Abstract
Increasing evidence has demonstrated the miRNAs’ action in cancerogenesis and tumor progression. Here, we explored the role and underlying mechanism of miR-3918 during glioma malignancy. miR-3918 and EGFR expression was detected in glioma tissues and tissues by RT-qPCR. The proliferative and migratory rate of glioma cells was assessed through CCK8 and Scratch wound-healing migration assay. Xenograft tumor mouse models were established for in vivo verification. A series of bioinformatics analysis coupled with luciferase reporter assays verified the targeted binding between miR-3918 and EGFR. Expression analyses demonstrated that miR-3918 was poorly expressed in glioma tissues while EGFR abundantly expressed. MiR-3918 overexpression impaired the proliferative and migratory capacities of glioma cells by inactivating PI3K/AKT and ERK pathways. Meanwhile, miR-3918 overexpression also retarded the growth of glioma xenograft. Mechanically, miR-3918 targeted EGFF which was further validated by the correlation of miR-3918 and EGFR expression in glioma tissues. When overexpressed, EGFR can restore the inactivated PI3K/AKT and ERK pathways caused by miR-3918 and influence the glioma cell proliferation and migration. Our findings are the first report that miR-3918/EGFR axis arrested the tumorigenesis of glioma via regulating PI3K/AKT and ERK pathways.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glioma ranks the first among central nervous system neoplasms, mainly originating from neuroglial progenitor cells (Gusyatiner and Hegi 2018). This disease is characterized by high aggressiveness and account for the highest morbidity and lethality in all intracranial tumors (Colquhoun 2017). In spite of surgery and chemoradiotherapy, clinical outcome of glioma patients remains unfavorable (Xu et al. 2020). For decades, molecular targeted therapy demonstrates advantages for cancer therapy. Accordingly, unraveling the underlying mechanism of glioma progression will facilitate glioma treatment at the molecular level.
MicroRNAs (miRNAs) are small non-protein coding RNAs which impact various genetic pathways triggering various cellular processes through post-transcriptional modification of miRNA targeted genes (Biswas 2018). Such biological features of miRNAs indicate that miRNA might play an important role in multitude physiological and pathophysiological conditions, including cancers (Acunzo et al. 2015). For example, miR-490-5p is responsible for inhibited proliferation of glioma cells because of its downregulation of CCND1 function (Zhao et al. 2018). An inactivation of PI3K/AKT signaling pathway regulated by miRNA-451 in glioma cells results in limited cell proliferation (Nan et al. 2018). Additionally, glioma cells with exogenous miRNA-451 successfully inhibited cancer progression in vitro and in vivo. Currently, the tumor-suppressive roles of miR-3918 have been described in hepatocellular carcinoma and gastric cancer (Yang et al. 2019; Fu et al. 2021). And yet, its biological function and action in glioma remained unknown.
EGFR (epidermal growth factor receptor) gene locates 7p11.2 and consists of 31 exons. It encodes a transmembrane glycoprotein belonging to the protein kinase superfamily. It has been well documented as a potent oncogene in different types of cancers including glioma (Nicholson et al. 2001; Harrison et al. 2020). Upregulation of EGFR can promote glioma cell malignant phenotypes and then gliomagenesis (Saadeh et al. 2018; Ribeiro et al. 2018). Its clinical significance in glioma has been also explored. However, the upstream mechanism of EGFR is not fully documented.
PI3K/AKT pathway is a typical oncogenic signaling cascade. Its hyperactivation confers tumor cell controlling malignant properties and promotes neoplasm transformation and tumor growth (Alzahrani 2019; Fresno et al. 2004; Chen et al. 2016). PI3K and AKT are the critical regulators of this cancer-related event. The phosphorylated PI3K and AKT triggered AKT translocation into the nucleus and induces phosphorylation of a group of effectors involved in proliferation and apoptosis of cells (Chai et al. 2018). In glioma, activated PI3K/AKT pathway enables glioma cell more proliferative and migratory capacities. In preclinical test, drugs targeting PI3K/AKT pathway exhibit potent anti-tumor potential (Li et al. 2016). Moreover, the ERK signaling pathway has been reported to play a crucial role in the progression of glioma [PMID: 32031702]. Therefore, further understanding of its action might be beneficial for the treatment of glioma.
