Abstract
Imatinib mesylate, a tyrosine kinase inhibitor, is the first choice in chronic myeloid leukemia treatment. However, resistance to imatinib may develop with time and in some cases, patients may not respond at all to imatinib. Progressive resistance to imatinib therapy is often due to mutations in the BCR/ABL region. Within the scope of our study 124 patients were evaluated via pyrosequencing between 2015 and 2020. In this regard, 32 patients who have a partial response and have no response to imatinib therapy were included in the study. In addition, next-generation sequencing (NGS) analysis was performed on 15 patients who were resistant to imatinib treatment according to the molecular follow-up reports. With pyrosequencing, 5 cases out of a total of 124 were found to be positive. This means that approximately 4.03% of the proportion is positive. But when we examined only 32 patients who have a partial response and have no response to imatinib therapy this rate is rising 15.6%. NGS analysis was performed with 15 patients who have no mutation with pyrosequencing of 32 patients and VUS (Variant of Uncertain Significance) mutation was detected in one. In this study, our aim was to determine the mutations of the BCR/ABL and to evaluate the mutations by NGS and pyrosequencing. Our study is important in terms of comparing the pyrosequencing with NGS mutation rates, drawing attention to the clinical importance of log reduction.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chronic myeloid leukemia (CML) is a myeloproliferative disease characterized by the t(9;22)(q34;q11) reciprocal chromosomal translocation and results in the formation of chimeric fusion gene BCR/ABL. The incidence of CML is 1-2/100.000 per year and it constitutes 15 percent of leukemias in adults. The mean age of the patients is 45 to 55 and the male/female ratio is 3/2 [1]. 95% of patients diagnosed with CML have a Philadelphia (Ph) chromosome. The presence of the Ph chromosome in the blood or bone marrow leads to the CML. The Ph chromosome occurs as a result of reciprocal translocation of the 9 to 22 chromosomes (Fig. 1). This chromosome leads to the emergence of a novel chimeric fusion gene called BCR/ABL. This gene product has tyrosine kinase activity. Tyrosine kinase enzymes break phosphorus from ATP and transfer it to tyrosine amino acid and play an important role in cell proliferation, apoptosis, and intracellular signal transduction (Fig. 2).
Philadelphia chromosome, t(9;22)(q34;q11). The BCR/ABL fusion protein is formed as a result of the reciprocal translocation of a part from chromosome 9 and a part from chromosome 22. This novel small chromosome 22 is called as Philadelphia chromosome. With the addition of the broken piece from chromosome 22 to chromosome 9, a 9 derivative chromosome is formed. Created with https://BioRender.com
Mechanism of action of Imatinib; Imatinib binds to the ATP binding site of BCR/ABL and prevents protein tyrosine phosphorylation, thus inhibiting the protein's enzyme activity. Created with https://BioRender.com
CML is characterized by three clinical stages and allows the treatment method to be followed according to the stage of the patient. These phases are called the chronic phase, accelerated phase, and the blast crisis. While mutations are rare in recently diagnosed chronic phase patients, the prevalence of mutations rises meaningfully in patients who developed resistance to TKI therapy and in advanced phase CML patients. Patients with CML require minimal residual follow-up according to the treatment response because of the recurrence prospect. Minimal residual disease (MRD) is defined as the presence of cells that have escaped treatment and caused relapse even though there is no clinical sign of leukemia. Because, while the patients with MRD negative display a very low rate of relapse, this rate increases significantly in MRD positivity [2].
In the treatment of CML, specific BCR/ABL protein tyrosine kinase inhibitor (TKI) imatinib mesylate is used. Imatinib, which is clinically effective for the treatment of CML disease firstly, closes the catalytic domain of ABL and acts at the molecular level by preventing ATP binding and therefore prevents cell activation and proliferation by preventing phosphorylation [3]. In treatment with TKI, the goal is to achieve a complete hematological response after 3 months, complete cytogenetic response after 6 months, and at least 3 log reductions in molecular disease after a year [4, 5]. Although imatinib is used effectively in the treatment of CML, the developing resistance to treatment in advanced phase patients is the most common handicap and was first identified in the phase 2 studies of imatinib. Several mechanisms are underlying this development of resistance; the best-known mechanism is the expansion of the BCR/ABL kinase region with mutations, hence impairing imatinib binding [3]. In addition, genomic amplification of BCR/ABL is among other mechanisms [6]. Besides ABL kinase region mutations, resistance to imatinib may develop independently of ABL mutations. The most important way of dealing with drug resistance and strengthening treatment may be to eliminate TKI resistance, thus reducing the leukemic disease burden. Developing recent strategies for determining drug resistance hence, expanding existing treatment areas is a critical need for the survival of CML patients. The mechanism of resistance contains clinical difficulties due to its multi-factor and heterogeneous nature. At this point, early diagnosis with high sensitivity tests such as next-generation sequencing (NGS) and timely treatment with next-generation therapeutics is significant to cope with resistance development [7].
