Abstract
Alzheimer's disease (AD) is one of the most severe neurodegenerative diseases observed in the elderly population. Although the hallmarks of AD have been identified, the methods for its definitive diagnosis and treatment are still lacking. Extracellular vesicles (EVs) have become a promising source for biomarkers since the identification of their content. EVs are released from multiple cell types and, when released from neurons, they pass from the brain to the blood with their cargo molecules. Hence, neuron-specific EV-resident microRNAs (miRNAs) are promising biomarkers for diagnosis of AD. This study aimed to identify altered miRNA content in small neuron-derived extracellular vesicles (sNDEVs) isolated from AD patients and healthy individuals. Furthermore, we examined the role of sNDEV-resident miRNAs in neuron-glia cellular interaction to understand their role in AD propagation. We identified 10 differentially expressed miRNAs in the sNDEVs of patients via next-generation sequencing and validated the most dysregulated miRNA, let-7e, with qRT-PCR. Let-7e was significantly increased in the sNDEVs of AD patients compared with those of healthy controls in a larger cohort. First, we evaluated the diagnostic utility of let-7e via ROC curve analysis, which revealed an AUC value of 0.9214. We found that IL-6 gene expression was increased in human microglia after treatment with sNDEVs of AD patients with a high amount of let-7e. Our study suggests that sNDEV-resident let-7e is a potential biomarker for AD diagnosis, and that AD patient-derived sNDEVs induce a neuroinflammatory response in microglia.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Alzheimer's disease (AD) is a chronic and progressive neurodegenerative disease, the most common type of dementia with symptoms including memory loss, behavioral instability, and difficulties in daily activities (Patients 2019). Accumulation of amyloid beta (Aβ) plaques and the formation of neurofibrillary tangles containing hyperphosphorylated tau, which are considered hallmarks of AD, lead to degeneration of synapses, inflammation, and neuronal cell death (Masters et al. 2015; Blennow et al. 2006; Dalvi 2012). Current diagnostic methods are limited to histopathological analysis, imaging techniques—including positron emission tomography (PET) and magnetic resonance imaging (MRI)—and detection of cerebrospinal fluid (CSF) biomarkers such as Aβ42 and Tau protein, neurogranin, and synaptic proteins (Sancesario and Bernardini 2018). Since the clinical symptoms of AD appear many years after the formation of pathological changes in the brain, early diagnosis is critical to determine the pathogenesis and treatment of AD. In recent years, blood-based biomarkers, differentially expressed in AD patients, have been identified since blood sampling is more accessible and less invasive than CSF (Chen et al. 2019a). Furthermore, since CSF is not suitable for routine tests due to challenges in sample collection, studies examining blood-based biomarkers, varying from proteins and nucleic acids to other biomolecules, have gained importance for diagnosing and evaluating AD progression.
Extracellular vesicles (EVs) are a promising source for blood-based biomarkers. They are released from almost every cell type and are found in all bodily fluids. EVs range from 30 nm to 1 µm diameter in size, and the EVs 30–150 nm in size are called small EVs (sEVs) (Jella et al. 2018; Zhang et al. 2019; Xiao et al. 2017; Liao et al. 2019). They contain cargo molecules, including proteins, lipids, and nucleic acids, that reflect the features of their cells of origin. They participate in critical functions such as cellular communication, transfer of biomolecules, and initiation of immune responses via transfer of cargo molecules to the target/recipient cells (Xiao et al. 2017; Giau and An 2016). These findings, microRNA (miRNA) transport via EVs, have been shown as a cellular interaction mechanism among cells (Valadi et al. 2007). EVs are highly enriched with these small, non-protein-coding RNA species, which participate in the regulation of gene expression by silencing the mRNAs of protein-coding genes (Paul et al. 2018; Cheng et al. 2014). Several studies have associated identified EV-resident miRNAs with neurodegenerative diseases in the central nervous system (CNS); importantly, the miRNA contents of EVs are altered in AD and other diseases (Chen et al. 2017; Ebrahimkhani et al. 2017; Leggio et al. 2017; Cheng et al. 2015; Yang et al. 2018; Lugli et al. 2015). Microglia are the brain's resident immune cells that mediate signaling between cells, phagocytic activity, immune responses, synapse formation, elimination of damaged cells, and brain development. In a disease state, microglia receive stimuli from the CNS cells that modulate their activity (proliferation ability, phagocytic activity, the release of inflammatory cytokines) for defense (Bachiller et al. 2018). A study demonstrated the transfer of miRNA by EVs from inflammatory glia to neurons (Prada et al. 2018). Since miRNAs in EVs manipulate target cell functions, examining small neuron-derived EV (sNDEV)-resident miRNA transfer from AD-neurons to glia may shed light on the progression and propagation of AD.
In the present study, we characterized dysregulation of miRNA expression in the sNDEVs of AD patients compared with healthy individuals. We identified 10 differentially expressed miRNAs in patient sNDEVs with next-generation sequencing (NGS) and validated the most significantly altered miRNA, let-7e. Further, we treated human microglia with AD patients’ sNDEVs containing a high level of let-7e and healthy controls’ sNDEVs to examine the sNDEV-resident miRNA-mediated cellular responses in target cells. The gene expression of IL-6 was increased in the AD patient sNDEV-treatment group, suggested that sNDEV-resident let-7e induced an inflammatory response in microglia in vitro.
