Abstract
Current evidence suggests that the human genome produces a large number of non-coding RNAs, including microRNAs and long non-coding RNAs (lncRNAs). Generally, lncRNAs are defined as RNA transcripts longer than 200 nucleotides that are not transcribed into proteins. In recent years, lncRNAs have been reported to play oncogenic roles in tumourigenesis. However, minimal research has been performed on the expression and clinicopathological significance of lncRNAs in papillary thyroid cancer (PTC). In the present study, we investigated not only the expression and clinicopathological significance of a novel lncRNA, NR_036575.1, in PTC tissues and adjacent non-cancerous tissues but also its potential function in TPC1 cells. The expression levels of the lncRNA NR_036575.1 in 83 pairs of PTC tissues and adjacent non-cancerous tissues were detected via quantitative real-time polymerase chain reaction. The relationships between the expression levels and clinicopathological characteristics of the lncRNA NR_036575.1 were analysed. In addition, we established two receiver operating characteristic (ROC) curves to assess the diagnostic value of NR_036575.1 expression. Cell Counting Kit-8 and transwell assays were used to assess cell proliferation and migration, respectively. The expression levels of the lncRNA NR_036575.1 were significantly higher in PTC tissues than in adjacent non-cancerous tissues. High NR_036575.1 expression was associated with extrathyroidal extension (ETE) (P = 0.011) and tumour size (P = 0.006). The ROC curves indicated that NR_036575.1 could potentially serve as a biomarker for identifying PTC and related, non-cancerous diseases (sensitivity, 80.7 %; specificity, 88 %), as well as for differentiating between PTC with or without ETE (sensitivity, 57.8 %; specificity, 86.7 %). NR_036575.1 knock-down significantly inhibited the proliferation and migration of TPC1 cells. Our findings are the first to describe lncRNA NR_036575.1 overexpression in PTC. NR_036575.1 expression was associated with both ETE and tumour size. In addition, NR_036575.1 modulation could regulate TPC1 cell proliferation and migration. The results of our study suggest that NR_036575.1 could be applied as a potential biomarker and a novel therapeutic target for PTC patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Thyroid cancer is a common malignancy that has exhibited a dramatically increasing incidence rate over the past several decades. Papillary thyroid cancer (PTC) is the most common histological subtype of thyroid cancer, constituting 80–85 % of all thyroid malignancies [1]. While disease-specific mortality is low for PTC patients, persistent and recurrent disease rates remain significant [2]. A few clinicopathological features, such as lymph node metastasis, extrathyroidal extension (ETE), larger tumour size (≥3 cm), and advanced tumour-node-metastasis (TNM) stage, have been associated with the necessity for extensive operations or a poor prognosis. PTC patients with ETE usually experience incomplete resection due to invasion into surrounding structures, as well as a high incidence of lymph node metastasis and recurrence [3]. ETE has also been studied as an independent prognostic factor of survival [4, 5]. Thus, exploring potential predictive biomarkers of ETE in PTC patients is becoming increasingly important.
Long non-coding RNAs (lncRNAs) are RNAs consisting of more than 200 nucleotides [6]. Similarly to microRNAs, lncRNAs do not encode proteins [7]. Originally, lncRNAs were considered to represent spurious transcriptional noise without a biological function due to their incapacity to encode proteins [8]. Recently, accumulating evidence suggests that lncRNAs have multiple functions in tumours over a wide range of biological processes, such as proliferation, apoptosis, and cell cycle arrest [9]. In our previous study (GSE66783) [10], we found that the novel lncRNA NR_036575.1 was significantly overexpressed in PTC tissues compared with adjacent non-cancerous tissues; our microarray assay results indicated that the expression of this lncRNA was 18.91-fold greater in PTC tissues than in adjacent non-cancerous tissues (P < 0.05). However, the expression, clinicopathological significance, and function of NR_036575.1 in PTC tissues and cell lines require further investigation on a large scale. The gene for NR_036575.1 is located at Chr19: 18360760-18366229, on the negative strand. The transcript of NR_036575.1 is 687 nucleotides in length. In the present study, we investigated the expression of NR_036575.1 in PTC tissues and adjacent non-cancerous tissues and then analysed the potential relationships between NR_036575.1 levels and clinicopathological features.