In addition to the primary response seen in patients who may not respond to TKIs and developed independently of the BCR/ABL mutation, one may experience recurrence after the first response. This is called secondary resistance. The development of resistance triggers the BCR/ABL kinase activity, worsening the prognosis by progressing from the chronic phase to the blastic phase, and the changes of treatment decrease after the transition to the blastic phase. Therefore, it has become significant to prevent resistance before it develops [8]. If first-line drug resistance develops in the treatment of imatinib mesylate, second-generation drug treatment is initiated and nilotinib and dasatinib are second-generation tyrosine kinase inhibitors used in the treatment of imatinib-resistant CML [9].
In this study, we examined the relationship between BCR/ABL mutations and response to imatinib treatment in terms of its clinical significance. We focused on the importance of the mutation detection sensitivity of the molecular methods in terms of the planning therapy. We aimed to detect different variants in the ABL gene using NGS that may be associated with resistance in imatinib-resistant patients whose mutations could not be detected by pyrosequencing.
Materials and methods
Establishing the patient groups
The study group consists of patients diagnosed with CML in the Hematology Department at Erciyes University and they are thought to have developed resistance after imatinib treatment was initiated. Between 2015 and 2020, 124 patients who applied to Erciyes University Medical Genetics Department for BCR/ABL mutation screening were evaluated in this retrospective and prospective study. Pyrosequence analysis results were evaluated from files on 32 patients who have a partial response and have no response to imatinib therapy (< 3 logs) retrospectively, while NGS analysis was conducted prospectively on 15 patients whose pyrosequencing results did not reveal a mutation, but who were deemed to be resistant based on molecular follow-up reports. After all, 32 patients were included in this study. Individuals under 18 years of age were excluded from this study. The present study was approved by Erciyes University Clinical Research Ethics Committee. All participants gave written informed consent in accordance with the Declaration of Helsinki.
Methods
The 14 hotspot mutation points were examined with pyrosequencing in CML patients for determining the imatinib resistance status. Besides, NGS analysis was performed with 15 patients who have no mutation with pyrosequencing to specify the ABL gene status. The pathogenicity classification of the variants was performed based on the ACMG (American College of Medical Genetics and Genomics) 2015 guide [10]. Method details of this article are included in the supplementary file.
Statistical analysis
The results were analyzed statistically and p < 0.05 was accepted significantly. Statistical analysis details of this article are included in the supplementary file.
Results
In all patients, treatment was launched with imatinib. In 20 patients with resistance to imatinib, treatment was continued with nilotinib and in 12 patients with dasatinib. The parameters of age and gender that may be effective on relapse were examined. These independent variables had no significant effect on relapse-independent survival. However, the majority of positive patients were male and detected in the chronic phase. Demographic and clinical characteristics of patients are demonstrated in Table 1.
All samples were evaluated according to the limit of detection (LOD) value for each mutation (Table 2). No mutation, if mutation frequency < LOD, there is mutation, if mutation frequency > LOD + 3%, and potential low-level mutation if mutation frequency ≥ LOD and ≤ LOD + 3%.
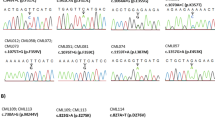
As a result, 5 cases out of a total of 124 were found to be positive by pyrosequencing. This means that approximately 4.03% of the proportion is positive. When just 32 patients with a partial response and no response to imatinib therapy were analyzed, the mutation detection rate increased to 15.6%. 2 of them were positive for T315I (ACT > ATT) mutation (Fig. 3), 2 of them for E255K mutation, and one of them was found to be positive for both F359V and F359C mutations. We detected a missense, heterozygous c.1370G > A (R457H) (NM_005157) variant of unknown clinical significance in exon 8 of the ABL gene in only 1 of 15 patients studied via NGS. The patient with VUS (Variant of Uncertain Significance) detected is in chronic phase, in the early 20 s, and male.