Materials and Methods
Patients and Plasma Collection
This study was approved by the clinical research ethics committee of Dokuz Eylul University (approval date: August 21, 2017; protocol number: 399-SBKAEK); written informed consent was provided by all participating volunteers or legal custodians according to the principles of the Declaration of Helsinki. AD patients (n = 23) and healthy controls (n = 28) were recruited by neurologists at the Department of Neurology, Dokuz Eylul University (Izmir, Turkey). AD patients were diagnosed according to the Diagnostic and Statistical Manual of Mental Disorders, fourth edition (DSM-IV) and the National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer's Disease and Related Disorders Association (NINCDS-ADRDA) diagnostic criteria. The healthy controls were chosen from individuals who did not have any neurological disorders. The results were validated in another set of AD patients. The clinical characteristics of AD patients and healthy subjects are given in Table 1. Peripheral blood was collected into anticoagulant-free tubes for the study and centrifuged at 3000 rpm for 10 min at 4 °C. The plasma was carefully obtained and stored in 0.5 ml aliquots at − 80 °C until use.
Isolation of sNDEVs from Human Plasma
Total EVs were isolated from 1 ml of plasma of AD patients and healthy controls using a Norgen Plasma/Serum Exosome Purification Mini Kit (Norgen Biotek Corp, Canada) according to the manufacturer's instructions. After total EV isolation, sNDEVs were enriched based on the method of Mustapic et al. (Mustapic et al. 2017). Briefly, total EV suspension was incubated with 8 µg of mouse anti-human CD171 (L1CAM neural cell adhesion protein) biotinylated antibody (clone 5G3; eBioscience, San Diego, CA) in 100 μl of 3% bovine serum albumin (BSA; 1:3.33 dilution of BSA 10% solution in phosphate-buffered saline [PBS; Thermo Scientific, USA]) for 60 min at 4 °C. After incubation, 80 μl of Streptavidin Plus UltraLink resin (Pierce/Thermo Scientific, USA) in 120 μl of 3% BSA was added to the mixture, incubated at 4 °C for 30 min. Following centrifugation at 200 × g at 4 °C for 10 min, the pellet was resuspended in 200 μl of 0.05 M glycine–HCl (pH 3.0) and incubated at 4 °C for 10 min. Then, the mixture was centrifuged again at 4500 × g at 4 °C for 10 min. Finally, the supernatant was transferred to a new tube containing 30 μl of 1 M Tris–HCl (pH 8.0).
Western Blot Analysis
For protein isolation, sNDEVs were lysed with mammalian protein extraction reagent (M-PER, Thermo Scientific, USA) for 10 min with vigorous vortexing, and protein concentrations were measured with a BCA assay. For western blot, 10 µg protein samples from EVs were separated using 10% SDS-PAGE gel and transferred to polyvinylidene fluoride (PVDF) membranes (Sigma-Aldrich, USA), blocked with 3% milk in phosphate-buffered saline with Tween (TBS-T) for 1 h at room temperature and incubated with primary antibodies at 4 °C overnight. The next day, membranes were incubated with secondary antibodies for 1 h at room temperature and washed with TBS-T. All the primary and secondary antibodies used were given in Supplementary Table 1. Images were captured with a UVP Gel Imager System (CA, USA) using a SuperSignal West Dura ECL reagent (Thermo Scientific, USA). The images were analyzed with ImageJ software (National Institutes of Health, USA).
Quantification and Size Profiling of sNDEVs
sNDEVs isolated from AD patients and healthy controls were quantified by EXOCET assay (System Biosciences, USA) according to the manufacturer's protocol. Briefly, samples (50 µg protein equivalent of sNDEVs) were lysed with lysis buffer and incubated for 5 min at 37 °C. To eliminate debris, they were centrifuged at 1500 × g for 5 min. For the EXOCET assay, 50 µl of debris-free supernatant and 50 µl reaction buffer were incubated for 20 min. The samples were read at 405 nm by a spectrophotometer, and the concentration of the samples was calculated with the standard curve. For size profiling, nanoparticle tracking analysis (NTA) was performed. The samples were profiled using the NanoSight NS300 particle size analyzer (Malvern Instruments, UK). For this process, the samples were diluted at 1:100 in PBS, and analysis was conducted in triplicate.
RNA Isolation, Library Generation and Small RNA Sequencing
RNAs were isolated from sNDEVs using exoRNeasy Serum/Plasma Midi Kit (QIAGEN, Germany) according to the manufacturer's instructions. The RNA concentration was measured using the NanoDrop spectrophotometer. The miRNA profiling of sNDEVs was performed via small RNA sequencing by Norgen Biotek Corp (Canada). Briefly, the total RNA isolated from sNDEVs was quantified by Bioanalyzer (ranging from 10.3 to 21 ng/µl). A Norgen Biotek Small RNA Library Prep Kit (Norgen Biotek, Canada) was used to generate sequencing libraries. Small RNA sequencing was carried out on an Illumina NextSeq 500 platform (Illumina, CA, USA). Approximately 10 million reads were conducted on RNA-sequencing data sets with 50 base-pair paired-end reads.
Bioinformatics Analysis
Raw reads were quality-checked with FastQC, and then 3′-end adapter sequences were removed using Cutadapt v2.4 (Martin 2011) and further aligned to the reference genome (GRCh38, Ensembl release 97) with miRDeep2 v0.1.2 (Martin 2011). Hairpin and mature miRNA sequences for Homo sapiens were obtained from miRBase v22 (Pathan et al. 2017) on quantifying the known miRNAs in our samples. Reads per kilobase of transcript per million mapped reads (RPKM) were calculated with the rpkm function of the R package edgeR. Gene expression analysis was performed using miRDeep2. Differential expression analysis was performed in the R environment v3.5.1 with the limma-voom utility of Bioconductor package limma (Mitchell et al. 2008). The thresholds of |log2FoldChange|≥ 1 and p-value ≤ 0.05 were applied to determine differentially expressed miRNAs. Target prediction of up- and downregulated miRNAs (p-value < 0.05) was performed using the R package multiMiR (Théry et al. 2018). Heat map visualization and hierarchical clustering of differentially expressed miRNAs were conducted using the pheatmap package in the R computing environment.