Methods
Eighty-three fresh PTC and adjacent non-cancerous tissues were collected from patients who underwent thyroid surgery at the First Hospital of China Medical University between 2009 and 2015. All of the specimens were immediately frozen in liquid nitrogen and stored at −80 °C until later use. The diagnosis of PTC was pathologically confirmed either intra- or postoperatively. None of the patients recruited for this study had previously undergone head and neck irradiation or oncological surgery. Written informed consent for the experimental use of the surgical specimens was obtained from all of the patients, according to the hospital’s ethical guidelines.
Total RNA isolation and qRT-PCR
Total RNA was extracted from the tumour tissues and adjacent non-cancerous tissues using RNAiso Plus (TaKaRa, Japan). A reverse transcription kit (RR036A, Takara, Japan) was used to transcribe total RNA and produce cDNA. Then, qRT-PCR was performed with SYBR Premix Ex Taq II (TaKaRa, Japan) and a LightCycler 480 system (Roche, USA). The qRT-PCR cycling profile was as follows: 95 °C for 30 s, followed by 50 cycles of 95 °C for 5 s and 60 °C for 30 s. The following sequences were used: NR_036575.1 S: 5′-CCCCGTGAGTTACACAGGAA-3′, AS: 5′-TCATTCACGGATCCCATGGAC-3′; GAPDH S: 5′-CAGGAGGCATTGCTGATGAT-3′, AS: 5′-GAAGGCTGGGGCTCATTT-3′. The double-standard curves method was used to calculate the relative expression of NR_036575.1.
Cell culture and transfection
The Nthy-ori 3-1 cell line was obtained from the European Collection of Cell Culture (ECACC, England). The TPC1 cell line was a gift from Professor Meiping Shen (Department of General Surgery, The First Affiliated Hospital of Nanjing Medical University, Nanjing, Jiangsu). IHH-4 cells were obtained from the Health Science Research Resources Bank (Osaka, Japan). The Nthy-ori 3-1 cells were maintained in RPMI 1640 supplemented with 10 % foetal bovine serum (FBS), the TPC1 cells were maintained in DMEM with 15 % FBS, and the IHH-4 cells were maintained in a 1:1 mixture of RPMI 1640 and DMEM, supplemented with 10 % FBS. The NR_036575.1 siRNA and the negative control (NC) were purchased from GenePharma (Shanghai, China). Three siRNAs were initially designed and used in our study. However, preliminary qRT-PCR results indicated that both siRNA-1 and siRNA-2 had a significantly higher silencing efficiency than siRNA-3. Thus, siRNA-1 and siRNA-2 were chosen for use in the cellular physiological function assays. The sequences of siRNA-1, siRNA-2, and siRNA-3 were 5′-UCAUCGGGUCUGUGGUCAUTT-3′, 5′-GAUCCGUGAAUGACAGAUUTT-3′, and 5′-GCUGGUGUGAAUUACAAUATT-3′, respectively. The sequence of the NC was 5′-UUCUCCGAACGUGUCACGUTT-3′. The TPC1 cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol.
Cell proliferation assay
Cell proliferation was assessed using the Cell Counting Kit-8 (CCK-8) assay (Dojindo, Kumamoto, Japan); 3 × 103 TPC1 cells per well were seeded onto a 96-well plate in a final volume of 100 μl and were then transfected with NR_036575.1 siRNA or the NC. The samples were evaluated at 0, 24, 48, and 72 h after gene transfection; 10 μl of CCK-8 solution was added to each well, and the cells were then incubated for 3 h at 37 °C. Absorbance was measured at 450 nm to calculate the number of viable cells.
Cell migration assay
A transwell assay was used to assess cell migration (Corning Co. Ltd., USA). At 24 h after being transfected with NR_036575.1 siRNA or the NC, a total of 1 × 105 cells in 100 μl of serum-free medium were added to each well. After 6 h of incubation at 37 °C, the cells that had migrated through the filters were fixed with methanol and stained with 0.5 % crystal violet; cells in five random fields were counted under a microscope at 100× magnification. Each experiment was performed in triplicate.
