Abstract
Diffuse glioma is the deadliest form of brain cancer, and the median survival of grade IV glioma (glioblastoma, GBM) is no more than 2 years even with maximal surgical resection followed by radiotherapy and chemotherapy, which are now the standard of care for GBM. Glioma shares common characteristics with most malignant tumours, such as invasiveness, rapid progression, resistance to various therapies and inevitable recurrence, while it also has its own unique features, such as high aggressiveness and immunotherapy resistance, which can be, respectively, attributed to epithelial-mesenchymal transition (EMT) and the immunosuppressive microenvironment. Here, we calculated the EMT score of glioma using The Cancer Genome Atlas (TCGA), the Chinese Glioma Genome Atlas (CGGA) and the Gene Expression Omnibus (GEO) datasets and validated its prognostic value. Then, we investigated its role in the glioma immune microenvironment, identified the enriched EMT-related immune genes and determined their specific biological functions in glioma. Furthermore, clinical relevance analysis showed the translational value of these EMT-related immune genes. In short, our findings reveal a critical link between EMT and the glioma immune microenvironment and offer important clues for further investigation of the underlying molecular mechanism.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Diffuse glioma, the most prevalent and life-threatening malignant brain tumour, has always been notorious for its diffuse and infiltrative nature, which yields devastating clinical outcomes (Ferlay et al. 2019). According to histological criteria, adult diffuse glioma is classified into oligodendroglioma, oligoastrocytoma, astrocytoma and glioblastoma and is graded into WHO II to IV stages, respectively (Molinaro et al. 2019). Among them, glioblastoma is the most common (accounting for approximately 55% of gliomas) and also the deadliest glioma (with a dismal median survival of 14–16 months) (Wen and Kesari 2008). Despite decades of research and aggressive treatment, only negligible benefit has been gained in the clinical practice of patient care (Bray et al. 2018). Because of the poor prognosis of glioma after the initial diagnosis, it is urgently needed to better understand the molecular pathways and specific molecules that coordinate the aggressive and malignant nature of brain tumours. EMT, a reversible cellular programme that transitions epithelial cells into mesenchymal cell states, is known to be critical for malignant progression (Dongre and Weinberg 2019). In the context of neoplasia, EMT confers on neoplastic cells increased invasive and metastatic potential and a stronger resistance against elimination by several common treatment methods (Lambert et al. 2017; Shibue and Weinberg 2017). Owing to the extremely aggressive and invasive nature of glioma, it has been gradually appreciated that EMT in glioma likely contributes significantly to gliomagenesis and remodelling of the glioma microenvironment (Lu et al. 2012). When EMT is induced in neoplastic cells, the resulting mesenchymal carcinoma cells can in return respond to other non-neoplastic cells, such as by modifying the activities and representation of those cells in the tumour microenvironment (TME) (Gajewski et al. 2013), specifically, various subtypes of adaptive innate and adaptive immune cells involved in tumourigenesis and progression. Some of these cells are anti-neoplastic cells (natural killer (NK) cells, the cytotoxic CD8+ T cells and classically activated M1 macrophages), while others are pro-neoplastic cells (alternately activated M2 macrophages, regulatory T cells (Treg cells) and myeloid-derived suppressor cells (MDSCs)) (Kerkar and Restifo 2012). With the emergence of novel immunotherapies that are revolutionary therapeutic regimens for cancers (Mellman et al. 2011), identifying the underlying regulatory mechanism of these two groups of immune cells that act in distinct manners (immune suppression and immune activation) is of great significance, and EMT may play a crucial role in this process.
Recent clinical immunotherapies have been discovered in a variety of tumours, and immune checkpoint blockade therapy is now the standard of care for treatment of non-small cell lung cancer, melanoma and a growing list of other malignant tumours (Gong et al. 2018). However, this revolutionary therapeutic advance did not benefit glioma patients, even though data from preclinical models favoured immunotherapy as a feasible approach for glioma (McGranahan et al. 2019). Accordingly, the focus of glioma immunology research is moving to the development of strategies that aim at various resistance mechanisms. Currently, studies of glioma have revealed some key resistance mechanisms of the anti-glioma immune response. Examples include the specific immunoprivileged site of glioma (Jackson et al. 2019), the scarce glioma infiltrating lymphocyte population with highly expressed immune checkpoints and the existence of a large number of immunosuppressive cells called tumour-associated macrophages (TAMs) (Hambardzumyan et al. 2016). In general, the anti-glioma immune response resistance mechanisms have been revealed to occur at all phases of anti-tumour immunity, including the prevention of the initiation of the response (intrinsic immunoresistance), deactivation of tumour-infiltrating immune cells (adaptive immunoresistance) and protection of the tumour from elimination by the immune system (acquired immunoresistance) (Bauer et al. 2014; Chongsathidkiet et al. 2018; Grossman et al. 2011; Woroniecka et al. 2018). Although there are a large number of factors that have been shown to cause the immune resistance of glioma, the mechanisms by which EMT contributes to glioma immune resistance are largely unknown, and answering this question may represent a novel research direction.
