Abstract
Biosensors based on microbial cells have been developed to monitor environmental pollutants. These biosensors serve as inexpensive and convenient alternatives to the conventional lab based instrumental analysis of environmental pollutants. Small monomeric naturally occurring fluorescent proteins (fp) can be exploited by converting them as small biosensing devices for biomedical and environmental applications. Moreover, they can withstand exposure to denaturants, high temperature, and a wide pH range variation. The current study employs newly identified novel fluorescent protein HriGFP from Hydnophora rigida to detect environmental contaminants like heavy metals and organo-phosphorous (pesticide) compounds such as methyl parathion. The HriGFP was initially tested or its expression in bacterial systems (Gram positive and Gram negative) and later on for its biosensing capability in E coli (BL21DE3) for detection of heavy metals and methyl parathion was evaluated. Our results indicated the discrete and stable expression of HriGFP and a profound fluorescent quenching were observed in the presence of heavy metals (Hg, Cu, As) and methyl parathion. Structural analysis revealed heavy metal ions binding to HriGFP via amino acid residues. In-silico-analysis further revealed strong interaction via hydrogen bonds between methyl parathion phosphate oxygen atoms and the amino group of Arg119 of HriGFP. This study implies that HriGFP can act as a biosensor for detecting harmful carcinogenic pesticide like methyl parathion in water resources in the vicinity of heavily pesticide impregnated agricultural lands and heavy metal contaminated water bodies around industrial areas.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Environmental pollution is increasing at an alarming level with the rapid progress in industrial development consequently affecting human health. Despite serious measures to combat environmental pollution, large amount of industrial wastes and pollutants are released into the environment. More than 70% of the pesticides used worldwide are either organo-phosphorous compounds or their derivatives [14]. Pesticides bearing organo-phosphorous compounds like methyl parathion (MP) have been widely used in the agriculture sector in order to increase food production. However, their persistent residual lethal amounts pose a serious threat to human health and food safety [8, 15]. Methods like Gas Chromatography Mass Spectroscopy (GC–MS), High-Performance Liquid Chromatography (HPLC) and Gas Chromatography, Inductively coupled plasma mass spectrometry (ICP-MS) despite being accurate are laborious, costly, and require specialized laboratories which makes them inaccessible to remote agricultural areas, especially in developing countries [3, 18]. Colorimetric or electrochemical detection of MP is based on the Enzymatic biosensors are not suitable for the field application as they are unstable [11]. Since these methods are also non-portable and hence an easy to use cost-effective portable biosensor system will be highly beneficial for on spot detection of methyl parathion and other environmental contaminants like heavy metals such as arsenic in the lands of predominantly agricultural countries and their water sources to safeguard the environment for larger public health interest. For instance, alarmingly high concentrations of arsenic have been reported from Pakistan recently (Podgorski et al. 2017) which certainly have great impact on human health due to potential arsenic poisoning. Therefore, there is a dire need to devise a robust approach for rapid detection of such environmental pollutants using cost-effective and point-of-care rapid detection approaches.
Different microbial biosensors and whole cell bio-reporters (WCB) have been developed for the quantitative estimation of such environmental pollutants [4, 9] including GFP based biosensors have been reported against heavy metals [5, 13]. The GFP-like coral proteins exhibit a broad spectral diversity ranging from blue, green, yellow, and red FPs [2]. The expression and stability of these wild type FPs can be further optimized by amino acids substitutions possessing favorable optical and biophysical properties. These modifications may ensure better rate of maturation, monomeric forms with reduced sensitivity to pH [1, 16] and the stability of such stable monomeric FPs have increased their demand to be used as biosensors.
