migration of pDNA and BSA AuNcs decreased when conjugated to the peptide and finally remained in the wells due to charge neutralization of the complexes. As reported earlier, chimeric peptide began to neutralize the negative charges of plasmids at ratio of 0.5 and the complete shift of pDNA band was seen when peptide concentration was increased up to ratio of 1 [7]. Unlike pDNA/HNH complex, the shift of BSA AuNcs is observed at higher concentrations of peptide (up to 1.0 μg).
Gel retardation assay of ternary complex (pDNA/HNH/BSA AuNcs) followed by hoechst staining of pDNA and using 520/740 emission filters for monitoring of the migration of hoechst and BSA AuNcs. Photograph of gel under 360 nm UV (without filter) showed blue emission of hoechst in the wells due to the complete shift of pDNA with the peptide, same result obtained with monitoring of hoechst using 520 nm emission filter (Fig. 2d). Using 740 nm emission filters, decrease in the migration of BSA AuNcs in the gel until a clear shift in migration was observed (Fig. 2e). Increase in the ratios of ternary complex (ratio = 0.5 to 15) is associated with increasing peptide concentration as provided in Table 1. Based on the results, although shift of pDNA occurred at low ratios but only at higher peptide concentrations (up to ratios 7.7), the complete shift of the BSA AuNcs was observed. The average ζ-potential of the HNH peptide, free BSA AuNcs and the complexes was also assessed (Fig. S3). The change in the ζ-potential of the complexes reveals that electrostatic interactions have been established.
Size distribution and morphology of both dual and ternary complexes were determined by dynamic light scattering analysis and transmission electron microscope, respectively (Fig. 3a–f). The DLS analysis of the pDNA/HNH complexes showed significant decrease in size of nanoparticles with increasing ratio, point out histone H1 as a condensing motif worked appropriately (Fig. 3a). As shown in Fig. 3a, the complexes with ratios between 1 and 30 showed a size range between 130–40 nm. TEM images indicated that HNH in complex with pDNA can form condensed semi-spherical particles with a size around 80 nm (Fig. 3b) similar to previous report [7]. Unlike the pDNA/HNH complexes, binding of peptide to BSA AuNcs induces a strong particle aggregation. An increase of the peptide concentration led to a continuous shift of the complex size from 50 to 3000 nm (Fig. 3c, d; S4). Size distribution analysis of ternary complex revealed that at certain ratios, the pDNA/HNH/BSA AuNcs complex is formed. As shown in Fig. 3e, at ratios below 2, the particles size is near to the size of BSA AuNcs (10 nm) which indicates peptide concentration is not enough to bind both pDNA and AuNcs. With increasing ratio (up to = 2), the size of complexes rises from 100 to 800 nm which indicates the formation of the complexes. In TEM images, different size of the aggregated complexes can be seen (Fig. 3f; S4).
The investigation of photoluminescence (PL) properties of the BSA AuNcs/HNH complexes and pDNA/HNH/BSA AuNcs complexes in comparison to free BSA AuNcs revealed a significant enhancement in fluorescence intensity (Fig. 4a, c). The increased PL intensity might be related to the phenomenon of aggregation-induced emission (AIE) which is in agreement with DLS results on formation of larger particles. The demonstration of the AIE effect involving electrostatic interaction between AuNcs stabilized by negative charge ligands and different cationic polymers, reported in several studies [18,19,20,21]. In a study, monodisperse and stable self-assembled particles using AuNcs protected by GSH and cationic polymers (PAH, PEI) were reported. The cross-linking between AuNcs led to a fourfold fluorescence enhancement compared to free AuNcs [22]. Although the increased PL (Fig. 4b, d) was observed for both complexes, a perfect linear correlation between different peptide concentrations in the ternary complexes and fluorescence increase (as BSA AuNcs/HNH complexes) was not observed demonstrating that the binding capacity of peptide to the AuNcs is reduced due to its binding to pDNA. The same result also can be concluded with comparing the size distribution of complexes in Fig. 3c, e. Increase of the peptide concentration in BSA AuNcs/HNH complexes led to a shift of size to 3000 nm which is obviously more than the increasing size of the ternary complexes. The fluorescence stability of the ternary complex was also assessed in 10 days (Fig. 5). No significant change in the fluorescence intensity indicates the high photostability of the complexes.