To explore the relationship between EMT of glioma and the adaptive changes of its immune microenvironment, we uncovered the underlying molecular mechanism. In this study, by applying single sample gene set enrichment analysis (ssGSEA) (Barbie et al. 2009) to TCGA, CGGA and GEO datasets, we calculated the EMT scores, determined their clinical relevance and validated their prognostic value in glioma. Then, we revealed the close relationships between EMT scores and the glioma immune microenvironment. Using gene expression correlation analysis, we selected EMT-related immune genes and enriched relevant signalling pathways. We found that all top 26 EMT-related immune genes played critical roles in oncobiology. Importantly, we have validated their significance with regard to clinical outcomes of glioma patients.
Materials and Methods
Data Collection
TCGA RNAseq data in fragments per kilobase of transcript per million mapped reads (FPKM) of lower-grade glioma (LGG) and GBM, as well as relevant clinical information, were downloaded from the GDC data portal (https://portal.gdc.cancer.gov/). We obtained data on WHO grade, histology type, IDH mutation status and 1p/19q codeletion status of TCGA submitters from the study by Ceccarelli et al. (2016). Gene expression data of thousands of glioma patients were obtained from CGGA (http://www.cgga.org.cn/), including one microarray dataset of 301 patients and two RNAseq datasets of 325 and 693 patients, respectively (Ceccarelli et al. 2016). Additionally, 11 microarray gene expression profiling datasets of glioma (GSE4290, GSE4412, GSE7696, GSE16011, GSE35158, GSE50161, GSE52009, GSE54004, GSE60898, GSE61374, GSE107850) were available at GEO (https://www.ncbi.nlm.nih.gov/gds/). Duplicate genes were excluded except their max values.
Single Sample Gene Set Enrichment Analysis
The EMT gene set was extracted from the Gene Ontology (GO) database based on the term ‘positive regulation of epithelial to mesenchymal transition’ (GO: 0010718). The ESTIMATE immune and stromal gene sets were obtained from the R (v3.5.2) package estimate (Yoshihara et al. 2013) (v1.0.13). The enrichment scores were processed by the python (v3.6.8) package gseapy (v0.9.13).
Immune Cell Abundance Estimate
ABsolute Immune Signal (ABIS) (Monaco et al. 2019), Estimate the Proportion of Immune and Cancer cells (EPIC) (Racle et al. 2017) and Immune Cell Abundance Identifier (ImmuCellAI) (Miao et al. 2019) were used to estimate the abundance of immune cells in TCGA RNAseq gene expression data.
GO Enrichment Analysis
Spearman’s correlation analyses were performed between EMT score and gene expression and between the immune score and gene expression in TCGA (GBM and LGG), CGGA (3 datasets) and 12 GEO glioma datasets. P values were adjusted by the Benjamini and Hochberg methods. Genes correlated with both EMT and the immune score (|rho| > =0.3, FDR < 0.05) in more than 12 of the 16 datasets were selected. The R package clusterProfiler (Yu et al. 2012) (v3.10.1) was used for GO enrichment analysis.
Statistical Analysis
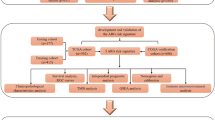
Kaplan–Meier curves with log rank test and Cox proportional hazards regression were generated by R packages survival (v 3.1–8) and survminer (v0.4.6). The significance of the difference between two groups was calculated by the two-sided Wilcoxon rank sum test. Spearman’s correlation was applied to evaluate the correlation between two groups of continuous values. All statistical analyses above were performed by R (v3.5.2). P values <0.05 were considered significant (Fig. 1).