Herein, we first demonstrated the biofunctional role and underlying mechanism of miR-3918 during glioma tumorigenesis. Our findings may offer novel therapeutic targets for glioma therapy.
Methods
Clinical Specimen
Eighteen glioma tissues and corresponding uncancerous tissues were gained from Wuhan Red Cross Hospital between 2018 and 2020. All patients signed informed consents. Ethical approval was acquired from Wuhan Red Cross Hospital.
Cells and Transfection
Human glioma cell lines (A172, SHG-44 and U251) were obtained from Shanghai Institute of Cell Life Sciences Resource Center (Shanghai, China), Normal human astrocytes (NHA) from Lonza, Basel, Switzerland. Glioma cells (DMEM medium) and NHA (RPMI-1640) were maintained under a 5% CO2 atmosphere at 37 °C.
MiR-3918 mimic and mimic NC, EGFR overexpression plasmids pcDNA3.1-EGFR (EGFR-OE), pcDNA3.1 (Empty vector), agomiR-NC, and ago-miR-3918 were purchased from Biorun, Biotech Co. Ltd., Wuhan, China. The oligonucleotides and constructs were introduced into SHG-44 and U251 cells (1 × 105) with Lipo3000 (Invitrogen, USA) as instructed by the manufacturer for 48 h. RT-qPCR was performed to test the transfection efficiency.
CCK8 Assay
1 × 105 cells were cultivated in 96-well plates for 24 h, 48 h, 72 h, and 96 h. At the indicated time, 10 μl CCK8 solution (GLPBIO, USA) was supplemented into each well for another 2 h. The plates were read at 450 nm using a microplate reader.
Scratch Wound-Healing Migration Assay
Cell migration was tested with scratch wound healing assay. Briefly, 1 × 105cells were cultivated in 6-well plates until above 85% confluence. Artificial wound was made with a 200-μl pipette tip across the center of the well. The cells were continuously cultured in fresh medium for additional 48 h. Cell monolayers were photographed at 0 h and 48 h.
Luciferase Reporter Assay
Amplified sequence of 3′UTR EGFR mRNA (wide-type, WT) and the corresponding mutant fragment (MUT) were fused into pmirGLO luciferase reporter vector by Hunan Fenghui Biotechnology Co., Ltd, China to construct pmirGLO-EGFR-WT and pmirGLO-EGFR-MUT vectors. Next, pmirGLO-EGFR-WT or pmirGLO-EGFR-MUT along with miR-3918 mimic or mimic NC were introduced into about 80% confluent SHG-44 and U251 cells. After 48h, the firefly and Renilla luciferase intensities were read using Luciferase Reporter System (Promega, USA). Luciferase activity = Firefly luciferase activity/Renilla luciferase activity.
RT-qPCR
Total RNA extraction was done with TRIzol reagent (Beijing Leigeng Biotechnology Co., Ltd., Beijing, China). Three micrograms RNA was subjected to reverse transcription to cDNA with the Quantitect Reverse Transcription Kit and miRNA miScript Reverse Transcriptase Kit (Qiagen, USA). The expression of miR-3918 and EGFR mRNA was quantified in the AB ViiA7 system (Thermo Fisher Scientific, USA) with SYBR gGreen quantitative PCR kit (Takara, Japan). The data was normalized to GAPDH. The primers are listed in Table 1.
