Abstract
Non-muscle myosin IIA heavy chain (MYH9) has been implicated in many physiological and pathological functions including cell adhesion, polarity, motility to cancer. However, its role in melanoma remains unexplored. The aim of our study was to evaluate the role of MYH9 in melanoma tumor development and metastasis and further to find out the potential underlying mechanisms. In this study, we evaluated the in vitro migratory and invasive properties and in vivo tumor development and metastasis in C57BL/6 mice by silencing MYH9 in B16F10 melanoma cells. Knocking down MYH9 enhanced migration and invasiveness of B16F10 cells in vitro. Furthermore, MYH9 silencing accelerated tumor growth and metastasis in melanoma subcutaneous and intravenous mouse models. Next, oncogenes analysis revealed epithelial–mesenchymal transition and Erk signaling pathway are being regulated with MYH9 expression. Finally, MYH9 silencing in B16F10 cells modulates the tumor microenvironment by manipulating the leukocytes and macrophages infiltration in tumors. These findings established the opposing role of MYH9 as a tumor suppressor in melanoma suggesting specific MYH9 based approaches in therapeutics.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Melanoma is a highly aggressive cancer originates in melanocytes (pigment producing cells), primarily resides in the skin. Melanoma represents around 4% of all skin cancers; however, it contributes most to skin cancer-related mortality (approximately 80%). Melanoma can metastasize in any organ such as lung, liver, lymph nodes and gastrointestinal tract [1, 2]. Tumor metastasis/spread of cancer cells to different organs throughout the body is one pf the primary cause of cancer-related deaths (estimated ~ 90%) worldwide [3]. Cancer cells metastasis is a complex and multistep process that involves the penetration of cancer cells through the basement membranes and extracellular matrix to invade adjacent tissues and metastasize via the circulatory system to extravasate, attach, proliferate and form a new tumor in distant tissue [4, 5].
Non-muscle-myosin-II (NMII) is one of the members of myosin superfamily that is represented by fifteen different classes. NMII is the conventional two-headed myosin, which is found to involved in the regulation of several cellular functions such as cell division, locomotion, polarity, adhesion and morphology in various non-muscle cells. Vertebrates express three non-muscle myosin II heavy chain isoforms. viz., myosin-IIA, -IIB and -IIC. Non-muscle myosin IIA heavy chains are also known as Myosin-9 (MYH9) [6, 7]. Besides the identified role of MYH9 in cell division, adhesion and migration, it is also found to contribute to the regulation of cancer cells tumorigenic properties [8,9,10,11]. Highly invasive ER positive breast cancer cells MCF-7 and -6 showed a higher MYH9 level compared to non-invasive cells. These invasive properties were downregulated by blebbistatin and MYH9 silencing treatment in MCF7/6 cells [12] and in MDA MB-231 cells, MYH9 silencing reduces spreading and migration [13] which indicate MYH9 as tumor inducer in breast cancer. In human non-small cell lung cancer (NSCLC), immunohistochemical evaluation of MYH9 indicates its correlation with adenocarcinoma differentiation and invasion [14]. Another study indicated that NSCLC (adenocarcinoma) cases lacking expression of either MYH9 or vimentin showed good recovery even without adjuvant chemotherapy with uracil–tegafur [15].
Despite the reports in other cancers, the role of MYH9 in melanoma development and metastasis is unclear. Understanding the role of MYH9 in regulating tumor cell proliferation migration, invasion, and colonies formation at a distant site could provide a better insight into melanoma development and metastasis. Data from the present study show that MYH9 suppress tumorigenesis and metastasis in murine melanoma cells tumorigenesis.
Materials and methods
Animals
All animal protocols were approved by the Institute of Life Sciences Animal Ethics Committee. All animals used in the experiments were of C57BL/6 background and 6–8 weeks of age.
Cell lines and culture
B16F10 were maintained in DMEM medium supplemented with 10% FBS (Origin: United States) (Gibco), streptomycin (100 μg/ml) and penicillin (100 μg/ml). Cells were maintained in an incubator with a humidified atmosphere containing 5% CO2 at 37 °C.