Results
Clinical Relevance of EMT Scores
First, we extracted EMT-related genes from the GO dataset, and then the enrichment scores for these genes were calculated. Based on the extent of EMT-related gene expression, we clustered gliomas into two major groups by median value. Second, using TCGA datasets, we showed that high stromal and immune signatures were correlated with high glioma grades (Fig. 2a). To validate this result, CGGA microarray, CGGA RNAseq batch 1, CGGA RNAseq batch 2, GSE4290, GSE7696, GSE50161, GSE52009 and GSE54004 were used, and similar results were found (Fig. 2b–i). Furthermore, compared to normal brain tissues, different grades of glioma, including LGG and GBM, showed higher EMT scores (Fig. 2e–g). Together, these findings suggest that EMT scores increase with the malignant progression of glioma. Considering the underlying significance of EMT, which represents the acquisition of cancer invasiveness and metastasis abilities, we hypothesised that EMT scores may be a prognostic marker of glioma.
Different EMT scores exist between different grades of glioma and normal brain tissues. (a–i) Quantification of EMT scores in non-tumour brain and gliomas of grade II, grade III and grade IV using different datasets, TCGA (a), CGGA microarray (b), CGGA RNAseq batch 1 (c), CGGA RNAseq batch 2 (d), GSE4290 (e), GSE7696 (f), GSE50161 (g), GSE52009 (h) and GSE54004 (i)
The Prognostic Value of EMT Scores in Glioma
Using TCGA datasets, we revealed that high EMT scores were associated with decreased survival in patients with glioma, while low EMT scores were associated with increased survival in glioma patients (Fig. 3a). Next, we repeated the analysis in other datasets (CGGA microarray, CGGA RNAseq batch 1, CGGA RNAseq batch 2, GSE4412-GPL96) and we obtained consistent results (Fig. 3b–e). The results above were confirmed by univariate and multivariate Cox proportional hazard regression analyses (Fig. 3f). It is worth noting that all survival differences between the high ETM score group and the low ETM score group were significant in various datasets except for the CGGA microarray dataset. Therefore, we believe that EMT scores can serve as a reliable prognostic marker in patients with glioma.
Kaplan–Meier survival curves and Cox regression of EMT scores in glioma. (a–e) Correlation between patient survival and EMT scores (high or low) in glioma based on datasets from TCGA, CGGA microarray, CGGA RNAseq batch 1, CGGA RNAseq batch 2 and GSE4412-GPL96. The log-rank test was used for statistical analysis. (f) The unadjusted and adjusted (by age, gender, histology and grade) hazard ratio (HR) and P value of the Cox regression of (a–e)
The Role of EMT in the Glioma Immune Microenvironment
Recently, several reports have indicated that neoplastic cells expressing markers associated with EMT show increased resistance to elimination by components of the adaptive and innate immune systems (Akalay et al. 2013; Dongre et al. 2017; Kudo-Saito et al. 2009; Terry et al. 2017). However, whether these EMT-induced immunosuppressive effects also exist and play important roles in glioma remains largely unknown. Therefore, to further explore the relationship between EMT scores and the glioma immune microenvironment in a panoramic way, we not only enriched the ESTIMATE immune scores and stromal scores of gliomas but also calculated the abundance of various immune cells of the glioma microenvironment using three different tools, ABIS, EPIC and ImmuCellAI. Through Spearman correlation analysis, we found that high EMT scores were associated with high immune scores and stromal scores, which suggested that these glioma tissues have abundant non-neoplastic cells (immune cells and stromal cells) in their TME (Fig. 4a). Next, when we analysed the relationship between EMT scores and the immune cell subtypes, two opposite correlation patterns were revealed, namely positive correlation (cell subtypes that include macrophages, cancer-associated fibroblasts, monocytes, etc.) and negative correlation (cell subtypes that include CD8 T cells, CD4 naive T cells, B cells, etc.) (Fig. 4a). The correlation coefficients between EMT scores and immune scores, stromal scores and other main immune cells were fairly high (immune scores (ESTIMATE), rho = 0.74; stromal scores (ESTIMATE), rho = 0.8; macrophages (EPIC), rho = 0.66; macrophages (ImmuCellAI), rho = 0.48; CD8 T cells (EPIC), rho = −0.61; CD4 naive (ImmuCellAI), rho = −0.55) (Fig. 4b-g). It is known that TAMs are closely related to an immunosuppressive TME, while abundant CD8 T cells and CD4 T cells often signify a favourable prognosis (Hambardzumyan and Gutmann 2016; Hilf et al. 2019; Keskin et al. 2019). Together, our results suggest that the EMT of glioma may exert an immunosuppressive effect on the glioma microenvironment.