Statistical analysis
All statistical analyses were performed using SPSS 20.0 (IBM, SPSS, Chicago, IL, USA). The Mann–Whitney U test was used to compare the expression of the lncRNA NR_036575.1 in the PTC and adjacent non-cancerous tissues. The Chi-square test was applied to assess the relationship between NR_036575.1 expression and clinicopathological characteristics. ROC curves were established to evaluate the diagnostic value of NR_036575.1 for differentiating between PTC and non-cancerous tissues and for identifying PTC with or without ETE. The t test was used for between-group comparisons. A difference was considered significant if the P value was <0.05.
Results
NR_036575.1 was overexpressed in PTC tissues and TPC1 cells
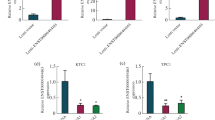
The expression of NR_036575.1 in 83 pairs of PTC and adjacent non-cancerous tissues was detected and analysed using qRT-PCR. NR_036575.1 expression was significantly elevated in human PTC tissue samples compared with paired adjacent non-cancerous tissue samples (Fig. 1a, P < 0.01). The median fold-change value was 7.73. The expression of NR_036575.1 was detected in two PTC cell lines (i.e. TPC1 and IHH-4) and in a normal human thyroid follicular epithelial cell line (i.e. Nthy-ori 3-1). We found that NR_036575.1 expression was higher in TPC1 cells than in Nthy-ori 3-1 cells (Fig. 1b).
Expression of NR_036575.1 in PTC tissues and PTC cell lines. a NR_036575.1 expression in 83 pairs of PTC and adjacent non-cancerous tissues. The expression level of NR_036575.1 was significantly higher in PTC tissues than in the paired, adjacent non-cancerous tissues, as determined by qRT-PCR. b NR_036575.1 expression in TPC1, IHH-4, and Nthy-ori 3-1 cells. The expression level of NR_036575.1 was significantly higher in TPC1 PTC cells than in Nthy-ori 3-1 normal human thyroid follicular epithelial cells. *P < 0.05, **P < 0.01
Correlations of NR_036575.1 expression with PTC clinicopathological features
To further understand the significance of NR_036575.1 in PTC, we examined the correlations of NR_036575.1 expression with the clinical pathological features of PTC and adjacent non-cancerous tissues. The median fold-change value of lncRNA NR_036575.1 expression in PTC tissues compared with adjacent non-cancerous tissues was 7.73, and this was used as a cut-off value. All of the patients were divided into the following two groups: high NR_036575.1 expression (≥7.73; n = 42) and low NR_036575.1 expression (<7.73; n = 41). The clinicopathological features of the 83 PTC patients are shown in Table 1. The large difference in NR_036575.1 expression between the PTC and adjacent non-cancerous tissues was significantly associated with tumour size (P = 0.006) and ETE (P = 0.011) in PTC (Table 1).
Diagnostic value of NR_036575.1
Next, we explored the diagnostic value of NR_036575.1. ROC curves were applied to evaluate whether NR_036575.1 could serve as a biomarker for predicting benign or malignant status. The area under the curve (AUC) was 0.915 (95 % CI 0.874–0.951, P < 0.001), suggesting that NR_036575.1 could be applied as a promising biomarker for PTC (Fig. 2a). The sensitivity and specificity values were 80.7 and 88 %, respectively, when the cut-off value was 0.34. The cut-off values for predicting benign or malignant status refer to the relative expression in the PTC and adjacent non-cancerous tissues, as determined by qRT-PCR. Moreover, we found that NR_036575.1 might be a diagnostic biomarker for ETE (Fig. 2b). The AUC was 0.704 (95 % CI 0.588–0.820, P = 0.001), and the sensitivity and specificity values were 57.8 and 86.7 %, respectively, when the cut-off value was 11.19. The cut-off values for predicting ETE refer to the fold-change in expression between the PTC and adjacent non-cancerous tissues, as determined by qRT-PCR.