We have shown in our previous studies that HriGFP can be successfully expressed [10], purified using E. coli expression system like many other GFP-like proteins e.g., GFP. Being a highly stable monomer FP [6], HriGFP (133 amino acids) is small enough to be used as a biosensor. Moreover, being small fluorescent protein HriGFP may also be superior over larger proteins because they would have a minimal impact on host strain metabolism allowing more resources to be committed to the production of the recombinant target protein. In the present study. the expression potential of HriGFP in Bacillus megaterium was evaluated initially [7], thereafter, the biosensing capability of these novel fluorescent proteins for detection of environmental pollutants like heavy metals (Hg, Cu, As), pesticides (methyl parathion) was studied
Material and Methods
Materials
All the materials used for this study are of analytical grade. Stock solutions of Copper, Mercury was prepared from their respective chloride salts (Sigma), whereas Arsenite solution was prepared form sodium arsenite (NaAsO2 Sigma). The glassware used was first cleaned by treatment with nitric acid (15%) for 24 h to remove trace elements, followed by sterilization after autoclaving at 121 °C for 20 min.
Expression of HriGFP in E. coli and Bacillus megaterium
The HriGFP gene was cloned into pET28a+ vector and transformed into E. coli BL21 (DE3) cells. The vector pET28a+ has a T7 lac promoter which was induced using Isopropyl β-d-1-thiogalactopyranoside. IPTG act as a molecular mimic of allolactose and initiate transcription of lac operon and trigger expression of the inserted genes under the control of this Lac operon (in this case HriGFP gene). The HriGFP pet28+ construct was directly transformed in E. coli (BL21DE3 cells), however, for transformation in Bacillus megaterium the gene was sub-cloned into a shuttle vector pHIS1522 (MoBiTec, Goettingen, Germany). The transformed cells were then streaked on agar plates with 30 µl induced culture with 0.3 mM IPTG 0.0.1% Kanamycin was added as a resistant marker in the medium. The plates were incubated at 37 °C overnight and portion of these plates was visualized under fluorescent microscope on a glass slide.
Structure Prediction of HriGFP
Structure prediction of the HriGFP was done by homology modeling. The three dimensional model was predicted using MODELLER (v9.21, local version). The predicted model was further evaluated by PROCHECK. Autodock Vina 1.1.2 was used for docking of HriGFP with MP.
In-Silico Prediction of Metal Interaction with HriGFP
The predicted HriGFP structure was submitted to COACH meta-server which showed specific aminoacid motifs interaction with divalent cations like Magnesium, Zinc, and Manganese.
Heavy Metals Biosensing Ability Test
The maximum contaminant level (MCL) for the presence of these heavy metals in drinking water is 1.0 mg/L, 2 µg/L, and 0.01 mg/L. Therefore, biosensing of these bacteria was tested by exposing the overnight culture with OD600 0.8–1 to different concentrations (50–200 µg/L) of heavy metals such as Mercury and Arsenic. The biosensing ability for copper was tested at various ranges from 25 g/L to 200 µg/L as the EPA standard for copper is relatively high.These cultures were also tested against different concentrations of monovalent cations like Sodium and potassium (100–300 µg/L). Deionized water and bacterial cell unexposed to heavy metals were used as a control. The decrease in florescence was quantified by using a fluorometer (Modulus Luminometer/Fluorometer (Turner BioSystems, Sunnyvale, CA) with appropriate filters).
Methyl Parathion Detection
The inoculum was prepared in 500 ml LB broth with Kanamycin (100 μg/mL). The culture was divided in two halves. Induction with 0.15 mM IPTG was performed with the one half of the culture when the O.D. 600 nm reached 0.6. The flasks were kept on incubator for 4–5 h at 37 °C with 100 rpm shaking. The cultures were spin down at 13000 rpm and dissolved in 10 ml of lysis buffer (500 mM NaCl, 0.1% TritonX, 10% glycerol, Tris 7.4 pH) and kept on ice. The half culture in lysis buffer was sonicated and only supernatant was collected where as other half was sonicated and cell lysate was kept, then treated with MP. Different concentrations of the methyl parathion working solutions were prepared from the MP stock solution (0.5%). 900 µl of the working MP solution was set as blank in the fluorimeter (BIORAD). 100 µl of induced and uninduced culture with HriGFP were added in the same cuvet with MP alternatively and the fluorescence intensity was determined.