A total of 8 patients in the group were in the treatment-unresponsive zone (< 1 log, 10%). Only 3 of them were detected as positive by pyrosequencing. Only 2 of 24 patients with the partial cytogenetic response (> 1 log) were found to be positive with pyrosequencing (Table 3).
The cut-off point of the log reduction was set to be 3 and the log reduction < 3 or log reduction ≥ 3 was compared with mutation or no mutation with Fisher’s exact test. In all of our patients with mutations, the log reduction was determined less than 3, and this relationship is statistically significant (p = 0.007, p < 0.05). Also, when we compared the log reduction values with the presence or absence of mutations in itself with the non-parametric Mann–Whitney U test, a lower log decrease was observed in the mutation group (p = 0.001, p < 0.05). This value is statistically significant.
Discussion
As a consequence of reciprocal translocation between chromosomes 9 and 22, the BCR/ABL fusion gene is formed, called the Philadelphia chromosome which leads to CML [11]. The product of this gene has increased tyrosine kinase activity and proliferates in the cell and stimulates the differentiation, cellular signaling pathways, and growth factors [12]. Imatinib is a TKI, used for the first-line therapy of CML, but today, resistance to TKI treatment still makes both the patients and the physicians anxious. In 2006, Wei et al. studied 30 early chronic phases (CP) CML patients to examine the relationship between BCR/ABL mutations and imatinib resistance type using the conventional sequencing technique. In that study, it was reported that pre-treatment screening was not financially effective as no mutations were detected before imatinib treatment and not all imatinib resistance was caused by the BCR/ABL mutation, but patients with signs of increased disease burden should be investigated for BCR/ABL mutations. Because it has been demonstrated that increased BCR/ABL mRNA levels detected by RT-qPCR (real-time quantitative polymerase chain reaction) were associated with the occurrence of a mutant clone [13]. Chien et al. studied the relationship between log reduction and mutation status and reported that low log reduction of BCR/ABL was related to ABL mutations [14]. Similarly, in our study, low log reduction was found to be highly correlated with the development of mutation. During TKI treatment, the persistence of minimal residual disease occurs in the majority of patients and may require lifelong treatment. In the study conducted in 2019, it is thought that a cause of persistence may be due to the BCR/ABL1 gene, and it is emphasized that more research should be designed and performed to evaluate the transcription rate of the gene and especially the cellular quantity [15]. According to current evidence, the deep molecular response is varied, and patients with undetectable disease may have varying levels of residual disease burden [16]. RT-qPCR, which measures log reduction of BCR-ABL1 in blood or bone marrow, is considered the gold standard to assess response to TKI therapy [17].
Depending on the medical condition of the patient, considering the conditions it is in and the other concomitant diseases, the choice of the appropriate tyrosine kinase inhibitor is critical and will increase the benefit of TKI by reducing the negative effects. Further, another critical issue is the mutation detection method. Generally, NGS and Sanger have been compared in the literature, and it was determined that NGS is a more sensitive technique and BCR/ABL kinase domain mutations can be detected at very low levels and at early stages with high rates via NGS. By resolving clonal complexity and recognizing compound mutations, NGS can detect the mutation character more accurately. In this sense, novel researchers aim to the routine use of NGS instead of Sanger Sequencing. But some problems can arise, such as proper analysis, sound bioinformatics, and financial distress [18,19,20,21,22,23].
Molecular detection of mutations is critical for overcoming resistance, increasing survival rates, improving patient prognoses, and achieving remission. In this context, molecular analysis and monitoring of the patients are becoming more important because depending on the type of mutation, the treatment options may also change [24]. Since BCR/ABL point mutations are prognostic determinants of CML, their importance cannot be ignored and most studies have focused on point mutations in the kinase domain. It was indicated that resistance mechanisms generally developed as a result of point mutations and changed the biochemical properties of imatinib binding points [6]. The T315I-mutation is substantial because it is the most frequent mutation and it is considered resistant to the second-line drugs too, which are produced for imatinib-resistant patients. T315I mutation is associated with increased oncogenic activity and disease progression. As well as T315I mutations, P-loop mutations are thought to be associated with high imatinib resistance and poor prognosis [9, 24]. In some patients, TKI resistance is found without known resistance mutations, but this is almost rare as the majority of patients with chronic phase CML are followed long term [25].