The Cytotoxicity and Cellular Uptake of Nanocomplexes
The cytotoxicity of different concentrations of HNH peptide, synthesized BSA AuNcs and the complexes after 24 h was investigated by MTT assay. As shown in Fig. 6, BSA AuNcs at concentrations ranging from 6 to 50 µg/µl decreased the cell viability, while chimeric peptide up to a concentration of 1000 ng/µl had no significant effect on viability of the cells, similar to a previous report [7]. Similar to the Au nanoparticles, the intracellular ROS generation by AuNcs is one of the important mechanisms of toxicity. Here, HNH peptide could partly counteract the toxicity effect posed by AuNcs, which explains the low toxicity of AuNcs within the ternary complexes. The toxicity of BSA AuNcs is studied on different cell lines including cervix cancer cells, liver cancer cells, melanoma cells, and human umbilical vein endothelial cells (HUVECs). The results indicated that the decline of cell viabilities observed by the AuNCs depend on varying degrees in the dose and treatment time, and the type of cell lines. In the study, HUVECs have shown more toxicity due to their higher intake of AuNCs [23]. Based on the result, the uptake potential of HEK 293 T cells might play an important role in the toxicity of AuNCs.
To determine uptake of the complexes, HEK 293 T cells were exposed to BSA AuNcs and ternary complexes in different ratios, then analyzed by olympus fluorescence microscope with two different wavelengths [(ex: 469 nm, em: 525 nm) and (ex: 585 nm, em: 647 nm)] (Figs. 7, 8). The results demonstrated that although free BSA AuNcs can enter the cells, non-covalent association with HNH peptide increases the rate and efficiency of AuNcs uptake into the living cells. The complex with N/P ratio 2.0 revealed the most internalization ratio into the cells (Fig. 7a). At higher ratios, most complexes are observed on the surface of the cells due to their larger size as shown in Fig. 7b. Control cells including the cells treated to pDNA/HNH complexes and untreated cells did not show any fluorescence (Fig. 8a–c). Interestingly, all treated cells displayed significantly both green and red fluorescence arising from BSA scaffold and AuNcs, respectively. It should be mentioned rather than HNH role in DNA binding in our nanocomplexes, BSA could have a minimum potential in more plasmid loading. Several studies reported serum albumin as an important choice of material in various tracing/delivery systems due to its non-toxic nature, biodegradability and cost effectiveness. Use of BSA as gene carrier still remained a challenge due to its negative surface charge. One possible way for application of BSA in the gene delivery systems is through modification of the negatively charged groups of albumin with cationic groups, thereby transforming it into cationic BSA [24,25,26,27,28,29]. Moreover, non-covalent attachment of cationic BSA Au–Ag Ncs with pDNA through its surface charge modification with ethylenediamine for the delivery of therapeutic suicide gene in HeLa cancer cells has been shown [30]. Therefore, it may be concluded presence of a cationic peptide (HNH) in the current nanocomplex could improve gene loading capacity through neutralization of BSA surface negative charge and enhancing of DNA binding capacity of BSA.
Fluorescence microscopy images of transfected cells with a N/P = 2 and b N/P = 3.8 shown in 4 different panel. In each panel: right top is excited by 469 nm and detected in 525 emission wavelength, left top is excited by 585 nm and detected in 647 emission wavelength, right bottom shows bright field and overlay is indicated in left bottom
Fluorescence microscopy images of transfected cells with a BSA AuNcs (cells are shown in 3 different panel; in each panel: right is excited by 469 nm and detected in 525 emission wavelength, middle is excited by 585 nm and detected in 647 emission wavelength, left shows bright field). b Fluorescence microscopy images of transfected cells with pDNA/HNH complexes and c cells control
Based on the result obtained from the size distribution analysis, cytotoxicity assay and bioimaging, the best ratio of the ternary complex for cell transfection was selected. The highest transfection efficiency was achieved for ternary complex with N/P = 2.0 (Fig. 9a), which showed 14-fold increase in transfection efficiency than higher ratios (Fig. 9b). The decreased efficiency of transfection at higher ratios might be due to the low uptake rate of the complexes as discussed in Fig. 7b. The pDNA/HNH peptide complex with ratio of 6.0 was used for verification of the transfection efficiency of ternary complexes. The decreased efficiency of transfection at ratio 2.0 compared to pDNA/HNH peptide complex with ratio 6.0 could be another evidence of toxicity of gold nanoclusters.
Simultaneous tracking and delivery of plasmid DNA by the nanocomplexes of graphene quantum dots (GQDs) and the recombinant peptides showed efficient tracking and enhanced internalization of plasmid into the cells with good biocompatibility [31]. Generally, our designed biomimetic peptides with ability of packaging, endosomal escape and cellular nuclear targeting allow us to prepare the promising nanocarriers for improved theranostics applications.