Statistical Analysis
Statistical analysis was performed with Graph-pad Prism 7.0 b. All the biosensing experiments were done in three biological replicates. The statistical significance was determined using the Mann–Whitney test with P < 0.05 considered significant.
Results
Expression of HriGFP in E. coli and Bacillus megaterium
HriGFP was successfully transformed and expressed in in E. coli and Bacillus megaterium as shown in both direct visualization of streaks under UV as well as under fluorescent microscopy Fig. 1.
Prediction of HriGFP Structure
The HriGFP structure was modeled by using MODELLER (v9.21, local version) with the template 2otb chain A. The HriGFP predicted structure comprises of 7 beta sheets and the structure resembles to beta barrel like other GFP molecules (Fig. 2a). Two different fluorescence tripartite motif IYG and KYG have been shown in the structure (Fig. 2b). Ramachandran analysis was performed using PROCHECK indicating 83.3% residues in the most favored region, 12.3% in the maximum allowed region, and 3.5% in the generously allowed region. Only 1 residue Arg91 lies in the outlier region. The structure assessment analysis showed that the predicted structure is of good quality.
a Predicted three-dimensional structure of HriGFP using Modeller (v9.21, local version) representing beta sheets in yellow and the coiled coils in green b Three-dimensional model of HriGFP showing the fluorescence motifs IYG at position 28–30 (orange) and KYG at position 61–63 (cyan). c Ramachandran plot of the 3D models of HriGFP. Red region represents the most favored region, yellow indicates the allowed region, light yellow indicates generously allowed region, and white indicates the disallowed region (Color figure online)
Interaction of HriGFP with Metal Ions
The HriGFP interaction with metal ions was further explored at structural level. The predicted HriGFP 3D structure was submitted to COACH meta-server for potential protein ligand interaction. Interestingly strong interaction was observed with divalent ions like Magnesium (Fig. 3). However, other metal ions like zinc and Manganese also interact with the HriGFP (Table 1). The interaction of the HriGFP to magnesium (Mg2+) is similar to the RNA polymerase VP1 (2yi9A). Two residues Arg12 and Asp25 are involved in this interaction. The oxygen atom of the Asp25 showed ionic interaction with magnesium ions, whereas the two nitrogen atoms of Arg12 also strongly interact with the divalent cation (Fig. 3).
Structure-based prediction of HriGFP interaction with the divalent cations by COACH a The divalent cation Magnesium represented in red interacts with Arg12 and Asp25 amino acid residues of HriGFP. b Zoomed in image of the interaction of the N atom of Arg12 and the oxygen atom of Asp25 with Mg2+. c The oxygen atom of the Asp25 amino acid residue interacts strongly via H-bond the Magnesium cation with 2.81A° distance (Color figure online)
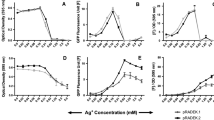
Heavy Metal Detection Using HriGFP
The HriGFP interaction observed with divalent cations like Magnesium, Manganese, and Zinc further prompted us to determine its interaction with heavy metals. Decrease in fluorescence intensity was observed with the increase in divalent heavy metal ions concentration like mercury and copper (Fig. 4a–b). However, strongest difference was observed in case of copper (Fig. 4a–b). Trivalent arsenite showed fluorescence quenching at lower 50 μg/L concentration. Decrease in fluorescence quenching was observed with the increase of arsenite concentration indicating that the HriGFP cannot detect trivalent ions efficiently (Fig. 4c). Moreover, HriGFP exposure to monovalent ions did not show any change in fluorescence intensity (Fig. 4d). Thus, we propose that Copper and Mercury might be interacting similarly to that of the predicted Magnesium ion interaction. Comparison of fluorescence quenching was carried out in order to evaluate the metal ions effectively detected by the HriGFP. Strongest fluorescence quenching was observed in the case of copper as opposed to mercury and arsenic. (Fig. 4e).