Western Blots
Cells were treated by RIPA Lysis Buffer (Thermofisher, USA) and incubated on ice for 10–20 min. After centrifugation, the obtained supernatant was quantified by Pierce BCA Protein Assay Kit (Abcam, USA). Ten μg protein samples was loaded on 10% SDS-PAGE and then run into PVDF membrane at 35 V and 250 mA overnight. The obtained membrane was incubated in blocking buffer at 4 °C for 1 h before 24 h inoculation at 4 °C overnight with diluted anti-PI3K antibody (PI3 Kinase p85 (19H8) Rabbit mAb, 1:1000, Cat no. 4257, Cell Signaling Technology, USA), anti-p-PI3K antibody (phospho-PI3 Kinase p85 (Tyr458)/p55 (Tyr199) (E3U1H) Rabbit mAb, 1:1000, Cat no. 17366, Cell Signaling Technology, USA), anti-AKT antibody (1:1000, Cat no. 4691, Cell Signaling Technology, USA), anti-p-AKT antibody (1:1000, Cat no. 4060, Cell Signaling Technology, USA), anti-ERK1/2 antibody (p44/42 MAPK Rabbit mAb, 1:1000, Cat no. 4695, Cell Signaling Technology, USA), anti-p-ERK1/2 antibody (phospho-p44/42 MAPK Rabbit mAb, 1:1000, Cat no. 4370, Cell Signaling Technology, USA), anti-EGFR antibody (1:1000, Cat no. GTX121919, GeneTex, USA), or anti-GAPDH antibody (1:1000, Cat no. GTX100118, GeneTex, USA). Following, the diluted 1.7-µl secondary antibodies were used to detect the blot on membrane. Chemiluminescence was used to visualize the protein band.
In Vivo Experiments
In vivo assays were carried out on ten healthy female nude BALB/c mice, aging 4 weeks and average weighting about 20 g. They were obtained from Shanghai Experimental Animal Center (Chinese Academy of Sciences, China). All mice were kept at 25 C with 50% humility and feed normally until sacrifice. All protocol was approved by the Animal Care and Use Committee of Wuhan Red Cross Hospital. SHG-44 cells were transfected with only Lipo3000 as control, agomiR-NC and agomiR-3918. There was no significant difference in miR-3918 expression between the control group and agomiR-NC group. However, agomiR-3918 remarkably enhanced miR-3918 expression compared with the other two groups (Fig. S1A). Therefore, SHG-44 cells (1 × 107 cells/mice) carrying agomiR-NC and agomiR-3918 were administered subcutaneously into the flank of the mice (n = 5). Tumor growth was monitored at every 4 days. The weight was measured at 28 days when xenograft tumors were obtained from euthanized mice. Tumor volumes = tumor length × width2.
Statistical Analysis
Statistical analysis was conducted with GraphPad Prism 9.0. Data are represented as mean ± SD. Statistical significance (p < 0.05) was calculated with unpaired t-test for two-group comparisons and ANOVA for multiple comparison. Correlations between miR-3918 and EGFR in glioma tissues were ascertained using Pearson’s method with statistical significance (p < 0.05).
Results
MiR-3918 Weakens the Proliferation and Migration of Glioma Cells by Inactivating the PI3K/AKT and ERK Signaling Pathways
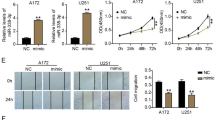
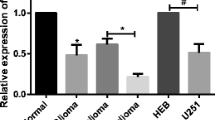
To interrogate the biofunctionality of miR-3918 in glioma, RT-qPCR was applied to detect its expression in glioma tissues and cells. As described in Fig. 1A, miR-3918 expression was lower in glioma tissues compared with that in normal tissues. Similarly, its expression was diminished in glioma cells compared with MHA cells (Fig. 1B). MiR-3918 expressed as fold decrease in SHG-44 and U251 cells; accordingly, we chose both glioma cells to establish miR-3918 overexpressing glioma cells by introduction of miR-3918 mimic into cells. Outcome from RT-qPCR analysis confirmed the upregulation of miR-3918 in SHG-44 and U251 cells (Fig. 1C). Next, we performed the CCK8 and scratch wound healing assays to compare the cell proliferation and migration between glioma cells transfected with miR-3918 mimic or mimic NC. As shown in Fig. 1D, upregulation of miR-3918 led to low proliferative rate of glioma cells. Meanwhile, the migratory capacity of SHG-44 and U251 cells was also diminished along with miR-3918 mimic transfection (Fig. 1E). The PI3K/AKT and ERK signaling are highly correlated with the occurrence of glioma. Therefore, we further tested the PI3K/AKT and ERK signaling. As shown in Fig. 1F–G, levels of phosphorylated PI3K, AKT, and ERK1/2 were lessened in miR-3918 overexpressing SHG-44 and U251 cells compared with the control cells. Collectively, miR-3918 suppresses glioma cell proliferation and migration via PI3K/AKT and ERK signaling pathways.