Conclusion
According to the results, the photostable peptide–cluster hybrid nanocomplex with sizes around 100–150 nm could be prepared in a very simple and fast procedure. The conjugation of AuNcs to chimeric peptide with ability of DNA packaging, endosomal disruption and cellular nuclear targeting could allow efficient tracking and enhance the transfection efficiency of luciferase gene in human embryonic kidney 293 cells.
References
Gonçalves, G. A. R., & Paiva, R. D. M. A. (2017). Gene therapy: Advances, challenges and perspectives. Einstein (Sao Paulo), 15(3), 369–375.
Wiethoff, C. M., & Middaugh, C. R. (2003). Barriers to nonviral gene delivery. Journal of pharmaceutical sciences, 92(2), 203–217.
Patil, S. D., Rhodes, D. G., & Burgess, D. J. (2005). DNA-based therapeutics and DNA delivery systems: A comprehensive review. The AAPS journal, 7(1), E61–E77.
Robbins, P. D., & Ghivizzani, S. C. (1998). Viral vectors for gene therapy. Pharmacology & Therapeutics, 80(1), 35–47.
Yin, H., Kanasty, R. L., Eltoukhy, A. A., Vegas, A. J., Dorkin, J. R., & Anderson, D. G. (2014). Non-viral vectors for gene-based therapy. Nature Reviews Genetics, 15(8), 541–555.
Sadeghian, F., Hosseinkhani, S., Alizadeh, A., & Hatefi, A. (2012). Design, engineering and preparation of a multi-domain fusion vector for gene delivery. International Journal of Pharmaceutics, 427(2), 393–399.
Alipour, M., Hosseinkhani, S., Sheikhnejad, R., & Cheraghi, R. (2017). Nano-biomimetic carriers are implicated in mechanistic evaluation of intracellular gene delivery. Scientific Reports, 7, 41507.
Majidi, A., Nikkhah, M., Sadeghian, F., & Hosseinkhani, S. (2016). Development of novel recombinant biomimetic chimeric MPG-based peptide as nanocarriers for gene delivery: Imitation of a real cargo. European Journal of Pharmaceutics and Biopharmaceutics, 107, 191–204.
Alipour, M., Majidi, A., Molaabasi, F., Sheikhnejad, R., & Hosseinkhani, S. (2018). In vivo tumor gene delivery using novel peptideticles: pH-responsive and ligand targeted core–shell nanoassembly. International Journal of Cancer, 143(8), 2017–2028.
Cheraghi, R., Nazari, M., Alipour, M., Majidi, A., & Hosseinkhani, S. (2016). Development of a targeted anti-HER2 scFv chimeric peptide for gene delivery into HER2-positive breast cancer cells. International Journal of Pharmaceutics, 515(1–2), 632–643.
Sahoo, H. (2012). Fluorescent labeling techniques in biomolecules: A flashback. RSC Advsances, 2(18), 7017–7029.
Jung, D., Min, K., Jung, J., Jang, W., & Kwon, Y. (2013). Chemical biology-based approaches on fluorescent labeling of proteins in live cells. Molecular BioSystems, 9(5), 862–872.
Lin, C. A. J., Lee, C. H., Hsieh, J. T., Wang, H. H., Li, J. K., … Shen, J. L. (2009). Synthesis of fluorescent metallic nanoclusters toward biomedical application: Recent progress and present challenges. Journal of Medical and Biological Engineering, 29(6), 276–283.
Sadeghan, A. A., Soltaninejad, H., Hosseinkhani, S., Hosseini, M., Ganjali, M. R., & Asadollahi, M. A. (2018). Fluorescence enhancement of silver nanocluster at intrastrand of a 12C-loop in presence of methylated region of sept 9 promoter. Analytica Chimica Acta, 1038, 157–165.
Soltaninejad, H., Sadeghan, A. A., Hosseinkhani, S., Asadollahi, M. A., Hosseini, M., & Ganjali, M. R. (2019). Application of intercalating molecules in detection of methylated DNA in the presence of silver ions. Methods and Applications in Fluorescence, 7(3), 035005.
Guan, G., Zhang, S. Y., Cai, Y., Liu, S., Bharathi, M. S., … Low, M. (2014). Convenient purification of gold clusters by co-precipitation for improved sensing of hydrogen peroxide, mercury ions and pesticides. Chemical Communications, 50(43), 5703–5705.