Biosensing of heavy metals by HriGFP: a–c Quenching of fluorescence in response to heavy metals like copper, mercury, and arsenic. d Fluorescence showing no change in its intensity on exposure to monovalent cations like Sodium and potassium. e Comparison of fluorescence quenching in percentage relative to different metals. The strongest fluorescence quenching was observed in the case of copper followed by mercury and arsenite. The error bars indicate ± SD. P value was calculated by students t test. *represents < 0.05 ** < 0.01*****0.0001
Docking of HriGFP with MP
The predicted HriGFP structure was further docked to the MP (Fig. 5). One of the most accepted target for organo-phosphorous (OP) compounds is Gly x Ser x Gly motif. However, different proteins that lack serine at their active site bind to OP via lysine and tyrosine [12]. However, our docking analysis revealed strong interaction of Arg119 with the MP. The amino group of Arginine interacts via two hydrogen bonds to the oxygen atoms 1 and 3 of the phosphate group. However, different residues like Pro130, Pro128, Ile67, Arg117, Leu116, Asp68, Lys70, and Glu126 (Fig. 5) interact to MP via hydrophobic interaction.
Structure-based prediction of HriGFP interaction with the methyl parathion a The Methyl parathion has been represented in cyan and is interacting with Arg119. b Zoomed in image of the interaction of the two H atom of the amino group of Arg119 with the two oxygen atoms of the phosphate group of methyl parathion. c The two oxygen atoms of methyl parathion interact via two H-bonds with atomic distance of 3.07 and 3.03A° to the H- atoms of the Arg119 amino acid residue. The other residues Ile67, Asp68, Lys70, Leu116, Arg117, Glu126, Pro128, and Pro130 represented in red interact hydrophobically with the MP
Biosensing of Methyl parathion
The biosensing ability of the HriGFP producing cells was tested with and without induction (Fig. 6). A significant increase in the fluorescence intensity was observed with the corresponding decrease in the concentration of methyl parathion. Thus methyl parathion can be detected from 500 mg to 0.05 mg/L (1.89 mM- 189 μM) concentration.
Comparison of fluorescence intensity between the induced and uninduced HriGFP producing cells after treatment with methyl parathion at different concentrations ranging from 500–0.05 mg/L concentrations. The error bars indicate ± SD. P value was calculated by students t test *represents < 0.05 ** < 0.01*****0.0001
Discussion
HriGFP is a phylogenetically unique fluore scent protein isolated from H.rigida [10]. In this study, the application of HriGFP was investigated. Three dimensional predicted structure of HriGFP demonstrated its resemblance to beta barrel that is typical feature of GFP-like molecules [17]. HriGFP possess two indigenous tripartite motifs IYG and KYG (Fig. 2). The ability of the beta barrel structure bearing FPs descends form the chromophore protected inside the beta barrel comprising of 11 beta sheets [17]. However, on the contrary the HriGFP structure bears 7 beta sheets and the two fluorescent motifs IYG and KYG of HriGFP are exposed (Fig. 2).
Conclusion
Thus, HriGFP can be used efficiently for the detection of heavy metals and harmful organo-phosphorous compounds based pesticides like methyl parathion.
References
Ai, H.-W., Baird, M. A., Shen, Y., Davidson, M. W., & Campbell, R. E. (2014). Engineering and characterizing monomeric fluorescent proteins for live-cell imaging applications. Nature Protocols,9, 910.
Alieva, N. O., Konzen, K. A., Field, S. F., Meleshkevitch, E. A., Hunt, M. E., Beltran-Ramirez, V., et al. (2008). Diversity and evolution of coral fluorescent proteins. PLoS ONE,3(7), e2680.
Batista, B. L., Rodrigues, J. L., Nunes, J. A., Tormen, L., Curtius, A. J., & Barbosa, F., Jr. (2008). Simultaneous determination of Cd, Cu, Mn, Ni, Pb and Zn in nail samples by inductively coupled plasma mass spectrometry (ICP-MS) after tetramethylammonium hydroxide solubilization at room temperature: comparison with ETAAS. Talanta,76(3), 575–579.