MYH9 lentiviral infection
High-titer MYH9 lentiviral particles from Sigma (TRCN0000310913) were used for MYH9 silencing in B16F10 cells cultured in 96 well plates were infected with control shRNA, MYH9 shRNA in DMEM. shRNA infected cells were left overnight at 37 °C and 5% CO2. The medium was changed next day and was replaced with fresh medium for 24 h. After 24 h of resting MYH9 transfected cells colonies were selected from cells grown in growth medium with 1 µg/ml puromycin. The puromycin selected colonies were evaluated for MYH9 silencing using western blotting and qPCR for MYH9 expression.
Cell invasion assay
The cell invasion assay was performed using 6-well transwell units (Costar) with an 8-µm pore size polycarbonate filter. The transwells were coated with 20 µg/ml type I collagen and incubated at 4 °C overnight. After washing with PBS, the wells were seeded with 2 × 105 cells/ml in incomplete DMEM medium and the bottom chambers were filled with 500 µl DMEM containing 5% FBS. Cells migrate for overnight at 37 °C and 5% CO2 and cells on the bottom side of the membrane were stained with 0.5% crystal violet dye in 10% formalin for 30 min and air dried. Migrated cells were counted using a Carl Zeiss inverted microscope (Carl Zeiss, Germany). Six independent areas per filter were counted, and the mean number of migrated cells was calculated.
Cell proliferation assay
Cell viability assay was performed to check the proliferation rate of B16F10 cells and cells were seeded in 96-well cell culture plastic plates at a density of 5 × 103 cells/well. After incubation for 48 h, 100 μl of 3-(4,5-Dimethylthiazol-2-Yl)-2,5-Diphenyltetrazolium Bromide (5 mg/ml) was added for 4 h and farmazan crystals were dissolved by adding dissolving solution (DMSO: isopropanol; 1:1) for 30 min. The absorbance of each well was acquired at 570 nm with the help of ELISA plate reader.
Clonogenic assay
B16F10 cells clonogenic assay was performed to measure the growth ability of single B16F10 cell to grow into a colony in vitro. Briefly, after transfecting B16F10 cells with MYH9 shRNA or scrambled control cells were seeded in complete DMEM media at a density of 1 × 103 cells in six well plates. The plates were incubated for 2 weeks at 37 °C and then stained with 0.1% crystal violet. Colonies with > 50 cells were counted manually.
Wound healing assays
Cell migration ability of B16F10 cells on 5% FBS treatment was detected by scratch assay. Cells were seeded in 12 well plates at the density of 3 × 105 cells/well and starved for 6–8 h at 90–100% confluency. Further, a straight scratch was created with 200 μl sterile pipette tip and then supplemented with DMEM medium alone and with 5% FBS for 24 h at 37 °C. Migration images were captured using an inverted microscope (CarlZeiss, Germany). The scratch wound widths were measured under a microscope and the relative percentage of wound closure was determined by comparing to control cells.
Quantitative PCR
Total RNA was isolated using Trizol (Invitrogen) and further purified by pure link RNA Mini Kit (Ambion life technologies) from cells. Further, 2 μg of RNA was reverse transcribed and cDNA was synthesized using a High-Capacity cDNA Reverse Transcription Kit (Applied biosystems). Real-time PCR amplification was performed using SYBR green (Applied biosystems) and QuantStudio 6 Flex Real-Time PCR (Applied Biosystem). A complete list of PCR primers is shown in Supplementary Table S1. All data were normalized to the housekeeping gene GAPDH.
Immunoblot analysis
Total protein was extracted using RIPA buffer (Cell Signaling) with protease buffer and 40 μg protein was separated on 10% SDS-PAGE. Resolved proteins were transferred to nitrocellulose membranes and blocked with 5% non-fat milk in TBS-T, followed by overnight incubation with diluted antibody (Supplementary Table S2) in blocking buffer at 4 °C. After washing with TBST, the membrane was incubated at room temperature for 1 h with secondary antibodies conjugated to HRP (Amersham). Membranes were visualized with enhanced chemiluminescence, followed by exposure to film.