Upregulation of miR-3918 inhibits glioma cell growth and migration in vitro by activating the PI3K/AKT and ERK signaling pathways. A qRT-PCR analysis for miR-3918 expression in glioma tissues. **P < 0.01. B qRT-PCR analysis for miR-3918 expression in MHA and different glioma cell lines. *P < 0.05, **P < 0.01 vs. NHA. C qRT-PCR analysis for miR-3918 expression in SHG-44 and U251 cells transfected with miR-3918 mimic or mimic NC. D CCK8 analysis for the proliferation of control and miR-3918 overexpressing SHG-44 and U251 cells. E Representative images and the quantification data of scratch wound-healing assay for control and miR-3918 overexpressing SHG-44 and U251 cells at 0 and 24 h after wound scratch. F, G Western blot for the expression of the proteins associated with PI3K/AKT and ERK signaling pathways in SHG-44 and U251 cells transfected with miR-3918 mimic or mimic NC. C–G Mimic, miR-3918 mimic. *P < 0.05, **P < 0.01 vs. mimic-NC
MiR-3918 Retards Tumor Growth of Glioma Cells In Vivo
We continuously analyze the role of miR-3918 in glioma tumorigenesis in vivo. A total of 1 × 105 SHG-44 cells transfected with agomiR-NC and agomiR-3918 were given subcutaneously on the right flank of mice. As shown in Fig. 2A, tumor volume was markedly reduced in mice subjected to exogenous injection of agomiR-3918 and agomiR-NC. Furthermore, the weights of the tumors formed by agomiR-3918 mimic SHG-44 cells were reduced compared with those formed by NC cells. Altogether, miR-3918 suppressed the tumor growth of glioma in vivo
EGFR is the Target of miR-3918 in Glioma Cells
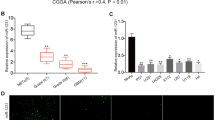
GSE10429 downloaded from GEO DataSets was used to identify the upregulated genes in GBM samples with adj.P < 0.05 and logFC > 1; at the same time, starBase and TargetScan were used to predict the downstream targeting genes of miR-3918. By Venny 2.1.0 analysis, 107 genes were overlapped from the results of GSE10429, starBase, and TargetScan (Fig. 3A). After STRING analysis, EGFR and CCND1 involving cell proliferation and PI3K-Akt pathway were confirmed (Fig. 3B). As shown in Fig. 3C, EGFR expression showed a significant upregulation in clinical tissues, compared with CCND1 expression. Therefore, EGFR interested us. The sequence of EGFR 3'UTR can recognize the complementary sequence on miR-3918 seed site (Fig. 3D). To examine this direct binding relationship, luciferase reporter assays were applied to examine the luciferase activity in SHG-44 and U251 cells cotransfected with EGFR 3′UTR-WT luciferase reporter construct (WT) and EGFR 3′UTR-MUT luciferase reporter construct (MUT) along with miR-3918 mimic or NC. As expected, miR-3918 mimic treatment considerably diminished the EGFR 3'UTR-WT-mediated luciferase activity while it showed no effect on EGFR 3′UTR-WT-mediated activity (Fig. 3E). After that, we further tested the EGFR expression in SHG-44 and U251 cells. Outcome from RT-qPCR analysis demonstrated significant upregulation of EGFR expression in both glioma cells (Fig. 3F). More interesting, miR-3918 displayed a negative correlation with EGFR expression in glioma tissues (Fig. 3G). Thusly, EGFR is a direct target of miR-3918 in glioma cells.
Identification of EGFR as a miR-3918 target in glioma cells. A The 107 genes were predicted to be targeted by miR-3918 and upregulated in GBM samples. GSE10429, mRNA microarray from GEO DataSets. starBase and TargetScan, two online tools to predict the targets of miR-3918. B EGFR and CCND1 were predicted to be associated with cell proliferation and PI3K-Akt signaling pathway. C. RT-qPCR analysis of EGFR and CCND1 in clinical tissues. **P < 0.01. D Predicted binding sites of EGFR WT 3′-UTR to miR-3918. E MiR-3918 mimics and EGFR 3′-UTR reporter wild-type (WT) or mutated (MT) were co-transfected in SHG-44 and U251 cells. Relative dual-luciferase activity was measured 48 h after transfection by illuminometer. **P < 0.01 vs. mimic-NC. F The mRNA expression of EFGR in SHG-44 and U251 cells were detected by qRT-PCR. **P < 0.01 vs. NHA. G The negatively correlation between EFGR expression level and miR-3918 expression level from glioma cohort was analyzed. **P < 0.01.