Govindaraju, S., Ankireddy, S. R., Viswanath, B., Kim, J., & Yun, K. (2017). Fluorescent gold nanoclusters for selective detection of dopamine in cerebrospinal fluid. Scientific Reports, 7, 40298.
Luo, Z., Yuan, X., Yu, Y., Zhang, Q., Leong, D. T., Lee, J. Y., et al. (2012). From aggregation-induced emission of Au (I)–thiolate complexes to ultrabright Au (0)@ Au (I)–thiolate core–shell nanoclusters. Journal of the American Chemical Society, 134(40), 16662–16670.
Le Guével, X., Perez Perrino, M., Fernández, T. D., Palomares, F., Torres, M. J., … Blanca, M. (2015). Multivalent glycosylation of fluorescent gold nanoclusters promotes increased human dendritic cell targeting via multiple endocytic pathways. ACS Applied Materials & Interfaces, 7(37), 20945–20956.
Dou, X., Yuan, X., Yu, Y., Luo, Z., Yao, Q., Leong, D. T., et al. (2014). Lighting up thiolated Au@ Ag nanoclusters via aggregation-induced emission. Nanoscale, 6(1), 157–161.
Wu, Z., Liu, J., Gao, Y., Liu, H., Li, T., … Zou, H. (2015). Assembly-induced enhancement of Cu nanoclusters luminescence with mechanochromic property. Journal of the American Chemical Society, 137(40), 12906–12913.
Yahia-Ammar, A., Sierra, D., Mérola, F., Hildebrandt, N., & Le Guével, X. (2016). Self-assembled gold nanoclusters for bright fluorescence imaging and enhanced drug delivery. ACS Nano, 10(2), 2591–2599.
Dong, L., Li, M., Zhang, S., Li, J., Shen, G., … Tu, Y. (2015). Cytotoxicity of BSA-stabilized gold nanoclusters. In Vitro and In Vivo Study. Small, 11(21), 2571–2581.
Chen, Q., & Liu, Z. (2016). Albumin carriers for cancer theranostics: A conventional platform with new promise. Advanced Materials, 28(47), 10557–10566.
Elsadek, B., & Kratz, F. (2012). Impact of albumin on drug delivery: New applications on the horizon. Journal of Controlled Release, 157(1), 4–28.
Larsen, M. T., Kuhlmann, M., Hvam, M. L., & Howard, K. A. (2016). Albumin-based drug delivery: Harnessing nature to cure disease. Molecular and Cellular Therapies, 4(1), 3.
Han, J., Wang, Q., Zhang, Z., Gong, T., & Sun, X. (2014). Cationic bovine serum albumin based self-assembled nanoparticles as siRNA delivery vector for treating lung metastatic cancer. Small (Weinheim an der Bergstrasse, Germany), 10(3), 524–535.
Fischer, D., Bieber, T., Brüsselbach, S., Elsässer, H. P., & Kissel, T. (2001). Cationized human serum albumin as a non-viral vector system for gene delivery? Characterization of complex formation with plasmid DNA and transfection efficiency. International Journal of Pharmaceutics, 225(1–2), 97–111.
Zhang, T., Song, X., Kang, D., Zhang, L., Zhang, C., … Jin, S. (2015). Modified bovine serum albumin as an effective charge-reversal platform for simultaneously improving the transfection efficiency and biocompatibility of polyplexes. Journal of Materials Chemistry B, 3(23), 4698–4706.
Dutta, D., Chattopadhyay, A., & Ghosh, S. S. (2016). Cationic BSA templated Au–Ag bimetallic nanoclusters as a theranostic gene delivery vector for HeLa cancer cells. ACS Biomaterials Science & Engineering, 2(11), 2090–2098.
Ghafary, S. M., Nikkhah, M., Hatamie, S., & Hosseinkhani, S. (2017). Simultaneous gene delivery and tracking through preparation of photo-luminescent nanoparticles based on graphene quantum dots and chimeric peptides. Scientific Reports, 7(1), 1–14.
Acknowledgement
The financial support of this work was provided by Research council of Tarbiat Modares University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing financial interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zohrabi, T., Hosseinkhani, S. Ternary Nanocomplexes of Metallic Nanoclusters and Recombinant Peptides for Fluorescence Imaging and Enhanced Gene Delivery. Mol Biotechnol 62, 495–507 (2020). https://doi.org/10.1007/s12033-020-00260-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-020-00260-0