Potential target genes and associated Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of differentially expressed EV-resident miRNAs were determined with DIANA-miRPath v3.0 (http://snf-515788.vm.okeanos.grnet.gr/). In addition, Gene Ontology (GO) functional enrichment analyses and comparison of all EV-resident miRNAs with those in EV databases were performed with FunRich software (Pathan et al. 2017). In this step, a p-value of < 0.05 was considered significant.
Validation of EV-resident let-7e by qRT-PCR
In sNDEVs, the most differentially expressed miRNA, let-7e-5p, was further validated by quantitative reverse transcription polymerase chain reaction (qRT-PCR) using the miScript SYBR Green PCR Kit (QIAGEN, Germany). Briefly, 40 ng of total RNA from EVs was used as input for cDNA reaction using the miScript II RT Kit (QIAGEN, Germany) according to the manufacturer's protocol. 2 μl of cDNA was used for a qPCR reaction carried out on a LightCycler 480 II (Roche, Switzerland). Caenorhabditis elegans miR-39 was used as an external control for EVs from patients and healthy controls, as cel-miR-39 has no mammalian homologue (Mitchell et al. 2008). The Ct values were normalized to the reference gene miR-39, and the fold changes were calculated using the formula 2−ΔΔCt. The primers used in the validation step are listed in Supplementary Table 2.
HMC3 Cell Culture
The human microglial cell line HMC3 was purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). HMC3 cells were maintained in Eagle's minimum essential medium (EMEM, Sigma-Aldrich, USA) supplemented with 10% exosome-depleted fetal bovine serum (FBS) and 1% penicillin–streptomycin (Thermo Scientific, USA) at 37 °C with 5% CO2.
Labeling and Cellular Uptake of EVs
EVs were labeled with PKH67 Green Fluorescent Cell Linker Kit for General Cell Membrane Labeling (Sigma-Aldrich, USA) according to the manufacturer's protocol. Labeled EVs (100 µg protein equivalent of EVs) were incubated with HMC3 cells for 4 and 24 h at 37 °C in the incubator. The uptake of EVs by HMC3 cells was visualized with a fluorescence microscope (Olympus IX71, Japan).
RNA Isolation from Cell Culture and RT-qPCR
Total RNA from sNDEV-treated HMC3 cells was isolated using the miRNeasy Mini Kit (QIAGEN, Germany) following the manufacturer's protocol. First, isolated RNA was quantified using the NanoDrop. Then, 1 µg RNA was used for cDNA synthesis using the miScript II RT Kit (QIAGEN, Germany). Next, the GoTaq qPCR Master Mix Kit (Promega, USA) was used for qPCR using amplification steps consisting of one cycle at 95 °C for 2 min, 45 cycles at 95 °C for 10 s, and one cycle at 60 °C for 1 min on an ABI 7500 system (Applied Biosystems, USA). The primers used in this step are listed in Supplementary Table 3. Finally, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a housekeeping gene to normalize mRNAs.
Statistical Analysis
Statistical analyses and construction of receiver operating characteristic (ROC) curves were performed with GraphPad Prism 8.0 (GraphPad Software Inc., CA, USA). ROC analysis was performed to determine the diagnostic value and predictive power of differentially expressed miRNAs. All data are expressed as the mean ± standard error of the mean (SEM). The Mann–Whitney test was used to compare differences between two groups for statistical significance. A p-value < 0.05 was considered a statistically significant difference.
Results
EV Isolation and Characterization
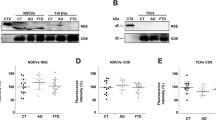
EVs were isolated from 1 ml blood from AD patients and healthy controls using commercial kits; sNDEVs were then enriched using an L1CAM antibody. Immune purification of EVs was confirmed by western blotting for the presence of the EV surface marker, tetraspanin CD63, and the absence of non-EV calnexin protein supported by β-actin as the positive internal control. Our results confirmed CD63 expression and lack of calnexin and β-actin in the EVs of both AD patients and healthy controls (Fig. 1a). The quantification of the sNDEV sample was done using EXOCET. There was no significant difference between sNDEVs from AD patients and healthy controls (Fig. 1b). Further analysis performed with NTA showed that the distribution of isolated sNDEVs from AD patients (the right panel) and healthy controls (the left panel) peaked at the diameter of 75.5 nm and 61.3 nm respectively, matching the definition of sEVs (Fig. 1c, Supplementary Fig. 1). Further characterization was performed to characterize EVs as recommended in the latest MISEV (minimal information for studies of extracellular vesicles) guideline (Théry et al. 2018). EVs were visually assessed with transmission electron microscopy (TEM) in our previous study (Arioz 2021). These findings indicate the success of EV isolation and are consistent with the characteristics of EVs.