Association between EMT scores and the glioma immune microenvironment. (a) Heatmap of Spearman’s correlation between EMT scores and immune scores, stromal scores and the abundance of various immune cells in the TME in GBM (n = 155) and LGG (n = 510) samples generated using the TCGA dataset. (b and c) Spearman correlation analysis between EMT scores and immune scores (ESTIMATE) (b) and stromal scores (ESTIMATE) (c). (d and e) Spearman correlation analysis between EMT scores and macrophages (EPIC) (d) and macrophages (ImmuCellAI) (e). (f and g) Spearman correlation analysis between EMT scores and CD8 T cells (EPIC) (f) and CD4 naive cells(ImmuCellAI) (g)
EMT-Related Immune Genes Enrichment, Analysis and its Clinical Relevance
To further investigate the molecular signatures behind the EMT and immune microenvironment of glioma, we performed Spearman’s correlation analyses between EMT scores and gene expression and between immune scores and gene expression in the TCGA, CGGA and GEO datasets. The genes associated with both EMT scores and immune scores in at least 15 of the 16 datasets were considered enriched and are depicted in Fig. 5a. We can see that most of these genes were previously reported to play different roles in glioma tumourigenesis and progression (Table 1). However, few studies have investigated the specific role of these genes in EMT and immune responses in gliomas. For example, CD44, a transmembrane molecule, is overexpressed in many tumours (Ishimoto et al. 2010; Jijiwa et al. 2011). The role of CD44 has been implicated in various malignant processes, including cell motility, tumour growth and angiogenesis (Naor 2016; Naor et al. 2008). In glioma, studies found that CD44 expression increased tumour cell invasion, proliferation and therapy resistance to decrease survival in glioma patients (Anido et al. 2010; Ariza et al. 1995; Liu et al. 2018; Mooney et al. 2016). However, whether it has dual roles in the EMT of glioma and regulates the glioma immune environment remains unclear. To explore which signalling pathways are involved in the EMT-related immune genes, we conducted GO enrichment analysis of the EMT-related immune genes (Fig. 5b). Specifically, in the biological process module of the GO analysis, the top four pathways that had the highest number of EMT-related immnue genes were all related to neutrophils, namely neutrophil mediated immunity, neutrophil activation, neutrophil activation involved in the immune response and neutrophil degranulation, suggesting that the neutrophil signalling pathways may be of significance in regulating both glioma EMT and the immune microenvironment. The other main pathways enriched with EMT-related immune genes included T cell activation, leukocyte migration, regulation of inflammatory response, leukocyte cell–cell adhesion, positive regulation of defence response, positive regulation of cytokine production, extracellular structure organization, extracellular matrix organization, response to molecule of bacterial origin, coagulation, haemostasis, blood coagulation, response to interferon-gamma, cellular response to interferon-gamma, platelet degranulation and the interferon-gamma-mediated signalling pathway. Through gene expression analysis, we detected an increased expression of EMT-related immune genes corresponding to GBM, IDH-WT glioma and 1p/19q non-codeleted glioma (Fig. 5c). Overall, our analysis incorporated the EMT-related immune genes and the relevant signalling pathways and their clinical significance, suggesting the potentially basic and clinical utility of EMT-related immune genes.
EMT-related immune genes and their relevant signalling pathways and clinical relevance. (a) Correlation between EMT-related immune genes and EMT scores and immune scores. The colour of the dots denotes EMT scores and the area of the dots denotes immune scores. The X-axis is different datasets, and the y-axis is gene names. (b) GO signalling pathway enrichment analysis of EMT-related genes identified in more than 12 of the 16 datasets. (c) Heat map of the expression of genes (from a) in GBM (n = 168) and LGG (n = 467) samples using the TCGA dataset
Association of EMT-Related Immune Genes with Clinical Outcomes in Glioma
To characterize the relationship between glioma EMT-related immune genes and clinical outcomes, we built a multivariate model in a forward fashion using relevant variables (age, gender, histology, WHO grade) using TCGA datasets (Table 2). In the univariate analysis, all top 26 EMT-related immune genes were positively associated with a significant increase in mortality of glioma patients, and the top five hazard genes were OSMR, CASP4, ANXA1, SP100 and BACE2. Moreover, using these 26 genes in the multivariate model adjusted by age, gender, histology and WHO grade, 18 EMT-related immune genes were positively associated with poor outcomes of glioma patients, and the top five HR genes were OSMR, BACE2, RAC2, IFI30 and TIMP1. Taken together, these data support the role of EMT-related immune genes in glioma pathogenesis.