MiR-3918 Enforced Expression Impairs Glioma Cell Proliferation and Migration by Targeting EGFR
To further validate that the miR-3918 inhibited malignant characteristics of glioma cells by targeting 3′UTR EGFR, EGRR-overexpressing vectors or miR-3918 mimic were introduced into SHG-44 and U251 cells. As illustrated in Fig. 4A, miR-3918 mimic transfection resulted in significantly reduced EGRR protein expression which was restored by additional transfection of EGFR-overexpressing vectors. Functionally, the inhibition of miR-3918 on glioma cell proliferation and migration was restored by upregulation of EGFR expression (Fig. 4B, C). Additionally, the p-AKT, p-PI3K, and p-ERK1/2 protein levels were upregulated in SHG-44 and U251 cells when EGFR was overexpressed, and these increased p-AKT, p-PI3K, and p-ERK1/2 protein levels were abolished by simultaneous cotransfection of miR-3918 and EGFR-overexpressing constructs (Fig. 4D, E). Thusly, our data highlighted miR-3918/EGFR axis regulates the PI3K/AKT and ERK signaling in glioma cells
Downregulation of EGFR is essential for the miR-3918-mediated inhibition of proliferation and migration in glioma cells. SHG-44 and U251 cells were transfected with empty vector, mimic-NC, EGFR-overexpression vector (EGFR-OE), miR-3918 mimic or EGFR-OE + miR-3918 mimic. A EGFR protein expression was tested by western blots. B The cell proliferation was assessed by CCK8 assays. C The cell migration was examined by scratch wound‐healing migration assay. D AKT, p-AKT, and PI3K, p-PI3K were detected by Western blots. E ERK1/2 and p-ERK1/2 were detected by Western blots. EGFR-OE, EGFR overexpression. mimic, miR-3918 mimic. **P < 0.01 vs. empty vector. $$P < 0.01 vs. mimic-NC. #P < 0.05, ##P < 0.01 vs. EGFR-OE + mimic
Discussion
In our current investigation, we found that miR-3918 was robustly expressed in glioma tissues and cells. Functionally, miR-3918 overexpression inhibited glioma cell proliferation and migration by inactivation of PI3K/AKT and ERK pathways in vitro. Xenograft tumor assays reiterated that miR-3918 retarded the glioma progression. Mechanistically, EGFR was verified as a novel target of miR-3918. Its enforced expression partially abolished the inhibition on glioma cell proliferation and migration resulted from miR-3918 overexpression. More importantly, EGFR overexpression reactivated the inactivated PI3K/AKT and ERK pathways by miR-3918 enforced expression. Collectively, our results provided innovative insights regarding therapeutic and diagnostic targets against glioma.
The significance of miRNAs to tumorigenesis and cancer progression was showed in a myriad of investigations. Recently, miR-3918 was poorly expressed in hepatocellular carcinoma and was a tumor suppressor for hepatocellular carcinoma (Yang et al. 2019). Additionally, Fu et al. reported that miR-3918 negatively modulates the oncogenic NF-κB pathway in gastric cancer (Fu et al. 2021). Correspondingly, a loss in the activity of miR-3918 led to uncontrollable proliferation and acquired stem cell–like characteristics contributing to tumor transformation. For the first time, we demonstrated the functionality and underlying mechanism of miR-3918 in glioma. In our present work, we also demonstrated the downregulation of miR-3918 in glioma tissues and cells. Functionally, miR-3918 overexpression inactivated the oncogenic PI3K/AKT and ERK pathways and impeded glioma cell proliferation and migration in vivo; likewise, it retarded tumor growth in vivo. As described above, miR-3918 played a tumoral suppressor in glioma progression.