Characterization of sNDEVs. (a) Isolated EVs were confirmed via western blot analyses of surface marker protein CD63 and endoplasmic protein calnexin and β-actin, not found in EVs. (Healthy/patient Exo: isolated EVs from healthy controls and AD patients). (b) The quantification of sNDEVs of healthy control and AD patients by EXOCET. (c) The size distribution of isolated sNDEVs from the healthy subjects on the left and the AD patients on the right by NTA
miRNA Profiling of sNDEVs
To identify differentially expressed miRNAs in AD patients, sNDEVs from both AD patients and healthy controls were analyzed by small RNA sequencing. NGS was performed on eight sNDEV samples from each group, and 4.6 to 10 million NGS reads were aligned to the reference human genome sequence. Here, only miRNAs with a minimum of one raw read count in at least three samples were considered as represented miRNAs in the EVs. A total of 117 miRNAs were identified in the sNDEVs of healthy controls and AD patients. While the levels of 22 miRNAs (18.8%) for the healthy group and 11 miRNAs (9.4%) for the AD patient group were specific to that group, 84 miRNAs (71.8%) were found in both groups (Fig. 2a). The list of group-unique miRNAs is given in Table 2. When our data set was compared to validated miRNAs in ExoCarta and Vesiclepedia, 103 miRNAs were already found in these EV databases (Fig. 2b). Among all miRNAs detected by NGS, the miR-10 and let-7 groups were the most representative miRNA families in our data set (Fig. 2c).
Profiling of EVs small RNA sequencing data. (a) Venn diagram analysis showing common and unique miRNAs detected in sNDEVs of healthy control and AD patient groups. (AD: Alzheimer's disease, HC: healthy control). (b) Comparison of sNDEV-resident miRNAs in our data set to the previously described miRNAs in the ExoCarta and Vesiclepedia databases. (c) The most representative miRNAs in sNDEVs. miR-10 family was the most expressed miRNA group based on NGS data
Differentially Expressed miRNAs in sNDEVs
Ten of the miRNAs were determined as differentially expressed between healthy controls and AD patients. Three miRNAs (hsa-let-7e-5p, hsa-miR-96-5p, hsa-miR-484) were found to be upregulated and seven miRNAs (hsa-miR-99b-5p, hsa-miR-100-5p, hsa-miR-30e-5p, hsa-miR-378i, hsa-miR-145-5p, hsa-miR-378c, hsa-miR-451a) were found to be downregulated; these differentially expressed miRNAs are listed in Table 3. Furthermore, the number of miRNAs and the RPKMs are given in Supplementary Tables 4 and 5. The heat map demonstrates the expression of differentially expressed miRNAs in each EV sample from AD patients and healthy controls (Fig. 3).
Heat map diagram and hierarchical clustering of differentially expressed 10 miRNAs in sNDEVs of healthy controls and AD patients by NGS. The diagram shows the LogFC expression level of dysregulated miRNAs. Fold changes were calculated as the expression in each sample (normalized count) relative to the average expression in the healthy control group. (C: healthy controls, P: patients with AD)
Pathway, Functional Enrichment, and Target Gene Analyses of Differentially Expressed miRNAs
KEGG and GO enrichment analyses were performed to identify differentially expressed miRNAs' biological functions and associated pathways. KEGG pathway analysis suggested pathways in which targets of the differentially expressed miRNAs are involved, including the apoptosis pathway, NF-κB signaling pathway, toll-like receptor signaling pathway, PI3K-Akt signaling pathway, and neurotrophin signaling pathway. These pathways could be prominent pathways involved in the development of AD (Fig. 4a). GO enrichment analysis was performed to explore the biological functions of the validated target genes of differentially expressed miRNAs based on three different categories: cellular component (CC), biological process (BP), and molecular function (MF). GO terms with a p-value < 0.05 were considered significant. The analysis found 30, 19, and 25 significant GO terms for CC, BP, and MF, respectively. Most of the target genes were enriched in the nucleoplasm for CC, positive regulation of transcription for BP, and protein binding for MF (Fig. 4b–d). Target genes of dysregulated miRNAs were also found with the TargetScan and miRTarBase databases, which provide computationally predicted and validated target mRNAs, respectively. Our results revealed 10 predicted and 10 experimentally validated target genes of exosome-related miRNAs that were deregulated in AD patient exosomes. Most target genes were associated with biological regulation and cellular, developmental, and metabolic processes and signaling (Fig. 5a, b).
KEGG and GO enrichment analysis. (a) KEGG pathway analysis on targets of dysregulated miRNAs. The top 25 most enriched pathways are shown. (KEGG: Kyoto Encyclopedia of Genes and Genomes database). (b–d) GO enrichment analysis on validated targets of dysregulated miRNAs based on cellular component, biological process, and molecular function. GO terms with a p-value < 0.05 were selected. (GO: Gene Ontology)
Validation of let-7e-5p with qRT-PCR
The differentially expressed let-7e-5p miRNA was further validated by qRT- PCR. Let-7e-5p was chosen based on its notable fold change and significant p-value. sNDEVs were isolated from an independent cohort of blood samples (20 AD patients and 14 healthy controls) to validate our results. qRT-PCR results confirmed that the let-7e-5p level was significantly upregulated in the sNDEVs of AD patients compared with those of healthy controls (Fig. 6a). In addition, the fold change was increased as 1.6 ± 0.3 standard deviations with a p-value of < 0.0001.