Many mechanisms such as quiescent CML stem cells, drug bioavailability, mechanism of drug efflux, and intracellular signal transmission independent of BCR/ABL mutations can also lead to imatinib resistance [26, 27]. Other mechanisms hypothesized to play a role in resistance development include gene amplification, gene rearrangements, epigenetic alterations, and miRNAs. MicroRNAs and epigenetic modifications are two examples of distinct ways that a cell can use to attain the same goal [28]. Cell-autonomous BCR-ABL1 kinase-independent genetic and epigenetic alterations and signals provided by the bone marrow (BM) microenvironment play a role in leukemic stem cell (LSC) persistence, innate or acquired resistance to TKIs, and also mortal blast crisis [29]. Knowing the patient’s mutational status, which might change over time, and the related therapeutic alternatives can help with therapeutic considerations. Individualizing the pharmacodynamics of mutations, as well as the unique side-effect profiles of second-generation TKIs, may support physicians make better treatment judgments [30].
As aforementioned, imatinib resistance is not always caused by BCR/ABL mutations. For this reason, our low positivity rates for imatinib resistance may be due to other reasons. Furthermore, to understand the molecular basis of resistance to drugs, mutations in the BCR/ABL fusion gene should be investigated in more detail with larger populations.
The exact cause of drug resistance is still unclear. Concerning this, in the study conducted by Gorre et al. in 2019, it has been emphasized that CML demonstrates population diversity, and the demographic features underlying CML are still not understood and clinical heterogeneity may be related to drug response [31]. Due to the heterogeneous nature of drug resistance, different studies are still needed in different populations. In this sense, we suggest revealing a perspective by examining the clinical characteristics and mutation profile of a group of population in this study we conducted.
In the present study, the mutation detection rate of pyrosequencing in imatinib-resistant patients was 15.6%, while this rate was 6.6% in the NGS method. According to the literature, the mutation detection rate has been reported as approximately 35% in NGS studies performed in imatinib-resistant patients [23]. Our mutation detection rate by NGS does not seem to be compatible with the literature. This can be attributed to the small scale of our patient group. Besides, the myeloid panel kit containing the non-translocated ABL1 gene used may be another reason. Because, the myeloid panel contains exons of the ABL1 gene encoding the kinase domain, but it may comprise an untranslated ABL1 substrate; hence, the mutation rate may decrease to an undetectable level even via NGS [21]. In the case of working with the appropriate kit, it may be possible to find treatment-resistant mutations that cannot be obtained with pyrosequence via NGS. The detection of novel resistance mutations which will contribute to the treatment is more possible with NGS.
In the literature, it is emphasized that many points are still not fully understood and many different studies are needed. Distinct studies are ongoing to overcome imatinib resistance, and this study will shed light on the next researches with the profiling of a different group of population. The literature was examined and the differences are observed between the findings obtained from the studies. These differences may arise from the characteristics of the patients, the inequality number of the patients in the study groups, and the different methods used to detect mutations.
The limitations of this study are the insufficient number of patients screened and the inability to access different demographic characteristics of the patients. Moreover, we could not detect 5 out of 8 patients who did not respond to treatment (< 1 log reduction) with both pyrosequencing and NGS. These patients are considered to be at high risk for disease progression and death. That’s why the mutation detection rate of the tests is extremely important.
In conclusion, less than a three-log reduction is highly associated with the development of drug resistance. In the case of less than one-log reduction of BCR/ABL, there is no response to the TKI therapy. While the drug treatment is planning by physicians, it is of great importance to take these situations into account. Because the drug can be changed according to the log reduction status which was calculated by RT-qPCR analysis. Also, mutation profile is another important point for the decision of the physicians. Identification of mutations that may occur outside of hot spots with appropriate methods in the early stages may improve individual treatment and alter clinical progression. Some patients acquire treatment resistance, but this cannot be identified at the molecular level, and the molecular response ranges of some patients are extremely varied. In this context, it is possible to say that individualized treatment can save a person’s life. This might be accomplished by studying the mutation profile, as well as other genetic and demographic data in larger populations and in more depth through further researches.
Only hot spot mutations can be examined with the pyrosequencing method. However, NGS method can explain the clinical symptoms and development of resistance to pharmacological response in patients by providing the possibility of detecting variants outside of these areas. Thus, we predict that a specific treatment can be developed for the patient by combining the data obtained by the NGS method with the patient's clinical findings and family history. The c.1370G > A variant, which we detected in our study, converted from arginine to histidine at codon 457 of the ABL gene and its clinical significance is uncertain in bioinformatics databases. It is also predicted to cause damaging effects according to many in silico programs. Therefore, this variant may enter the literature as pathogenicity that causes resistance in the future. In addition, other mechanisms that may cause drug resistance should be examined in detail. This will increase the remission for each patient.