Via a Watson-Crick base pairing mechanism, miRNAs recognize the 3′UTR mRNA and thereby exert its functionalities (Correia et al. 2019). In this way, miRNAs work as oncomirs or tumoral suppressors (Lee and Dutta 2009). Outcome from our bioinformatics target prediction of miR-3918, we focused EGFR. EGFR is a cell surface protein involved in tyrosine autophosphorylation and plays an important role in cell proliferation. Its oncogenicity was well recognized in certain malignancies (Nicholson et al. 2001; Harrison et al. 2020; Ciardiello and Tortora 2008). In glioma patients, upregulation of EGFR shows tight association with unfavorable clinicopathological characteristics, such as advanced clinical stage (Saadeh et al. 2018). Overactivation of EGFR triggers AKT signaling and thereby plays an important role in pro-proliferation and pro-metastasis properties in glioma (Zhang et al. 2020). Nevertheless, the upstream regulation of EGFR remained fully untangled. Consistent with previous studies, we found the upregulation of EGFR in glioma tissues. Luciferase reporter assay further coordinated the targeted relationship between EGFR and miR-3918, which supported the significant negative correlation between their expressions in glioma tissues. Further rescued assay demonstrated that EGFR overexpression can restore the anti-proliferative and anti-migratory behaviors caused by miR-3918 overexpression in glioma cells. More importantly, EGFR overexpression in glioblastoma multiform is responsible for hyperactivating PI3K/Akt/mTOR signaling. In line with previous studies, EGFR is found to enhance the phosphorylated PI3K, AKT, and ERK1/2 and rescued the loss of PI3K, AKT, and ERK1/2 activity resulted from miR-3918 enforced expression.
Of note, one mRNA might be recognized by many miRNAs. For example, miR-374b, miR-375, miR-450a-5p, and miR-133b are reported to bind EGFR mRNA 3′UTR and then in the onset and progression of glioma (Pan et al. 2019; Zhang et al. 2018; Liu et al. 2020; Dong et al. 2019). Therefore, EGFR might be involved in a complex regulatory network and impact the glioma onset and progression. Furthermore, the number of clinical samples was small.
In conclusion, miR-3918 overexpression constrained the proliferation and migration of glioma cells. MiR-3918 can target EGFR and inhibit the oncogenic function of EGFR via inactivating PI3K/AKT and ERK pathways. Our findings suggested that employment of miR-3918/EGFR axis might be a novel approach for glioma management.
Availability of Data and Material
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Acunzo M, Romano G, Wernicke D, Croce CM (2015) MicroRNA and cancer–a brief overview. Adv Biol Regul 57:1–9
Alzahrani AS (2019) PI3K/Akt/mTOR inhibitors in cancer: at the bench and bedside. Semin Cancer Biol 59:125–132
Biswas S (2018) MicroRNAs as therapeutic agents: the future of the battle against cancer. Curr Top Med Chem 18(30):2544–2554
Colquhoun A (2017) Cell biology-metabolic crosstalk in glioma. Int J Biochem Cell Biol 89:171–181
Chen H, Zhou L, Wu X et al (2016) The PI3K/AKT pathway in the pathogenesis of prostate cancer. Front Biosci (Landmark edition) 21:1084–1091
Chai C, Song LJ, Han SY, Li XQ, Li M (2018) MicroRNA-21 promotes glioma cell proliferation and inhibits senescence and apoptosis by targeting SPRY1 via the PTEN/PI3K/AKT signaling pathway. CNS Neurosci Ther 24(5):369–380
Correia De Sousa M, Gjorgjieva M, Dolicka D, Sobolewski C, Foti M (2019) Deciphering miRNAs' Action through miRNA Editing. Int J Mol Sci 20(24)
Ciardiello F, Tortora G (2008) EGFR antagonists in cancer treatment. New England J Med 358(11):1160–1174
Dong ZQ, Guo ZY, Xie J (2019) The lncRNA EGFR-AS1 is linked to migration, invasion and apoptosis in glioma cells by targeting miR-133b/RACK1. Biomed Pharmacother 118:109292