Diagnostic value of lncRNA NR_036575.1. a Diagnostic value of lncRNA NR_036575.1 for differentiating between PTC and non-cancerous tissues. The area under the curve (AUC) was 0.915 (95 % CI 0.874–0.951, P < 0.001). The sensitivity was 80.7 %, and the specificity was 88 %. b Diagnostic value of lncRNA NR_036575.1 for differentiating between PTC with and without ETE. The AUC was 0.704 (95 % CI 0.588–0.820, P = 0.001). The sensitivity was 57.8 %, and the specificity was 86.7 %
Knock-down of NR_036575.1 inhibits cell proliferation and migration in vitro
Three siRNAs were initially designed to silence NR_036575.1. However, after assessing their silencing efficiency using qRT-PCR, both siRNA-1 and siRNA-2 were found to have a silencing efficiency significantly higher than siRNA-3. Thus, siRNA-1 and siRNA-2 were chosen for use in the cellular physiological function assays (Fig. 3a). We conducted CCK-8 cell counting assays and transwell assays to determine the influence of NR_036575.1 in vitro. We observed that compared with the control treatment, NR_036575.1 knock-down in TPC1 cells led to a notable reduction in cell viability (Fig. 3b). NR_036575.1 knock-down in TPC1 cells also resulted in significantly decreased cell migration ability (Fig. 3c, P < 0.01). Collectively, these data suggested that NR_036575.1 contributes to the proliferation and migration of TPC1 cells.
NR_036575.1 knock-down inhibits cell proliferation and migration in TPC1 cells. a NR_036575.1 expression in TPC1 cells after siRNA transfection. The expression level of NR_036575.1 was significantly lower in the siRNA-1 and siRNA-2 groups than in the NC group. However, there were no significant differences between the NC and siRNA-3 groups. b NR_036575.1 knock-down inhibits cell proliferation. The CCK-8 cell counting assay was applied to analyse the proliferation of TPC1 cells. Cell proliferation was significantly inhibited in the NR_036575.1 siRNA groups. c NR_036575.1 knock-down inhibits cell migration. The transwell assay was applied to analyse the migration of TPC1 cells. Cell migration was significantly inhibited in the NR_036575.1 siRNA groups. *P < 0.05, **P < 0.01
Discussion
Thyroid cancer is the most common malignant tumour of the endocrine system, and its incidence has been steadily increasing over the past few decades [1, 11]. The proximity of the thyroid gland to the aerodigestive tract (i.e. larynx, trachea, pharynx, and oesophagus), recurrent laryngeal nerve, strap muscles, and carotid artery renders all these tissues susceptible to thyroid cancer invasion [12]. The direct invasion of these structures occurs in 7–31.9 % of PTC patients [3, 13]. Some investigators have proposed that ETE does not impact disease recurrence, even though ETE has previously been associated with large tumour size and multifocality [14, 15]. However, a large amount of evidence indicates that local invasion is a critical determinant of ultimate disease control, overall morbidity, and mortality [12]. ETE is also considered to be a risk factor associated with more extensive surgeries [16]. PTC patients with ETE usually have a worse prognosis than those without ETE [12, 17]. ETE is commonly evaluated preoperatively using ultrasound imaging and contrast-enhanced computed tomography; however, the predictive value of this approach is limited [18, 19]. Therefore, it is necessary to identify potential biomarkers for preoperatively predicting the occurrence of ETE.