Discussion
During malignancy progression, neoplastic cells undergo dynamic and reversible transformations between a spectrum of phenotypic states, the extremes of which are defined by the expression of epithelial and mesenchymal phenotypes (Dongre and Weinberg 2019; Nieto et al. 2016; Tam and Weinberg 2013). In the course of EMT, the interactions between cells and between cells and the extracellular matrix are reshaped, which results in the detachment of epithelial cells from other cells and their adhesive basement membrane. At the same time, a transcriptional programme is activated to promote mesenchymal fate. Thus, neoplastic cells can successfully eliminate their differentiated epithelial traits, including polarity, cell–cell adhesion and absence of motility, and acquire mesenchymal characteristics, including invasiveness, motility and the stemness of malignancies, which are all associated with high-grade malignancy (Polyak and Weinberg 2009; Thiery et al. 2009). Our studies on the role of EMT in gliomagenesis and progression validated these findings, suggesting that EMT may be a shared characteristic of malignancies. Here, we found that EMT scores increased with the malignant progression of gliomas in different datasets and validated this observation as an excellent indicator for the prognosis of glioma. Behind this clinical significance, various paracrine signalling factors have been reported to trigger the induction of an EMT programme by activating a corresponding diverse cohort of inter- and intracellular signalling cascades (Scheel et al. 2011). In response, an array of EMT-inducing transcription factors (TFs), such as SLUG, SNAIL, TWIST and ZEB1, becomes expressed and functionally activated (De Craene and Berx 2013). In addition, EMT can be activated by different extracellular signalling factors or their combinations, such as metabolic end products and immunoreactive products in the TME.
With regard to the glioma microenvironment, its immune characteristic is most distinctive for the extremely powerful immunosurveillance escape ability (Ito et al. 2019; Lim et al. 2018; Weller and Le Rhun 2019). Compared with melanoma, in which approximately 50% of patients respond to a combination blockade of CTLA-4 and PD-1 immunotherapy and the response is durable in approximately 75% of patients, in glioma only 10% of patients respond to immunotherapy (Jackson et al. 2019). Therefore, in the context of the revolutionary clinical success of immunotherapy for a variety of solid tumours and haematological tumours, it is urgent to determine the molecular pathways underlying glioma immune escape and immunotherapy resistance. In the central nervous system, the first line of immune defence of glioma is the blood–brain barrier. Then, a large number of TAMs (constituting more than 30% of infiltrating cells in GBM) in the TME exert powerful effects to induce immunosuppression. Furthermore, preclinical studies have revealed that the intracranial location of a tumour leads to systemic immunotolerance against relevant tumour antigens (Chongsathidkiet et al. 2018; Hambardzumyan et al. 2016). Regarding the intrinsic resistance in glioma, tumour-infiltrating lymphocytes are scarce, and a previous report showed that the CD4+ T cell counts dropped below 300 cells/mm3 in most GBM patients who were undergoing temozolomide treatment and radiotherapy (Grossman et al. 2011). By preventing internalization of the G protein–coupled receptor sphingosine-1-phosphate receptor 1, GBM and other brain malignancies can sequester a large number of functional T cells in the bone marrow (Chongsathidkiet et al. 2018). For the adaptive resistance of glioma, preclinical studies showed that T cells infiltrating glioma express a series of immune checkpoints and present a severe exhaustion signature. Even so, the response of glioma to checkpoint blockades is poor (Ito et al. 2019). There is no question that all these resistance mechanisms are involved in the different stages of glioma immunosuppression and are vital for guiding the clinical practices of glioma treatment. There are also other mechanisms, for example, activation of EMT enables carcinoma cells to express several immunoevasive and immunosuppressive molecules that make themselves resistant to CD8+ T cell cytoplasmic attack (Akalay et al. 2013). Moreover, EMT can induce the expression of TSP1 and TGFβ, which promote the formation of Treg cells, while EMT-induced expression of CCL2 and LCN2 can promote the formation of tolerance-inducing dendritic cells (DCs) (Kudo-Saito et al. 2013; Sangaletti et al. 2016). The expression of programmed cell death 1 ligand 1 (PD-L1) can be induced by EMT-specific TFs, while the expression of MHC class I can be decreased in mesenchymal neoplastic cells (Noman et al. 2017). Similarly, pro-tumorigenic M2 macrophages can be recruited into the TME through mesenchymal tumour cells (Hsu et al. 2014). Even though it is increasingly appreciated that EMT and tumour immunoresistance are inextricably linked in an array of malignant tumours, the interaction of EMT and the immunosuppressive glioma microenvironment is largely unknown, let alone the molecular mechanisms behind it. Here, however, we revealed the close relationship between EMT scores and the glioma immune microenvironment at the gene level.