Many more studies are needed with a large cohort that examines the response range, mutation profile, and the different demographic properties of the patients, and in this way, it is a promising idea that personal treatment planning can also be possible.
Data availability
All data are available upon request.
References
Faderl S, Talpaz M, Estrov Z, O’brıen S, Kurzrock R, Kantarjian HM. The biology of chronic myeloid leukemia. N Engl J Med. 1999;341(3):164–72.
Ross DM, Branford S, Seymour JF, Schwarer AP, Arthur C, Yeung DT, et al. Safety and efficacy of imatinib cessation for CML patients with stable undetectable minimal residual disease: results from the TWISTER study. Blood. 2013;122(4):515–22.
Linev AJ, Ivanov HJ, Zhelyazkov IG, Ivanova H, Goranova-Marinova VS, Stoyanova VK. Mutations associated with imatinib mesylate resistance—review. Folia Med (Plovdiv). 2018;60(4):617–23.
Izzo B, Gottardi EM, Errichiello S, Daraio F, Baratè C, Galimberti S. Monitoring chronic myeloid leukemia: how molecular tools may drive therapeutic approaches. Front Oncol. 2019;9:1–12.
Hantschel O. Chronic myeloid leukemia. HemaSphere. 2019;3(S2):47.
Melo JV, Chuah C. Resistance to imatinib mesylate in chronic myeloid leukaemia. Cancer Lett. 2007;249(2):121–32.
Yaghmaie M, Yeung CC. Molecular mechanisms of resistance to tyrosine kinase inhibitors. Curr Hematol Malig Rep. 2019;14(5):395–404.
Bavaro L, Martelli M, Cavo M, Soverini S. Mechanisms of disease progression and resistance to tyrosine kinase inhibitor therapy in chronic myeloid leukemia: an update. Int J Mol Sci. 2019;20(24):1–23.
Etienne G, Dulucq S, Huguet F, Schmitt A, Lascaux A, Hayette S, et al. Incidence and outcome of BCR-ABL mutated chronic myeloid leukemia patients who failed to tyrosine kinase inhibitors. Cancer Med. 2019;8(11):5173–82.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–23.
Kang ZJ, Liu YF, Xu LZ, Long ZJ, Huang D, Yang Y, et al. The philadelphia chromosome in leukemogenesis. Chin J Cancer. 2016;35(1):1–15.
Branford S, Rudzki Z, Walsh S, Parkinson I, Grigg A, Szer J, et al. Detection of BCR-ABL mutations in patients with CML treated with imatinib is virtually always accompanied by clinical resistance, and mutations in the ATP phosphate-binding loop (P-loop) are associated with a poor prognosis. Blood. 2003;102(1):276–83.
Wei Y, Hardling M, Olsson B, Hezaveh R, Ricksten A, Stockelberg D, et al. Not all imatinib resistance in CML are BCR-ABL kinase domain mutations. Ann Hematol. 2006;85(12):841–7.
Chien SH, Liu HM, Chen PM, Ko PS, Lin JS, Chen YJ, et al. The landscape of BCR-ABL mutations in patients with Philadelphia chromosome-positive leukaemias in the era of second-generation tyrosine kinase inhibitors. Hematol Oncol. 2020;38(3):390–8.
Baccarani M, Rosti G, Soverini S. Chronic myeloid leukemia: the concepts of resistance and persistence and the relationship with the BCR-ABL1 transcript type. Leukemia. 2019;33(10):2358–64. https://doi.org/10.1038/s41375-019-0562-1.
Mahon FX, Etienne G. Deep molecular response in chronic myeloid leukemia: the new goal of therapy? Clin Cancer Res. 2014;20(2):310–22.
Alikian M, Gale RP, Apperley JF, Foroni L, Alikian M. Molecular techniques for the personalised management of patients with chronic myeloid leukaemia. Biomol Detect Quantif. 2017;11:4–20. https://doi.org/10.1016/j.bdq.2017.01.001.
El Fakih R, Chaudhri N, Alfraih F, Rausch CR, Naqvi K, Jabbour E. Complexity of chronic-phase CML management after failing a second-generation TKI. Leuk Lymphoma. 2019;61(4):776–87.