PCR array
Changes in the expression of mouse oncogenes and tumor suppressor genes (Supplementary Table S3) were detected using RT2 Profiler Mouse oncogenes and tumor suppressor PCR Array (PAMM-502ZA; Qiagen). Total RNA was extracted by Trizol method as mentioned in the previous section. cDNA was synthesized using the RT2 First Strand Kit (Qiagen). RT2 SYBR Green qPCR Master Mix (Qiagen) was used for the reaction following the manufacturer’s instructions. Amplification and real-time analysis were performed by QuantStudio 6 Flex Real-Time PCR (Applied Biosystem).
In vivo tumorigenesis
C57BL/6 mice (4–6-week-old) were maintained in specific pathogen-free conditions. All experimental protocols were approved by the Animal Ethics Committee of the Institute of Life Sciences and all experiments were performed in accordance with the approved guidelines and regulations. For the subcutaneous tumor model, B16F0 cells transfected with MYH9 shRNA or control shRNA suspended with PBS at a concentration of 2 × 106 cells/200 μl and subcutaneously injected into C57BL/6 mice (n = 5 for each group) and the mice were observed for 2 weeks. The tumor volume was measured at the experimental endpoint.
For lung metastasis study, cells suspended in PBS at a concentration of 1 × 106 cells/200 μl and injected intravenously into C57BL/6 mice (n = 4 for each group). At the end of the experiment (after 3 weeks), mice were euthanized, and the tumor was collected. The B16F10 tumor specimen was fixed in 10% formalin, embedded in paraffin and sectioned at 5 µm for further histopathological studies.
Immunohistochemistry
Tumor tissue samples were fixed in 10% formaldehyde solution before being embedded in paraffin wax. Sections of 5 µm were mounted on positively charged slides. For immunohistochemical assay, 3% hydrogen peroxide in methanol was used for endogenous peroxidases blocking. These sections were probed with Ki-67, CD34, CD45 and F4/80 primary antibody (Supplementary Table S2) overnight at 4 °C. Tumor tissue sections slides were subsequently incubated with HRP tagged secondary antibody. Staining was visualized with DAB and counterstained with hematoxylin. The images were taken using a Leica microscope.
Hematoxylin and eosin (H&E) staining
Tumor tissue samples were fixed in 10% formaldehyde solution before being embedded in paraffine wax. Section of 5 µm were mounted on positively charged slides. Briefly, sections were deparaffinised in xylene and further rehydrated. Next, sections were stained in hematoxylin and differentiate in 1% acid alcohol. Further, eosin staining was done followed by dehydration and mounting. Staining was visualized and images were taken using Leica microscope.
Statistical analyses
All experiments were performed in triplicate. Data are presented as mean ± standard deviation (SD) and analyzed using Microsoft Excel and Graphpad Prism6.0 (GraphPad Software, La Jolla, CA). Statistical analyses were conducted between the controls and the treated experimental groups using Student’s t-test assuming two-tailed distributions, and differences were considered to be statistically with P ≤ 0.05.
Results
MYH9 knocking down induces migration and invasion in B16F10 cells in vitro
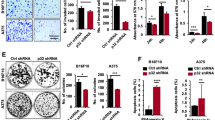
To study the role of MYH9 in B16F10 cell migration and invasion, we silenced MYH9 in B16F10 cells and our result showed reduction of MYH9 (Fig. 1a). Our results showed increased invasion through collagen in transwell chamber and migration in MYH9 shRNA transfected cells compared to control shRNA transfected B16F10 cells (Fig. 1b, c). Further, we evaluated colony formation and cell proliferation capacity and a decreased number of colonies and proliferation were observed in MYH9 silenced B16F10 cells compared to control (Supplementary Fig. 1a, b) suggesting the critical function of MYH9 in migration, invasion and colony formation of B16F10 cells.