Validation of hsa-let-7e in sNDEVs. (a) The expression level of let-7e-5p was analyzed in sNDEVs of healthy controls (n = 14) and AD patients (n = 20) using the cel-miR-39 for normalization. The dark-colored horizontal bar shows the median expression level of miRNA. ****p < 0.0001 (Mann–Whitney U test). (qRT-PCR: quantitative reverse transcription-polymerase chain reaction) (b) Receiver operating characteristic (ROC) curve analysis for hsa-let-7e-5p in sNDEVs to discriminate AD patients from healthy controls. AUC was used to determine AD discrimination level and calculated as 0.9214 (95% CI = 0.80–1.00) with a sensitivity of 85.7% and specificity of 100%. (AUC: area under the curve, CI: confidence interval)
Diagnostic Value of EV-Resident let-7e-5p for AD Patients
ROC analysis was performed to evaluate the diagnostic value and utility of sNDEV miRNAs in distinguishing AD patients from healthy controls; the AUC of let-7e-5p was calculated as 0.9214 (95% CI = 0.80–1.00). This miRNA's sensitivity and specificity were 85.7% and 100%, respectively, at the cutoff value of 1.857-fold (Fig. 6b). Interestingly, there was no correlation between the MMSE score and the sNDEV-resident let-7e-5p level.
Effects of sNDEV Uptake in Microglial Activation
To evaluate how sNDEVs affect microglia, we evaluated the cellular uptake of sNDEVs and their effects on the inflammatory response in vitro. First, we demonstrated the uptake of PKH67-labeled EVs by human microglia using fluorescence microscopy. After 24 h of incubation, the green fluorescence signal from labeled EVs became more intense in HMC3 cells (Fig. 7). After we confirmed the cellular uptake of EVs by microglia, we examined the role of sNDEV-resident miRNAs on the gene expression of inflammatory cytokines in HMC3 cells. The mRNA expression levels of IL-1β, IL-6, and TNF-α were examined with qRT-PCR. Of these genes, the level of IL-6 mRNA was significantly increased in AD patient sNDEV-treated HMC3 cells (Fig. 8).
sNDEV-resident let-7e on gene expression of inflammatory cytokine IL-6 in human microglial cells. HMC3 cells were incubated with sNDEVs containing a low let-7e for the HC group (n = 7) and a high let-7e for the AD group (n = 7) for 24 h. The control group (n = 9) was not treated with EVs. The gene expression level of IL-6 was analyzed with qRT-PCR. **p < 0.01 (Mann–Whitney U test). (HC: healthy control, AD: Alzheimer's disease, qRT-PCR: quantitative reverse transcription polymerase chain reaction)
Discussion
There are approximately 55 million people affected by AD worldwide, and the number of patients is predicted to increase in the forthcoming years, with 10 million new cases per year. (Rabbito et al. 2020). Since AD is a slowly progressing disease, and its clinical symptoms appear many years after the start of degeneration, early diagnosis and treatment strategies are crucial (Hane et al. 2017). Even though specific neuroimaging tools such as PET, MRI, and CSF biomarkers are used to diagnose AD, new studies focus on less invasive, repeatable, and sensitive blood-based biomarkers (Ash 2007). EVs are a new source in biomarker research for many diseases, including AD. Since EVs carry biomolecules reflecting the state of origin cells they release, their content can be considered a reporter for the disease (Zhang et al. 2019). miRNAs in EVs have become popular since the study by Valadi et al. indicating that miRNAs are enriched in EVs and are carried via these vesicles into recipient cells (Valadi et al. 2007).
Our study used a combination of sNDEVs and miRNAs to examine potential AD biomarkers. We isolated sNDEVs from the blood of healthy individuals and AD patients, and we analyzed differentially expressed miRNA profiles in them by small RNA sequencing for the first time. The analysis resulted in the identification of 117 miRNAs. The miR-10, let-7, and miR-30 families were the most abundant miRNA groups reported in previous vesicle studies (Gracia et al. 2017; Ogata-Kawata et al. 2014; Worst et al. 2019; Derkow et al. 2018; Gámez-Valero et al. 2019). Alteration of sEV miRNA expression in AD has been investigated for biomarker potential. These studies analyzed exosomes from serum or plasma. A small number of studies have investigated miRNA expression in neuron-derived exosomes (Liu et al. 2021a, b). One of the miRNAs shown to undergo changes in AD in these studies, miR-30e-5p, also showed a significant change in our study (Cheng et al. 2015; Manna et al. 2020).
Furthermore, miR-451 was also reported to increase in a study by Dong et al. (Dong et al. 2021), while its expression decreased in our study and the study by McKeever et al. (McKeever et al. 2018). miRNA profiling was not done in a single study performed on neuron-derived exosomes, but the level of selected miRNAs was examined by PCR.
Additionally, the relationship between miRNAs, whose expression is changed in AD, and clinical parameters has been examined. While miRNAs were observed to correlate with cognitive findings, some studies did not find any correlation between clinical findings and the expression of miRNAs (Liu et al. 2014). For example, there was no correlation between the let-7e level and clinical findings in our study.
Based on the NGS data set, we found dysregulation of 10 miRNAs in sNDEVs of AD patients (Table 3). Among these, three (let-7e, miR-96, and miR-484) were upregulated, and the rest (miR-99b, miR-100, miR-30e, miR-378i, miR-145, miR-378c, and miR-451a) were downregulated. The most significantly altered miRNA was let-7e. The let-7 miRNA family is one of the most highly expressed miRNAs in neuronal and glial cells. In previous studies, it was shown that the binding of let-7 miRNA members to toll-like receptors (TLRs) caused neuroinflammation by releasing inflammatory cytokines such as interleukin 6 (IL-6) and tumor necrosis factor alpha (TNF-α). At the same time, they trigger the activation of the NF-κB pathway, in the end leading to neurodegeneration (Fabbri et al. 2012). Another study indicated the variety and elevated levels of the let-7 miRNA family and their association with extracellular vesicles in AD (Derkow et al. 2018). It was also shown that the let-7 miRNA family was the most dysregulated miRNA group in AD EVs, and EV-resident let-7 miRNAs were found to be associated with neuronal inflammation (Chen et al. 2019a; Gámez-Valero et al. 2019). However, to the best of our knowledge, upregulation of let-7e miRNA in sNDEVs in AD is not well documented in the literature. Validation of let-7e in a different patient cohort with qRT-PCR supported the sequencing data that the level of let-7e in sNDEVs is significantly increased in AD patients.