Machova Polakova K, Kulvait V, Benesova A, Linhartova J, Klamova H, Jaruskova M, et al. Next-generation deep sequencing improves detection of BCR-ABL1 kinase domain mutations emerging under tyrosine kinase inhibitor treatment of chronic myeloid leukemia patients in chronic phase. J Cancer Res Clin Oncol. 2015;141(5):887–99.
de Lavallade H, Jackson S, Kizilors A, Etienne G, Huguet F, Guerci-Bresler A, et al. Prospective evaluation of ABL kinase domain mutational analysis by next-generation-sequencing in newly diagnosed CP CML patients undergoing first-line treatment with nilotinib alone or nilotinib + pegylated interferon-α2a in a prospective phase III trial. Blood. 2019;134(Supplement_1):664–664.
Soverini S, Abruzzese E, Bocchia M, Bonifacio M, Galimberti S, Gozzini A, et al. Next-generation sequencing for BCR-ABL1 kinase domain mutation testing in patients with chronic myeloid leukemia: a position paper. J Hematol Oncol. 2019;12(1):1–11.
Soverini S, Bavaro L, de Benedittis C, Martelli M, Iurlo A, Orofino N, et al. Prospective assessment of NGS-detectable mutations in CML patients with nonoptimal response: the NEXT-in-CML study. Blood. 2020;135(8):534–41.
Soverini S, Martelli M, Bavaro L, De Benedittis C, Papayannidis C, Sartor C, et al. Next-generation sequencing improves BCR-ABL1 mutation detection in Philadelphia chromosome-positive acute lymphoblastic leukaemia. Br J Haematol. 2021;193(2):271–9.
Koçkan B, Toptaş T, Atagündüz I, Tuğlular AT, Özer A, Akkiprik M. Molecular screening and the clinical impacts of BCR-ABL KD mutations in patients with imatinib-resistant chronic myeloid leukemia. Oncol Lett. 2018;15(2):2419–24.
Braun TP, Eide CA, Druker BJ. Response and resistance to BCR-ABL1-targeted therapies. Cancer Cell. 2020;37(4):530–42.
Apperley JF. Part I: mechanisms of resistance to imatinib in chronic myeloid leukaemia. Lancet Oncol. 2007;8(11):1018–29.
Azad NA, Shah ZA, Pandith AA, Rasool R, Rasool JA, Baba SM, et al. Analysis of ABL kinase domain mutations as a probable cause of imatinib resistance in chronic myeloid leukemia patients of Kashmir. Meta Gene. 2018;17:93–8. https://doi.org/10.1016/j.mgene.2018.05.003.
Fojo T. Multiple paths to a drug resistance phenotype: Mutations, translocations, deletions and amplification of coding genes or promoter regions, epigenetic changes and microRNAs. Drug Resist Updates. 2007;10(1–2):59–67.
Perrotti D, Silvestri G, Stramucci L, Yu J, Trotta R. Cellular and molecular networks in chronic myeloid leukemia: the leukemic stem, progenitor and stromal cell interplay. Curr Drug Targets. 2016;18(4):377–88.
Bixby D, Talpaz M. Mechanisms of resistance to tyrosine kinase inhibitors in chronic myeloid leukemia and recent therapeutic strategies to overcome resistance. Hematol Am Soc Hematol Educ Program. 2009;2009:461–76.
Gorre M, Sashidhar R, Annamaneni S, Digumarti R, et al. Demographic and clinical characteristics of chronic myeloid leukemia patients: a study on confined populations of southern India. Indian J Med Paediatr Oncol. 2019;40(1):70–6.
Acknowledgements
The results reported in this article were partially presented at the 7th International Congress of Pharmaceutical Chemistry: API Design, Synthesis, Production, and Standardization, on March 14–17, 2019.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
MD* developed the theory and concept of the study. HA designed the study. NK produced the first draft and completed the entire manuscript. NG carried out the NGS laboratory studies and performed the statistical analysis. HK analyzed the patient’s data. AY conducted the literature search. MK and SC provided clinical information of patients. MD evaluated the clinical results and contributed to the lettering of the manuscript. All the authors collected patient data and read and approved the final manuscript (*: corresponder).
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to disclose.
Ethical approval
This study was approved with the decision dated Dec 16th, 2020 and numbered 629 by Erciyes University Clinical Research Ethics Committee.
Informed consent
All participants gave written informed consent in accordance with the Declaration of Helsinki.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Karasu, N., Akalin, H., Gokce, N. et al. Detection of mutations in CML patients resistant to tyrosine kinase inhibitor: imatinib mesylate therapy. Med Oncol 38, 120 (2021). https://doi.org/10.1007/s12032-021-01571-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-021-01571-1