MYH9 silencing regulates migration and invasion of mouse melanoma cells in vitro. a Relative mRNA and immunoblot expression showing the level of MYH9 in control shRNA and MYH9 shRNA transfected B16F10 cells. b Invasion assay showing the number of invading cells through transwell chamber 12 h after stimulation with 5% FBS. c wound healing/migration assay at 0 h, 12 h and 24 h post wounding and percentage wound closure in control and MYH9 shRNA transfected B16F10 cells. All data are represented as mean ± SD. Student’s t-test was used for all statistical analyses (***P ≤ 0.001; **P ≤ 0.01)
MYH9 regulates oncogenes, EMT marker and downstream signaling molecules in B16F10 cells
The in vitro tumorigenesis and metastasis are regulated by various oncogenes, epithelial and mesenchymal markers. As in vitro assays revealed the increased migration and invasion in MYH9 silenced B16F10 cells. Next, we investigated the effect of MYH9 knocking down on the expression of different oncogenes involved with epithelial-to-mesenchymal transition (EMT) markers and signaling pathways. Increased expression of oncogenes (cyclin-D1 and cMyc) and mesenchymal markers (slug and twist), epithelial marker (E-cadherin) and decreased level of mesenchymal markers (snail and MMP9) was found in MYH9 silenced B16F10 cells than control (Fig. 2a, b).
MYH9 silencing regulates oncogenes and EMT markers expression in B16F10 cells. a qPCR showing the expression of oncogenes and EMT markers in control shRNA and MYH9 shRNA treated B16F10 cells. b Immunoblot showing the level of various oncogenes and EMT markers in control shRNA and MYH9 shRNA treated B16F10 cells. c Heat map showing changes in expression of oncogenes genes in MYH9 shRNA treated and control shRNA treated B16F10 cells. All data are represented as mean ± SD. Student’s t-test was used for all statistical analyses (**P ≤ 0.01, *P ≤ 0.05). n.s. not significant
Furthermore, we performed PCR array assay to explore the role of MYH9 silencing in B16F10 cells on the expression profile of different oncogenes and tumor suppressor makers. Our results showed a higher expression of oncogenes (Bcl2, Egf, Erbb2, JunD, Kras, Mdm2, Mos, Mycn, Raf1, Ret, Tnf, S100a4 and Ccnd1) and tumor suppressor genes (Hic1, Cdh1, Mgmt, Nf2 and Trp73) in MYH9 silenced B16F10 cells compared to control cells. However, lower expression of oncogenes (Met, Men1 and Ros1) and tumor suppressor genes (Runx3, Serpinb5 and Wt1) was observed in MYH9 silenced B16F10 cells compared to control cells (Fig. 2c), indicating MYH9 as tumor suppressor in B16F10 cells tumorigenesis.
Next, to find out the associated possible mechanism, we assessed the phosphorylation of Erk signaling pathways and found increased activation of Erk in MYH9 silenced B16F10 cells compared to control cells, suggesting its involvement with MYH9 signaling (Fig. 2b).
MYH9 silencing promotes murine melanoma cell tumor formation and lung metastasis in vivo
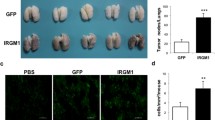
To further investigate the role of MYH9 in melanoma tumor formation in vivo, we injected control shRNA and MYH9 shRNA transfected B16F10 cells subcutaneously in C57BL/6 mice. After 15 days, mice were euthanized and our results showed increased tumor size with MYH9 silenced B16F10 cells compared to control (Fig. 3a–c) suggesting MYH9 as a tumor suppressor in melanoma.
MYH9 knockdown in mouse melanoma cells induce tumor development and modulate tumor microenvironment in vivo. a, b Representative images showing size and volume of tumors, c hematoxylin & eosin staining images (H&E) in C57BL/6 mice injected with control and transfected B16F10 cells subcutaneously. d Immunohistochemical analysis for proliferation (Ki-67), angiogenesis (CD34) and e leukocyte (CD45) and macrophage (F4/80) infiltration in subcutaneous tumors. All data are represented as mean ± SD. The student’s t-test was used for all statistical analyses (**P ≤ 0.01)
Next, to identify the role of MYH9 in melanoma cells lung metastasis, we injected control and MYH9 shRNA transfected B16F10 cells intravenously through tail vein in C57BL/6 mice and after 21 days mice injected with MYH9 shRNA B16F10 cells showed a large number of tumor foci in the lung compared to control (Fig. 4a, b) establishing the inhibitory role of MYH9 in melanoma metastasis.