Early studies showed that elevation of miR-96 may be related to neuroprotection and AD (Kinoshita et al. 2014). Furthermore, upregulation of miR-484 in plasma EVs, downregulation of miR-451a in CSF EVs, and a decrease in miR-100 in sNDEVs of AD patients were indicated in previous studies, consistent with our results (McKeever et al. 2018; Rani et al. 2017; Serpente et al. 2020). Thus, validation and combination of all these miRNAs as a potential biomarker panel might be a valuable tool for further studies and diagnosis.
Thus far, two studies have assessed the miRNA content of sNDEVs in AD. Cha and colleagues showed a decrease in miR-212 and miR-132 in sNDEVs of AD patients using qRT-PCR compatibly with the patients’ miRNA profile in brain tissues (Cha et al. 2019). Serpente et al. found four differentially expressed and validated miRNAs (miR-23a, miR-223, miR-100, miR-190a) in AD patients' sNDEVs with an miRNA panel (Serpente et al. 2020). Our study analyzed the complete miRNA profile of sNDEVs isolated from AD patients’ blood using small RNA sequencing, which has not been reported previously.
KEGG and GO enrichment analyses were assessed to explore associated pathways and biological functions of dysregulated miRNAs. KEGG pathway analysis sheds light on many pathways in AD development and progression. GO analysis specified important terms for three classes of functions. The top 10 GO terms for the cellular component included “nucleoplasm,” “cytosol,” “nucleus,” “membrane,” “cytoplasm,” and “extracellular exosome,” which are associated with EV structure as shown in the previous studies (Chen et al. 2018, 2019b; Yue et al. 2020). For the biological process, the most frequently expressed GO terms were associated with “regulation of transcription,” “translation,” “protein regulation,” “response to stimulus,” and “regulation of apoptotic process,” implying that these miRNAs in sNDEVs may be involved in the regulation of AD-related proteins. Analysis of molecular functions supported this finding, with GO terms such as “protein binding,” “RNA binding,” “DNA binding,” “enzyme binding,” and “miRNA binding.”
Providing cell-to-cell interaction is another essential function of EVs. AD causes neuroinflammation in the CNS that triggers microglial activation due to neurodegeneration. Against the CNS damage, microglia are activated as immune cells and trigger an inflammatory response by pro-inflammatory cytokine (IL-1β, IL-6, TNF-α) secretion further contributing to neuronal dysfunction, inducing neuronal cell death, and losing their phagocytic ability (Heneka et al. 2015; Calsolaro and Edison 2016). This process promotes the progression and propagation of the disease. Crosstalk between neuron and glial cells that causes inflammation in AD has motivated researchers to focus on cell-to-cell communication mediators such as EVs. Aside from neurotransmitters, it is known that neurons release EVs to interact with other cells in healthy and disease-state conditions (Caruso Bavisotto et al. 2019). In an in vitro study by Bahrini et al., it was shown that sNDEVs are involved in synapse elimination by promoting microglial phagocytosis (Bahrini et al. 2015). EVs manipulate target cells via cargo molecules, including miRNAs and proteins. It was proven that the delivery of dysregulated miRNAs to neural cells altered the gene expressions and affected brain functions (Gupta and Pulliam 2014). Another study showed that miR-132-enriched sNDEVs regulate brain vascular integrity as providers of neurovascular interaction (Xu et al. 2017). Hence, investigating sNDEV content transfer into neighbor/target cells could be a key factor in determining disease propagation routes and mechanisms. This study investigated whether AD-neurons could transfer miRNA content and affect inflammatory responses in microglia via sNDEV-resident miRNA let-7e. Incubation of microglia with the high level of let-7e-containing sNDEVs of AD patients elevated the gene expression of IL-6.
In summary, our study has shown the altered miRNA profile in sNDEVs of AD patients. Also, it proved that sNDEVs miRNA could contribute to the pathogenesis and progression of AD as a neuron-glia communicator. Based on NGS results, validation of the most dysregulated miRNA in sNDEVs suggested that let-7e may be a promising and valuable biomarker candidate for AD diagnosis, and sNDEV let-7e can trigger inflammatory responses in microglia. Although the current study was conducted on a limited number of patients, and this situation limits our study's significance, our findings indicate the potential of sNDEVs in the diagnosis of AD. Furthermore, the heterogeneity seen within the AD patient group might be related to the fact that some motor symptoms were seen in the follow-ups, which could be helpful for the distinction between AD and frontotemporal dementia. The alteration in the sNDEV miRNome could pave the way for less invasive routes of AD diagnosis and prognosis, since AD diagnosis is made by eliminating other possibilities. Lastly, new studies with a high number of participants, including different stages of AD and mild cognitive impairment, will increase the potential diagnostic value of sNDEVs and shed light on their significant role in neuron-glia interactions in AD.