In summary, our studies shed new light on the underlying mechanism of EMT and the adaptation of the glioma immune microenvironment. We have not only uncovered the clinical relevance of EMT scores and their excellent prognostic value in glioma patients, but also raised the possibility that EMT signalling pathways may be at the core of glioma immune resistance and tumour relapse from immunotherapy. We found that all top 26 EMT-related immune genes played critical roles in oncobiology. Importantly, we validated their significance with regard to clinical outcomes of glioma patients. Clinically, our findings contribute to the underlying biological basis for the interaction of EMT and the glioma immune environment.
References
Akalay I, Janji B, Hasmim M et al (2013) Epithelial-to-mesenchymal transition and autophagy induction in breast carcinoma promote escape from T-cell-mediated lysis. Cancer Res 73:2418–2427. https://doi.org/10.1158/0008-5472.Can-12-2432
Anido J, Sáez-Borderías A, Gonzàlez-Juncà A et al (2010) TGF-beta receptor inhibitors target the CD44(high)/Id1(high) Glioma-initiating cell population in human Glioblastoma. Cancer Cell 18:655–668. https://doi.org/10.1016/j.ccr.2010.10.023
Ariza A et al (1995) Role of CD44 in the invasiveness of glioblastoma multiforme and the noninvasiveness of meningioma: an immunohistochemistry study. Hum Pathol 26:1144–1147. https://doi.org/10.1016/0046-8177(95)90278-3
Barbie DA, Tamayo P, Boehm J et al (2009) Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462:108–112. https://doi.org/10.1038/nature08460
Bauer HC, Krizbai IA, Bauer H, Traweger A (2014) "you shall not pass"-tight junctions of the blood brain barrier. Front Neurosci 8:392. https://doi.org/10.3389/fnins.2014.00392
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68:394–424. https://doi.org/10.3322/caac.21492
Ceccarelli M, Barthel F, Malta T et al (2016) Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell 164:550–563 https://doi.org/10.1016/j.cell.2015.12.028
Chongsathidkiet P, Jackson C, Koyama et al (2018) Sequestration of T cells in bone marrow in the setting of glioblastoma and other intracranial tumors. Nat Med 24:1459–1468. https://doi.org/10.1038/s41591-018-0135-2
De Craene B, Berx G (2013) Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer 13:97–110. https://doi.org/10.1038/nrc3447
Dongre A, Rashidian M, Reinhardt F, Bagnato A, Keckesova Z, Ploegh HL, Weinberg RA (2017) Epithelial-to-Mesenchymal transition contributes to immunosuppression in breast carcinomas. Cancer Res 77:3982–3989. https://doi.org/10.1158/0008-5472.Can-16-3292
Dongre A, Weinberg RA (2019) New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol 20:69–84. https://doi.org/10.1038/s41580-018-0080-4
Ferlay J, Colombet M, Soerjomataram I et al (2019) Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer 144:1941–1953. https://doi.org/10.1002/ijc.31937
Gajewski TF, Schreiber H, Fu YX (2013) Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol 14:1014–1022. https://doi.org/10.1038/ni.2703
Gong J, Chehrazi-Raffle A, Reddi S, Salgia R (2018) Development of PD-1 and PD-L1 inhibitors as a form of cancer immunotherapy: a comprehensive review of registration trials and future considerations. Journal for immunotherapy of cancer 6:8. https://doi.org/10.1186/s40425-018-0316-z
Grossman SA, Ye X, Lesser G, Sloan A, Carraway H, Desideri S, Piantadosi S (2011) Immunosuppression in patients with high-grade gliomas treated with radiation and temozolomide. Clinical cancer research : an official journal of the American Association for Cancer Research 17:5473–5480. https://doi.org/10.1158/1078-0432.Ccr-11-0774