MYH9 knockdown increases mouse melanoma cell’s lung metastasis and modulate associated tumor microenvironment in vivo. a Representative images showing tumors, b hematoxylin & eosin staining images (H&E) in the lung after intravenous injection of control and transfected B16F10 cells. c Immunohistochemical analysis for proliferation (Ki-67), angiogenesis (CD34) and d leukocyte (CD45) and macrophage (F4/80) infiltration in intravenous tumors from C57BL/6 mice after injecting control and shRNA transfected B16F10 cells. e Schematic representation of MYH9 role in melanoma tumorigenesis
MYH9 silencing promotes proliferation, angiogenesis and modulates tumor microenvironment in melanoma cell tumors
The tumor microenvironment includes tumor cells and non-tumor cells such as infiltrating immune cells; and mutual interaction between these cells reprogram the tumor development. Therefore, we explored the role of MYH9 in immune cells infiltrating in melanoma tumors. In this study immunohistochemical analysis of subcutaneous tumor sections displayed that the MYH9 knocking down induces cell proliferation (Ki-67), tumor angiogenesis (CD34) and inflammatory cell recruitment (CD45) and macrophage (F4/80) than shRNA control tumors (Fig. 3d, e).
Similarly, in the metastasis study, lung tumor sections also showed high expressions of cell proliferation and angiogenesis markers. Further, leukocytes and macrophage infiltration were also high in MYH9 silenced B16F10 cell tumors compared to control tumors (Fig. 4c, d). Together, these results indicate the inhibitory role of MYH9 in B16F10 cells tumorigenesis by manipulating the tumor microenvironment in metastasis (Fig. 4e).
Discussion
MYH9 has various physiological and pathological functions; however, it’s role in melanoma is not known. In the present study, we showed for the first time that MYH9 acts as a tumor suppressor in melanoma. In detail, MYH9 silencing promotes various in vitro tumorigenic properties of B16F10 cells. Further, oncogenic and EMT markers were also upregulated in MYH9 silenced melanoma cells. Furthermore, large size of tumors in subcutaneous and large numbers of tumor foci in lung metastasis was observed with increase infiltration of leukocytes and macrophages in MYH9 silenced B16F10 tumors that indicated a critical role of MYH9 in the modulation of tumor microenvironment. Together, our findings demonstrated MYH9 as a tumor suppressor in mouse melanoma tumor development and metastasis.
MYH9 has many physiological functions such as cell adhesion, polarity, and motility as skeletal muscle development and differentiation, smooth muscle tension maintenance as a skeleton protein, podocyte cytoskeletal structure, and various motorized functions require the involvement of MYH9 [16, 17]. Besides these properties, it also modulates the invasion and metastasis of various cancers [13, 14, 18, 19]. To date, the role of MYH9 in cancer development is controversial; however, its role in tumorigenesis may depend on various cancer cell types. Various studies have shown that in vitro tumorigenic properties including cell proliferation, migration and invasion are mainly connected to cancer progression and metastasis in in vivo tumorigenesis [20,21,22,23]. In the present study, MYH9 knockdown in B16F10 mouse melanoma cells induced cell migration and invasion in vitro. MYH9 role in cancer is context dependent and act both as tumor promotor and suppressor in different cancers, our result is consistent with head and neck cancer where MYH9 silencing increase cell invasion [24]. Further, PCR array analysis showed higher expression of oncogenes Bcl2, Egf, Erbb2, JunD, Kras, Mdm2, Mos, Mycn, Raf1, Ret, Tnf, S100a4 and Ccnd1 in MYH9 silenced B16F10 cells indicates a tumor-suppressive role of MYH9 in melanoma cells. Interestingly, our results showed the expression of E-cadherin; a tumor suppressor gene is found to be associated with higher migration and invasion in B16F10 cells. Various studies showed that the invasive and metastatic cancers including breast cancer, prostate cancer, brain cancer and ovarian cancer express higher level of E-cadherin, indicating role of E-cadherin in induction metastasis in some cancers rather than inhibiting cancer progression [25,26,27,28,29,30].