Over 90 % of the mammalian genome represents non-coding RNAs (ncRNAs), which are transcribed but do not encode proteins. miRNAs and lncRNAs are the two main ncRNA types, and the deregulated expression of these RNAs is considered to play an important role in tumour development and progression [20]. Many miRNA biomarkers have been reported for the differential diagnosis of thyroid neoplasms using fine-needle aspiration biopsy (FNAB) samples. Pallante et al. found that miR-221, miR-222, and miR-181b had changes in expression ranging from 5- to 35-fold in FNAB samples of PTC compared with that in samples from other thyroid nodules [21]. Mazeh et al. found that miR-221 could be applied as a biomarker for differentiating benign and malignant thyroid nodules. The specificity of this marker was 100 %, and the negative and positive predictive values were 96 and 100 %, respectively [22]. These findings demonstrate that specific miRNAs could potentially serve as highly accurate diagnostic tools in preoperative FNAB samples. Recently, many studies have shown that lncRNAs are not only frequently dysregulated in various tumours but also have multiple functions in a wide range of biological processes [9]. Although the mechanisms of lncRNA involvement in tumourigenesis have not yet been thoroughly elucidated, a few studies have initiated research on the diagnostic value of lncRNAs in cancer. For example, the expression levels of three lncRNAs in serum, CUDR, LSINCT-5, and PTENP1, have been reported to be potential diagnostic markers of gastric cancer [23]. The lncRNA POU3F3 reportedly has a high diagnostic value for oesophageal squamous cell carcinoma [24]. However, studies on lncRNAs as potential biomarkers for and therapeutic targets of thyroid cancer are limited. The size of a PTC tumour is an important factor in TNM staging; large tumours are more likely to be aggressive [25]. In our study, we found that the lncRNA NR_036575.1 was significantly overexpressed in thyroid cancer tissues. In addition, the expression of NR_036575.1 was associated with tumour size and ETE. The lncRNA NR_036575.1 may be a diagnostic biomarker for thyroid cancer (sensitivity, 80.7 %; specificity, 88 %). The fold-change of NR_036575.1 expression in PTC tissues relative to that in adjacent non-cancerous tissues was a predictive marker for ETE (sensitivity, 57.8 %; specificity, 86.7 %). Thus, NR_036575.1 can be applied as a biomarker for thyroid cancer and ETE. Furthermore, we found that NR_036575.1 was overexpressed in PTC cell lines and NR_036575.1 knock-down inhibited TPC1 cell proliferation and migration in vitro.
This study has several limitations. First, the mechanisms of NR_036575.1 involvement in PTC remain incompletely understood. Second, large-scale studies with long-term follow-ups will be necessary to confirm our results.
In conclusion, we have provided the first description of the novel lncRNA NR_036575.1 being overexpressed in PTC, and we found that the level of NR_036575.1 expression was correlated with ETE and tumour size. NR_036575.1 may be a potential diagnostic biomarker and a novel therapeutic target for PTC.
References
Chen AY, Jemal A, Ward EM. Increasing incidence of differentiated thyroid cancer in the United States, 1988–2005. Cancer. 2009;115(16):3801–7.
Davies L, Welch HG. Thyroid cancer survival in the United States: observational data from 1973 to 2005. Arch Otolaryngol Head Neck Surg. 2010;136(5):440–4.
Wang Z, Zhang H, He L, Dong W, Li J, Shan Z, et al. Association between the expression of four upregulated miRNAs and extrathyroidal invasion in papillary thyroid carcinoma. Onco Targets Ther. 2013;6:281–7.
Nishida T, Nakao K, Hashimoto T. Local control in differentiated thyroid carcinoma with extrathyroidal invasion. Am J Surg. 2000;179(2):86–91.
McCaffrey TV, Bergstralh EJ, Hay ID. Locally invasive papillary thyroid carcinoma: 1940–1990. Head Neck. 1994;16(2):165–72.
Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–41.
Dang Y, Lan F, Ouyang X, Wang K, Lin Y, Yu Y, et al. Expression and clinical significance of long non-coding RNA HNF1A-AS1 in human gastric cancer. World J Surg Oncol. 2015;13:302.
Li Y, Chen W, He F, Tan Z, Zheng J, Wang W, et al. NEAT expression is associated with tumor recurrence and unfavorable prognosis in colorectal cancer. Oncotarget. 2015;6(29):27641–50.
Yu G, Yao W, Gumireddy K, Li A, Wang J, Xiao W, et al. Pseudogene PTENP1 functions as a competing endogenous RNA to suppress clear-cell renal cell carcinoma progression. Mol Cancer Ther. 2014;13(12):3086–97.
Lan X, Zhang H, Wang Z, Dong W, Sun W, Shao L, et al. Genome-wide analysis of long noncoding RNA expression profile in papillary thyroid carcinoma. Gene. 2015;569(1):109–17.
Davies L, Welch HG. Increasing incidence of thyroid cancer in the United States, 1973–2002. JAMA. 2006;295(18):2164–7.
Ark N, Zemo S, Nolen D, Holsinger FC, Weber RS. Management of locally invasive well-differentiated thyroid cancer. Surg Oncol Clin N Am. 2008;17(1):145–55.