Hambardzumyan D, Gutmann DH, Kettenmann H (2016) The role of microglia and macrophages in glioma maintenance and progression. Nat Neurosci 19:20–27. https://doi.org/10.1038/nn.4185
Hilf N, Kuttruff-Coqui S, Frenzel K et al (2019) Actively personalized vaccination trial for newly diagnosed glioblastoma. Nature 565:240–245. https://doi.org/10.1038/s41586-018-0810-y
Hsu DS, Wang H-J, Tai S-K et al (2014) Acetylation of snail modulates the cytokinome of cancer cells to enhance the recruitment of macrophages. Cancer Cell 26:534–548. https://doi.org/10.1016/j.ccell.2014.09.002
Ishimoto T, Oshima H, Oshima M et al (2010) CD44+ slow-cycling tumor cell expansion is triggered by cooperative actions of Wnt and prostaglandin E2 in gastric tumorigenesis. Cancer Sci 101:673–678. https://doi.org/10.1111/j.1349-7006.2009.01430.x
Ito H, Nakashima H, Chiocca EA (2019) Molecular responses to immune checkpoint blockade in glioblastoma. Nat Med 25:359–361. https://doi.org/10.1038/s41591-019-0385-7
Jackson CM, Choi J, Lim M (2019) Mechanisms of immunotherapy resistance: lessons from glioblastoma. Nat Immunol 20:1100–1109. https://doi.org/10.1038/s41590-019-0433-y
Jijiwa M, Demir H, Gupta S et al (2011) CD44v6 regulates growth of brain tumor stem cells partially through the AKT-mediated pathway. PLoS One 6:e24217. https://doi.org/10.1371/journal.pone.0024217
Kerkar SP, Restifo NP (2012) Cellular constituents of immune escape within the tumor microenvironment. Cancer Res 72:3125–3130. https://doi.org/10.1158/0008-5472.Can-11-4094
Keskin DB, Anandappa A, Sun J et al (2019) Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature 565:234–239. https://doi.org/10.1038/s41586-018-0792-9
Kudo-Saito C, Shirako H, Ohike M, Tsukamoto N, Kawakami Y (2013) CCL2 is critical for immunosuppression to promote cancer metastasis. Clinical & experimental metastasis 30:393–405. https://doi.org/10.1007/s10585-012-9545-6
Kudo-Saito C, Shirako H, Takeuchi T, Kawakami Y (2009) Cancer metastasis is accelerated through immunosuppression during snail-induced EMT of cancer cells. Cancer Cell 15:195–206. https://doi.org/10.1016/j.ccr.2009.01.023
Lambert AW, Pattabiraman DR, Weinberg RA (2017) Emerging biological principles of metastasis. Cell 168:670–691. https://doi.org/10.1016/j.cell.2016.11.037
Lim M, Xia Y, Bettegowda C, Weller M (2018) Current state of immunotherapy for glioblastoma nature reviews. Clin Oncol 15:422–442. https://doi.org/10.1038/s41571-018-0003-5
Liu Z, Kuang W, Zhou Q, Zhang Y (2018) TGF-beta1 secreted by M2 phenotype macrophages enhances the stemness and migration of glioma cells via the SMAD2/3 signalling pathway. Int J Mol Med 42:3395–3403. https://doi.org/10.3892/ijmm.2018.3923
Lu KV, Chang JP, Parachoniak C et al (2012) VEGF inhibits tumor cell invasion and mesenchymal transition through a MET/VEGFR2 complex. Cancer Cell 22:21–35. https://doi.org/10.1016/j.ccr.2012.05.037
McGranahan T, Therkelsen KE, Ahmad S, Nagpal S (2019) Current state of immunotherapy for treatment of glioblastoma. Curr Treat Options Oncol 20:24 https://doi.org/10.1007/s11864-019-0619-4
Mellman I, Coukos G, Dranoff G (2011) Cancer immunotherapy comes of age. Nature 480:480–489. https://doi.org/10.1038/nature10673
Miao Y-R, Zhang Q, Lei Q, Luo M, Xie G-Y, Wang H, Guo A-Y (2019) ImmuCellAI: a unique method for comprehensive T-cell subsets abundance prediction and its application in cancer immunotherapy bioRxiv. Adv Sci. https://doi.org/10.1101/872184