Further, we investigated the mechanism involved in MYH9 mediated suppression of in vitro tumorigenesis. In this study, we found higher activation of Erk. Next, to validate in vitro findings, we injected MYH9 silenced B16F10 cells subcutaneously and intravenously to check the role of MYH9 in tumor formation and metastasis. MYH9 silenced cells formed a large size of the tumor and a higher number of tumor foci in the lung that showed the tumor-suppressive role of MYH9 in melanoma. Similarly, in head and neck cancer, p53 expression was associated with MYH9 expression and MYH9 silencing induced tumor formation [24] and in another study, MYH9 silencing triggers invasive squamous cell carcinomas development [18].
To further find out the association of MYH9 in the modulation of the establishment of the tumor microenvironment, we evaluated tumor microenvironment in MYH9 silenced tumors and found higher proliferation, angiogenesis and infiltration of higher leukocytes and macrophages in MYH9 silenced B16F10 cells tumors. Tumor microenvironment plays a critical role in tumor establishment as the mutual communication between tumor and non-tumor cells provide a favorable environment to tumor cells growth [31,32,33,34]. In this study, MYH9 silenced melanoma cells promote the leucocytes and macrophages migration in tumor milieu that leads to the large size of tumor development and lung metastasis. Previous studies showed the role of macrophages in tumor invasion and metastasis of tumor cells [35, 36]. Certainly, these findings indicate that MYH9 suppresses tumorigenesis by manipulating the tumor microenvironment.
In conclusion, this study showed MYH9 silencing induces in vitro tumor cell properties and in vivo melanoma cells tumorigenesis and metastasis. Further, MYH9 regulates EMT-markers and the shaping of tumor microenvironment. However, further studies are required to elaborate on the MYH9 mediated tumor microenvironment manipulation and tumor formation. Collectively, our study indicates that MYH9 could be a promising therapeutic candidate for controlling the progression and metastasis of melanoma.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA: Cancer J Clin. 2018;68(1):7–30. https://doi.org/10.3322/caac.21442.
Whiteman DC, Green AC, Olsen CM. The growing burden of invasive melanoma: projections of incidence rates and numbers of new cases in six susceptible populations through 2031. J Investig Dermatol. 2016;136(6):1161–71. https://doi.org/10.1016/j.jid.2016.01.035.
Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331(6024):1559–644. https://doi.org/10.1126/science.1203543.
Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147(2):275–92. https://doi.org/10.1016/j.cell.2011.09.024.
Liu Q, Zhang H, Jiang X, Qian C, Liu Z, Luo D. Factors involved in cancer metastasis: a better understanding to “seed and soil” hypothesis. Mol Cancer. 2017;16(1):176. https://doi.org/10.1186/s12943-017-0742-4.
Bresnick AR. Molecular mechanisms of nonmuscle myosin-II regulation. Curr Opin Cell Biol. 1999;11(1):26–33. https://doi.org/10.1016/s0955-0674(99)80004-0.
Bondzie PA, Chen HA, Cao MZ, Tomolonis JA, He F, Pollak MR, et al. Non-muscle myosin-IIA is critical for podocyte f-actin organization, contractility, and attenuation of cell motility. Cytoskeleton (Hoboken). 2016;73(8):377–95. https://doi.org/10.1002/cm.21313.
Betapudi V, Gokulrangan G, Chance MR, Egelhoff TT. A proteomic study of myosin II motor proteins during tumor cell migration. J Mol Biol. 2011;407(5):673–86. https://doi.org/10.1016/j.jmb.2011.02.010.
Hirata N, Takahashi M, Yazawa M. Diphosphorylation of regulatory light chain of myosin IIA is responsible for proper cell spreading. Biochem Biophys Res Commun. 2009;381(4):682–7. https://doi.org/10.1016/j.bbrc.2009.02.121.
Beadle C, Assanah MC, Monzo P, Vallee R, Rosenfeld SS, Canoll P. The role of myosin II in glioma invasion of the brain. Mol Biol Cell. 2008;19(8):3357–68. https://doi.org/10.1091/mbc.E08-03-0319.
Sabbir MG, Dillon R, Mowat MR. Dlc1 interaction with non-muscle myosin heavy chain II-A (Myh9) and Rac1 activation. Biol Open. 2016;5(4):452–60. https://doi.org/10.1242/bio.015859.