Belkin, S. (2003). Microbial whole-cell sensing systems of environmental pollutants. Current Opinion in Microbiology,6, 206–212.
Chakraborty, T., Babu, P. G., Alam, A., & Chaudhari, A. (2008). GFP expressing bacterial biosensor to measure lead contamination in aquatic environment. Current Science,00113891, 94.
Chalfie, M., Tu, Y., Euskirchen, G., Ward, W. W., & Prasher, D. C. (1994). Green fluorescent protein as a marker for gene expression. Science,263, 802–805.
Glick, B. R. (1995). Metabolic load and heterologous gene expression. Biotechnology Advances,13, 247–261.
Gong, J., Wang, L., & Zhang, L. (2009). Electrochemical biosensing of methyl parathion pesticide based on acetylcholinesterase immobilized onto Au–polypyrrole interlaced network-like nanocomposite. Biosensors and Bioelectronics,24, 2285–2288.
Harms, H., Wells, M. C., & van der Meer, J. R. (2006). Whole-cell living biosensors—are they ready for environmental application? Applied Microbiology and Biotechnology,70, 273–280.
Idrees, M., Thangavelu, K., Sikaroodi, M., Smith, C., Sivaraman, J., Gillevet, P., et al. (2014). Novel fluorescent protein from Hydnophora rigida possesses green emission. Biochemical and Biophysical Research Communications,448, 33–38.
Lei, Y., Mulchandani, P., Wang, J., Chen, W., & Mulchandani, A. (2005). Highly sensitive and selective amperometric microbial biosensor for direct determination of p-nitrophenyl-substituted organophosphate nerve agents. Environmental Science & Technology,39, 8853–8857.
Lockridge, O., & Schopfer, L. M. (2010). Review of tyrosine and lysine as new motifs for organophosphate binding to proteins that have no active site serine. Chemico-Biological Interactions,187, 344–348.
Martinez AR, Heil JR, Charles TC (2018) Development of a GFP fluorescent bacterial biosensor for the detection and quantification of silver and copper ions. bioRxiv 2018:296079.
Mulchandani, A., Chen, W., Mulchandani, P., Wang, J., & Rogers, K. R. (2001). Biosensors for direct determination of organophosphate pesticides. Biosensors and Bioelectronics,16, 225–230.
Mulchandani, P., Chen, W., Mulchandani, A., Wang, J., & Chen, L. (2001). Amperometric microbial biosensor for direct determination of organophosphate pesticides using recombinant microorganism with surface expressed organophosphorus hydrolase. Biosensors and Bioelectronics,16, 433–437.
Mérola, F., Erard, M., Fredj, A., & Pasquier, H. (2016). Engineering fluorescent proteins towards ultimate performances: lessons from the newly developed cyan variants. Methods and Applications in Fluorescence,4, 012001.
Stepanenko, O. V., Stepanenko, O. V., Kuznetsova, I. M., Verkhusha, V. V., & Turoverov, K. K. (2013). Beta-barrel scaffold of fluorescent proteins: Folding, stability and role in chromophore formation. International review of cell and molecular biology: Elsevier.
Zambonin, C. G., Quinto, M., de Vietro, N., & Palmisano, F. (2004). Solid-phase microextraction–gas chromatography mass spectrometry: A fast and simple screening method for the assessment of organophosphorus pesticides residues in wine and fruit juices. Food Chemistry,86, 269–274.
Acknowledgements
We thank Dr. Patrick Gillevet, Clint and Masoume from George Mason University (Identification and closing was done there and expression photos are from there as well). This work was supported by the Higher education commission Grant No. 20–347 /NRPU/R&D/HEC/2014/1360 awarded to HB.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Saeed, S., Mehreen, H., Gerlevik, U. et al. HriGFP Novel Flourescent Protein: Expression and Applications. Mol Biotechnol 62, 280–288 (2020). https://doi.org/10.1007/s12033-020-00243-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-020-00243-1