Fu T, Ji K, Jin L et al (2021) ASB16-AS1 up-regulated and phosphorylated TRIM37 to activate NF-κB pathway and promote proliferation, stemness, and cisplatin resistance of gastric cancer. Gastric cancer: official journal of the International Gastric Cancer Association and the Japanese Gastric Cancer Association 24(1):45–59
Fresno Vara JA, Casado E, De Castro J, Cejas P, Belda-Iniesta C, González-Barón M (2004) PI3K/Akt signalling pathway and cancer. Cancer Treat Rev 30(2):193–204
Gusyatiner O, Hegi ME (2018) Glioma epigenetics: from subclassification to novel treatment options. Semin Cancer Biol 51:50–58
Harrison PT, Vyse S, Huang PH (2020) Rare epidermal growth factor receptor (EGFR) mutations in non-small cell lung cancer. Semin Cancer Biol 61:167–179
Li X, Wu C, Chen N et al (2016) PI3K/Akt/mTOR signaling pathway and targeted therapy for glioblastoma. Oncotarget 7(22):33440–33450
Lee YS, Dutta A (2009) MicroRNAs in cancer. Annu Rev Pathol 4:199–227
Liu Y, Yang L, Liao F, Wang W, Wang ZF (2020) MiR-450a-5p strengthens the drug sensitivity of gefitinib in glioma chemotherapy via regulating autophagy by targeting EGFR. Oncogene 39(39):6190–6202
Nan Y, Guo H, Guo L et al (2018) MiRNA-451 inhibits glioma cell proliferation and invasion through the mTOR/HIF-1α/VEGF signaling pathway by targeting CAB39 Human gene therapy. Clin Dev 29(3):156–166
Nicholson RI, Gee JM (2001) MEEGFR Harper cancer prognosis. European journal of cancer Oxford, England, 1990 37(Suppl 4)S 9–15
Pan DS, Cao P, Li JJ, Fan D, Song ZQ (2019) MicroRNA-374b inhibits migration and invasion of glioma cells by targeting EGFR. Eur Rev Med Pharmacol Sci 23(10):4254–4263
Ribeiro De Souza AL, Marra K, Gunn J et al (2018) Optimizing glioma detection using an EGFR-targeted fluorescent affibodY. Photochem Photobiol 94(6):1167-1171
Saadeh FS, Mahfouz R, Assi HI (2018) EGFR as a clinical marker in glioblastomas and other gliomas. Int J Biol Markers 33(1):22–32
Xu S, Tang L, Li X, Fan F, Liu Z (2020) Immunotherapy for glioma: current management and future application. Cancer Lett 476:1–12
Yang G, Wang X, Liu B et al (2019) circ-BIRC6, a circular RNA, promotes hepatocellular carcinoma progression by targeting the miR-3918/Bcl2 axis. Cell cycle (Georgetown, Tex.) 18(9):976–989
Zhao L, Tang X, Luo R, Duan J, Wang Y, Yang B (2018) MicroRNA-490-5P targets CCND1 to suppress cellular proliferation in glioma cells and tissue through cell cycle arrest. Curr Neurovasc Res 15(3):246–255
Zhang X, Niu W, Mu M, Hu S, Niu C (2020) Long non-coding RNA LPP-AS2 promotes glioma tumorigenesis via miR-7-5p/EGFR/PI3K/AKT/c-MYC feedback loop. J Exp Clin Cancer Res: CR 39(1):196
Zhang LX, Jin W, Zheng J et al (2018) MicroRNA-375 regulates proliferation and apoptosis of glioma cancer cells by inhibiting CTGF-EGFR signaling pathway. Bratislavske lekarske listy 119(1):17–21
Author information
Authors and Affiliations
Contributions
YH and HMW performed the experiments and data analysis. YH and HMW conceived and designed the study. YH and HMW made the acquisition of data. YH did the analysis and interpretation of data. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics Approval
The present study was approved by the Ethics Committee of Wuhan Red Cross Hospital (Wuhan, China). The processing of clinical tissue samples is in strict compliance with the ethical standards of the Declaration of Helsinki. All patients signed written informed consent.
Consent to Participate
All patients signed written informed consent.
Consent for Publication
Consent for publication was obtained from the participants.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Han, Y., Wang, H. MiR-3918 Inhibits Tumorigenesis of Glioma via Targeting EGFR to Regulate PI3K/AKT and ERK Pathways. J Mol Neurosci 72, 433–440 (2022). https://doi.org/10.1007/s12031-021-01952-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12031-021-01952-1