Derycke L, Stove C, Vercoutter-Edouart AS, De Wever O, Dolle L, Colpaert N, et al. The role of non-muscle myosin IIA in aggregation and invasion of human MCF-7 breast cancer cells. Int J Dev Biol. 2011;55(7–9):835–40. https://doi.org/10.1387/ijdb.113336ld.
Betapudi V, Licate LS, Egelhoff TT. Distinct roles of nonmuscle myosin II isoforms in the regulation of MDA-MB-231 breast cancer cell spreading and migration. Cancer Res. 2006;66(9):4725–33. https://doi.org/10.1158/0008-5472.can-05-4236.
Katono K, Sato Y, Jiang SX, Kobayashi M, Nagashio R, Ryuge S, et al. Prognostic significance of MYH9 expression in resected non-small cell lung cancer. PLoS One. 2015;10(3):e0121460. https://doi.org/10.1371/journal.pone.0121460.
Maeda J, Hirano T, Ogiwara A, Akimoto S, Kawakami T, Fukui Y, et al. Proteomic analysis of stage I primary lung adenocarcinoma aimed at individualisation of postoperative therapy. Br J Cancer. 2008;98(3):596–603. https://doi.org/10.1038/sj.bjc.6604197.
Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol. 2009;10(11):778–90. https://doi.org/10.1038/nrm2786.
Conti MA, Adelstein RS. Nonmuscle myosin II moves in new directions. J Cell Sci. 2008;121(Pt 1):11–8. https://doi.org/10.1242/jcs.007112.
Schramek D, Sendoel A, Segal JP, Beronja S, Heller E, Oristian D, et al. Direct in vivo RNAi screen unveils myosin IIa as a tumor suppressor of squamous cell carcinomas. Science. 2014;343(6168):309–13. https://doi.org/10.1126/science.1248627.
Xia ZK, Yuan YC, Yin N, Yin BL, Tan ZP, Hu YR. Nonmuscle myosin IIA is associated with poor prognosis of esophageal squamous cancer. Dis Esophagus. 2012;25(5):427–36. https://doi.org/10.1111/j.1442-2050.2011.01261.x.
Shin DH, Kim OH, Jun HS, Kang MK. Inhibitory effect of capsaicin on B16-F10 melanoma cell migration via the phosphatidylinositol 3-kinase/Akt/Rac1 signal pathway. Exp Mol Med. 2008;40(5):486–94. https://doi.org/10.3858/emm.2008.40.5.486.
Zhao K, Wei L, Hui H, Dai Q, You QD, Guo QL, et al. Wogonin suppresses melanoma cell B16-F10 invasion and migration by inhibiting Ras-medicated pathways. PLoS One. 2014;9(9):e106458. https://doi.org/10.1371/journal.pone.0106458.
Koszalka P, Pryszlak A, Golunska M, Kolasa J, Stasilojc G, Skladanowski AC, et al. Inhibition of CD73 stimulates the migration and invasion of B16F10 melanoma cells in vitro, but results in impaired angiogenesis and reduced melanoma growth in vivo. Oncol Rep. 2014;31(2):819–27. https://doi.org/10.3892/or.2013.2883.
Razak NA, Abu N, Ho WY, Zamberi NR, Tan SW, Alitheen NB, et al. Cytotoxicity of eupatorin in MCF-7 and MDA-MB-231 human breast cancer cells via cell cycle arrest, anti-angiogenesis and induction of apoptosis. Sci Rep. 2019;9(1):1514. https://doi.org/10.1038/s41598-018-37796-w.
Coaxum SD, Tiedeken J, Garrett-Mayer E, Myers J, Rosenzweig SA, Neskey DM. The tumor suppressor capability of p53 is dependent on non-muscle myosin IIA function in head and neck cancer. Oncotarget. 2017;8(14):22991–3007. https://doi.org/10.18632/oncotarget.14967.
Christiansen JJ, Rajasekaran AK. Reassessing epithelial to mesenchymal transition as a prerequisite for carcinoma invasion and metastasis. Cancer Res. 2006;66(17):8319–26. https://doi.org/10.1158/0008-5472.can-06-0410.