Data Availability Statement
The data sets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Abbreviations
- AD :
-
Alzheimer's disease
- Aβ :
-
Amyloid beta
- APP :
-
Amyloid precursor protein
- AUC :
-
Area under the curve
- BACE1 :
-
Beta-secretase 1
- cDNA :
-
Complementary DNA
- CNS :
-
Central nervous system
- CSF :
-
Cerebrospinal fluid
- EV :
-
Extracellular vesicle
- GO :
-
Gene ontology
- HC :
-
Healthy control
- IL :
-
Interleukin
- KEGG :
-
Kyoto Encyclopedia of Genes and Genomes
- L1CAM :
-
Neural cell adhesion protein
- miRNA :
-
MicroRNA
- MISEV :
-
Minimal information for studies of extracellular vesicles
- MMSE :
-
Mini-Mental State Exam
- MRI :
-
Magnetic resonance imaging
- mRNA :
-
Messenger RNA
- sNDEV :
-
Small neuron-derived extracellular vesicle
- NGS :
-
Next-generation sequencing
- NTA :
-
Nanoparticle tracking analysis
- PET :
-
Positron emission tomography
- ROC :
-
Receiver operating characteristic
- sEVs :
-
Small extracellular vesicles
- TEM :
-
Transmission electron microscopy
- TLR :
-
Toll-like receptor
- TNF :
-
Tumor necrosis factor
References
Arioz BI et al (2021) Proteome profiling of neuron-derived exosomes in Alzheimer's disease reveals hemoglobin as a potential biomarker. Neurosci Lett 755
Ash EL (2007) 27 - Dementia. In: Marshall RS, Mayer SA (eds) On Call Neurology, 3rd edn. W.B. Saunders, pp 401–417
Bachiller S et al (2018) Microglia in neurological diseases: A road map to brain-disease dependent-inflammatory response. Front Cell Neurosci 12(488)
Bahrini I et al (2015) Neuronal exosomes facilitate synaptic pruning by up-regulating complement factors in microglia. Sci Rep 5:7989–7989
Blennow K, de Leon MJ, Zetterberg H (2006) Alzheimer’s disease. Lancet 368(9533):387–403
Calsolaro V, Edison P (2016) Neuroinflammation in Alzheimer’s disease: Current evidence and future directions. Alzheimers Dement 12(6):719–732
Caruso Bavisotto C et al (2019) Extracellular vesicle-mediated cell-cell communication in the nervous system: Focus on neurological diseases. Int J Mol Sci 20(2):434
Cha DJ et al (2019) miR-212 and miR-132 are downregulated in neurally derived plasma exosomes of Alzheimer’s patients. Front Neurosci 13:1208–1208
Chen JJ et al (2017) Potential roles of exosomal microRNAs as diagnostic biomarkers and therapeutic application in Alzheimer’s disease. Neural Plast 2017:7027380
Chen W-X et al (2018) Analysis of miRNA signature differentially expressed in exosomes from adriamycin-resistant and parental human breast cancer cells. Biosci Rep 38(6):BSR20181090
Chen JJ et al (2019a) Exosome-encapsulated microRNAs as promising biomarkers for Alzheimer’s disease. Rev Neurosci 31(1):77–87
Chen W-X et al (2019b) Bioinformatics analysis of dysregulated microRNAs in exosomes from docetaxel-resistant and parental human breast cancer cells. Cancer Manag Res 11:5425–5435
Cheng L et al (2014) Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J Extracell Vesicles 3
Cheng L et al (2015) Prognostic serum miRNA biomarkers associated with Alzheimer’s disease shows concordance with neuropsychological and neuroimaging assessment. Mol Psychiatry 20(10):1188–1196
Dalvi A (2012) Alzheimer’s disease. Dis Mon 58(12):666–677
Derkow K et al (2018) Distinct expression of the neurotoxic microRNA family let-7 in the cerebrospinal fluid of patients with Alzheimer’s disease. PLoS One 13(7):e0200602
Dong Z et al (2021) Profiling of serum exosome MiRNA reveals the potential of a MiRNA panel as diagnostic biomarker for Alzheimer’s disease. Mol Neurobiol 58(7):3084–3094
Ebrahimkhani S et al (2017) Exosomal microRNA signatures in multiple sclerosis reflect disease status. Sci Rep 7(1):14293
Fabbri M et al (2012) MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA 109(31):E2110–E2116
Gámez-Valero A et al (2019) Exploratory study on microRNA profiles from plasma-derived extracellular vesicles in Alzheimer’s disease and dementia with Lewy bodies. Transl Neurodegener 8(1):31
Gracia T et al (2017) Urinary exosomes contain microRNAs capable of paracrine modulation of tubular transporters in kidney. Sci Rep 7(1):40601
Gupta A, Pulliam L (2014) Exosomes as mediators of neuroinflammation. J Neuroinflammation 11:68–68
Hane FT et al (2017) Recent progress in Alzheimer’s disease research, part 3: Diagnosis and treatment. J Alzheimers Dis 57(3):645–665
Heneka MT et al (2015) Neuroinflammation in Alzheimer’s disease. Lancet Neurol 14(4):388–405
Jella KK et al (2018) Exosomes, their biogenesis and role in inter-cellular communication, tumor microenvironment and cancer immunotherapy. Vaccines (Basel) 6(4)
Kinoshita C et al (2014) Rhythmic oscillations of the microRNA miR-96-5p play a neuroprotective role by indirectly regulating glutathione levels. Nat Commun 5(1):3823
Leggio L et al (2017) microRNAs in Parkinson's disease: From pathogenesis to novel diagnostic and therapeutic approaches. Int J Mol Sci 18(12)
Liao W et al (2019) Exosomes: The next generation of endogenous nanomaterials for advanced drug delivery and therapy. Acta Biomater 86:1–14