Park YM, Wang SG, Goh JY, Shin DH, Kim IJ, Lee BJ. Intraoperative frozen section for the evaluation of extrathyroidal extension in papillary thyroid cancer. World J Surg. 2015;39(1):187–93.
Ahn D, Sohn JH, Jeon JH, Jeong JY. Clinical impact of microscopic extrathyroidal extension in patients with papillary thyroid microcarcinoma treated with hemithyroidectomy. J Endocrinol Invest. 2014;37(2):167–73.
Shin JH, Ha TK, Park HK, Ahn MS, Kim KH, Bae KB, et al. Implication of minimal extrathyroidal extension as a prognostic factor in papillary thyroid carcinoma. Int J Surg. 2013;11(9):944–7.
Kim WY, Kim HY, Son GS, Bae JW, Lee JB. Clinicopathological, immunohistochemical factors and recurrence associated with extrathyroidal extension in papillary thyroid microcarcinoma. J Cancer Res Ther. 2014;10(1):50–5.
Radowsky JS, Howard RS, Burch HB, Stojadinovic A. Impact of degree of extrathyroidal extension of disease on papillary thyroid cancer outcome. Thyroid. 2014;24(2):241–4.
Kwak JY, Kim EK, Youk JH, Kim MJ, Son EJ, Choi SH, et al. Extrathyroid extension of well-differentiated papillary thyroid microcarcinoma on US. Thyroid. 2008;18(6):609–14.
Choi JS, Kim J, Kwak JY, Kim MJ, Chang HS, Kim EK. Preoperative staging of papillary thyroid carcinoma: comparison of ultrasound imaging and CT. AJR Am J Roentgenol. 2009;193(3):871–8.
Kentwell J, Gundara JS, Sidhu SB. Noncoding RNAs in endocrine malignancy. Oncologist. 2014;19(5):483–91.
Pallante P, Visone R, Ferracin M, Ferraro A, Berlingieri MT, Troncone G, et al. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr Relat Cancer. 2006;13(2):497–508.
Mazeh H, Mizrahi I, Halle D, Ilyayev N, Stojadinovic A, Trink B, et al. Development of a microRNA-based molecular assay for the detection of papillary thyroid carcinoma in aspiration biopsy samples. Thyroid. 2011;21(2):111–8.
Dong L, Qi P, Xu MD, Ni SJ, Huang D, Xu QH, et al. Circulating CUDR, LSINCT-5 and PTENP1 long noncoding RNAs in sera distinguish patients with gastric cancer from healthy controls. Int J Cancer. 2015;137(5):1128–35.
Tong YS, Wang XW, Zhou XL, Liu ZH, Yang TX, Shi WH, et al. Identification of the long non-coding RNA POU3F3 in plasma as a novel biomarker for diagnosis of esophageal squamous cell carcinoma. Mol Cancer. 2015;14:3.
Tuttle RM, Haddad RI, Ball DW, Byrd D, Dickson P, Duh QY, et al. Thyroid carcinoma, version 2.2014. J Natl Compr Canc Netw. 2014;12:1671–80.
Acknowledgments
This work was supported by the Liaoning BaiQianWan Talents Program (No. 2014921033), Natural Science Foundation of Liaoning Province (No. 2015020536), Science and Technology Project of Shenyang City (No. F16-205-1-41), the Liaoning Province PhD Start-up Fund (Nos. 20141042 and 201501008), and the National Natural Science Foundation of China (Nos. 81402208 and 81502319).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest in relation to the publication of this study. The authors have full control of all of the primary data and agree to allow the journal to review the data if needed.
Ethical standards
Our study was approved by the Ethics Committee of the First Affiliated Hospital of China Medical University, Shenyang, China.
Informed consent
Written informed consent was obtained from all study participants.
Rights and permissions
About this article
Cite this article
Sun, W., Lan, X., Wang, Z. et al. Overexpression of long non-coding RNA NR_036575.1 contributes to the proliferation and migration of papillary thyroid cancer. Med Oncol 33, 102 (2016). https://doi.org/10.1007/s12032-016-0816-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-016-0816-y