Nieman MT, Prudoff RS, Johnson KR, Wheelock MJ. N-cadherin promotes motility in human breast cancer cells regardless of their E-cadherin expression. J Cell Biol. 1999;147(3):631–44. https://doi.org/10.1083/jcb.147.3.631.
Kowalski PJ, Rubin MA, Kleer CG. E-cadherin expression in primary carcinomas of the breast and its distant metastases. Breast Cancer Res. 2003;5(6):R217–22. https://doi.org/10.1186/bcr651.
Putzke AP, Ventura AP, Bailey AM, Akture C, Opoku-Ansah J, Celiktas M, et al. Metastatic progression of prostate cancer and e-cadherin regulation by zeb1 and SRC family kinases. Am J Pathol. 2011;179(1):400–10. https://doi.org/10.1016/j.ajpath.2011.03.028.
Lewis-Tuffin LJ, Rodriguez F, Giannini C, Scheithauer B, Necela BM, Sarkaria JN, et al. Misregulated E-cadherin expression associated with an aggressive brain tumor phenotype. PLoS One. 2010;5(10):e13665. https://doi.org/10.1371/journal.pone.0013665.
Reddy P, Liu L, Ren C, Lindgren P, Boman K, Shen Y, et al. Formation of E-cadherin-mediated cell-cell adhesion activates AKT and mitogen activated protein kinase via phosphatidylinositol 3 kinase and ligand-independent activation of epidermal growth factor receptor in ovarian cancer cells. Mol Endocrinol. 2005;19(10):2564–78. https://doi.org/10.1210/me.2004-0342.
Kwak T, Drews-Elger K, Ergonul A, Miller PC, Braley A, Hwang GH, et al. Targeting of RAGE-ligand signaling impairs breast cancer cell invasion and metastasis. Oncogene. 2017;36(11):1559–722. https://doi.org/10.1038/onc.2016.324.
Schiavoni G, Gabriele L, Mattei F. The tumor microenvironment: a pitch for multiple players. Front Oncol. 2013;3:90. https://doi.org/10.3389/fonc.2013.00090.
Nasser MW, Wani NA, Ahirwar DK, Powell CA, Ravi J, Elbaz M, et al. RAGE mediates S100A7-induced breast cancer growth and metastasis by modulating the tumor microenvironment. Cancer Res. 2015;75(6):974–85. https://doi.org/10.1158/0008-5472.can-14-2161.
Furudate S, Fujimura T, Kambayashi Y, Kakizaki A, Hidaka T, Aiba S. Immunomodulatory effect of imiquimod through CCL22 produced by tumor-associated macrophages in B16F10 melanomas. Anticancer Res. 2017;37(7):3461–71. https://doi.org/10.21873/anticanres.11714.
Cassetta L, Fragkogianni S, Sims AH, Swierczak A, Forrester LM, Zhang H, et al. Human tumor-associated macrophage and monocyte transcriptional landscapes reveal cancer-specific reprogramming, biomarkers, and therapeutic targets. Cancer Cell. 2019;35(4):588–602.e10. https://doi.org/10.1016/j.ccell.2019.02.009.
Lin Y, Xu J, Lan H. Tumor-associated macrophages in tumor metastasis: biological roles and clinical therapeutic applications. J Hematol Oncol. 2019;12(1):76. https://doi.org/10.1186/s13045-019-0760-3.
Acknowledgements
This work was supported, in whole or in part, by Grants-in-Aid from the Department of Biotechnology (DBT), New Delhi Grant Numbers, 102/IFD/SAN/2237, BT/PR13045/BRB/10/1461/2015, Department of Science and Technology, Grant Number EMR/2016/003932 and Institute of Life Sciences, Bhubaneswar, India core fund. Satyendra Kumar Singh duly acknowledges the financial assistance from SERB N-PDF Grant (PDF/2017/001017), New Delhi, India.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Singh, S.K., Sinha, S., Padhan, J. et al. MYH9 suppresses melanoma tumorigenesis, metastasis and regulates tumor microenvironment. Med Oncol 37, 88 (2020). https://doi.org/10.1007/s12032-020-01413-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-020-01413-6