Liu CG et al (2014) MicroRNA-193b is a regulator of amyloid precursor protein in the blood and cerebrospinal fluid derived exosomal microRNA-193b is a biomarker of Alzheimer’s disease. Mol Med Rep 10(5):2395–2400
Liu CG et al (2021a) MicroRNA-135a in ABCA1-labeled exosome is a serum biomarker candidate for Alzheimer’s disease. Biomed Environ Sci 34(1):19–28
Liu CG et al (2021b) ABCA1-labeled exosomes in serum contain higher microRNA-193b levels in Alzheimer’s disease. Biomed Res Int 2021:5450397
Lugli G et al (2015) Plasma exosomal miRNAs in persons with and without Alzheimer disease: Altered expression and prospects for biomarkers. PLoS One 10(10):e0139233
Manna I et al (2020) Exosomal miRNAs as potential diagnostic biomarkers in Alzheimer's disease. Pharmaceuticals (Basel) 13(9)
Martin M (2011) CUTADAPT removes adapter sequences from high-throughput sequencing reads. EMBnet. J 17
Masters CL et al (2015) Alzheimer’s disease. Nat Rev Dis Primers 1:15056
McKeever PM et al (2018) MicroRNA expression levels are altered in the cerebrospinal fluid of patients with Young-Onset Alzheimer’s disease. Mol Neurobiol 55(12):8826–8841
Mitchell PS et al (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA 105(30):10513–10518
Mustapic M et al (2017) Plasma extracellular vesicles enriched for neuronal origin: A potential window into brain pathologic processes. Front Neurosci 11:278
Ogata-Kawata H et al (2014) Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One 9:e92921
Patients C (2019) Alzheimer's association 2019. Alzheimer's disease facts and figures. Alzheimer's and Dementia 15(3)
Pathan M et al (2017) A novel community driven software for functional enrichment analysis of extracellular vesicles data. J Extracell Vesicles 6(1):1321455
Paul P et al (2018) Interplay between miRNAs and human diseases. J Cell Physiol 233(3):2007–2018
Prada I et al (2018) Glia-to-neuron transfer of miRNAs via extracellular vesicles: a new mechanism underlying inflammation-induced synaptic alterations. Acta Neuropathol 135(4):529–550
Rabbito A et al (2020) Biochemical markers in Alzheimer's disease. Int J Mol Sci 21(6)
Rani A et al (2017) miRNA in circulating microvesicles as biomarkers for age-related cognitive decline. Front Aging Neurosci 9(323)
Sancesario GM, Bernardini S (2018) Diagnosis of neurodegenerative dementia: where do we stand, now? Ann Transl Med 6(17):340
Serpente M et al (2020) MiRNA profiling in plasma neural-derived small extracellular vesicles from patients with Alzheimer’s disease. Cells 9(6):1443
Théry C et al (2018) Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 7(1):1535750
Valadi H et al (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9(6):654–659
Van Giau V, An SS (2016) Emergence of exosomal miRNAs as a diagnostic biomarker for Alzheimer’s disease. J Neurol Sci 360:141–152
Worst TS et al (2019) miR-10a-5p and miR-29b-3p as extracellular vesicle-associated prostate cancer detection markers. Cancers 12(1):43
Xiao T et al (2017) The role of exosomes in the pathogenesis of Alzheimer’ disease. Transl Neurodegener 6:3
Xu B et al (2017) Neurons secrete miR-132-containing exosomes to regulate brain vascular integrity. Cell Res 27(7):882–897
Yang TT et al (2018) The serum exosome derived microRNA-135a, -193b, and -384 were potential Alzheimer’s disease biomarkers. Biomed Environ Sci 31(2):87–96
Yue B et al (2020) Characterization and transcriptome analysis of exosomal and nonexosomal RNAs in bovine adipocytes. Int J Mol Sci 21(23)
Zhang Y et al (2019) Exosomes: biogenesis, biologic function and clinical potential. Cell Biosci 9:19
Acknowledgements
The authors would like to thank Dr. Ben Cathcart for his critical reading of the manuscript, IBG Optical Imaging Core facility members, and Genc Laboratory research technician Nilsu Askin for their contribution.
Funding
This study was financially supported by The Scientific and Technological Research Council of Turkey (TUBITAK, Project No: 217S584).
Author information
Authors and Affiliations
Contributions
SG and GY designed the study. DYD, BT, MO designed and performed the experiments. DYD, BT, KUT, HU, GK, GY, and SG analyzed and interpreted the data. DYD, BT, KUT, and SG wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval
The study was carried out according to the principles of the Declaration of Helsinki, and it was approved by the clinical research ethics committee of Dokuz Eylul University (approval date: August 21, 2017; protocol number: 399-SBKAEK).
Consent to Participate
Written informed consents were provided by all the patients or legal custodians and the healthy controls.
Consent for Publication
All the authors agree to the publication of this work.
Competing Interests
The authors declare that no competing interests could be construed as a potential conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Durur, D.Y., Tastan, B., Ugur Tufekci, K. et al. Alteration of miRNAs in Small Neuron-Derived Extracellular Vesicles of Alzheimer's Disease Patients and the Effect of Extracellular Vesicles on Microglial Immune Responses. J Mol Neurosci 72, 1182–1194 (2022). https://doi.org/10.1007/s12031-022-02012-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12031-022-02012-y