Molinaro AM, Taylor JW, Wiencke JK, Wrensch MR (2019) Genetic and molecular epidemiology of adult diffuse glioma nature reviews. Neurology 15:405–417. https://doi.org/10.1038/s41582-019-0220-2
Monaco G, Lee B, Xu W et al (2019) RNA-Seq signatures normalized by mRNA abundance allow absolute deconvolution of human immune cell types. Cell Rep 26:1627–1640 e1627. https://doi.org/10.1016/j.celrep.2019.01.041
Mooney KL, Choy W, Sidhu S et al (2016) The role of CD44 in glioblastoma multiforme. J Clin Neurosci 34:1–5. https://doi.org/10.1016/j.jocn.2016.05.012
Naor D (2016) Editorial: interaction between hyaluronic acid and its receptors (CD44, RHAMM) regulates the activity of inflammation and Cancer. Front Immunol 7:39. https://doi.org/10.3389/fimmu.2016.00039
Naor D, Wallach-Dayan SB, Zahalka MA, Sionov RV (2008) Involvement of CD44, a molecule with a thousand faces, in cancer dissemination Seminars in cancer biology 18:260–267 https://doi.org/10.1016/j.semcancer.2008.03.015
Nieto MA, Huang RY, Jackson RA, Thiery JP (2016) EMT: 2016. Cell 166:21–45. https://doi.org/10.1016/j.cell.2016.06.028
Noman MZ, Janji B, Abdou A et al (2017) The immune checkpoint ligand PD-L1 is upregulated in EMT-activated human breast cancer cells by a mechanism involving ZEB-1 and miR-200. Oncoimmunology 6:e1263412. https://doi.org/10.1080/2162402x.2016.1263412
Polyak K, Weinberg RA (2009) Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer 9:265–273. https://doi.org/10.1038/nrc2620
Racle J, de Jonge K, Baumgaertner P, Speiser DE, Gfeller D (2017) Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. Elife. https://doi.org/10.7554/eLife.26476
Sangaletti S, Tripodo C, Santangelo A et al (2016) Mesenchymal transition of high-grade breast carcinomas depends on extracellular matrix control of myeloid suppressor cell activity. Cell Rep 17:233–248. https://doi.org/10.1016/j.celrep.2016.08.075
Scheel C, Eaton EN, Li SH-J et al (2011) Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell 145:926–940. https://doi.org/10.1016/j.cell.2011.04.029
Shibue T, Weinberg RA (2017) EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol 14:611–629. https://doi.org/10.1038/nrclinonc.2017.44
Tam WL, Weinberg RA (2013) The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med 19:1438–1449. https://doi.org/10.1038/nm.3336
Terry S, Savagner P, Ortiz-Cuaran S, Mahjoubi L, Saintigny P, Thiery JP, Chouaib S (2017) New insights into the role of EMT in tumor immune escape. Mol Oncol 11:824–846. https://doi.org/10.1002/1878-0261.12093
Thiery JP, Acloque H, Huang RY, Nieto MA (2009) Epithelial-mesenchymal transitions in development and disease. Cell 139:871–890. https://doi.org/10.1016/j.cell.2009.11.007
Weller M, Le Rhun E (2019) Immunotherapy for glioblastoma: quo vadis? Nat Rev Clin Oncol 16:405–406. https://doi.org/10.1038/s41571-019-0195-3
Wen PY, Kesari S (2008) Malignant gliomas in adults. N Engl J Med 359:492–507. https://doi.org/10.1056/NEJMra0708126
Woroniecka K, Chongsathidkiet P, Rhodin K et al (2018) T-cell exhaustion signatures vary with tumor type and are severe in Glioblastoma. Clin Cancer Res 24:4175–4186. https://doi.org/10.1158/1078-0432.Ccr-17-1846
Yoshihara K, Shahmoradgoli M, Martínez E et al (2013) Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 4:2612. https://doi.org/10.1038/ncomms3612
Yu G, Wang LG, Han Y, He QY (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16:284–287. https://doi.org/10.1089/omi.2011.0118
Acknowledgements
We thank the investigators and patients who participated in CGGA, TCGA and GEO for providing data.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors have no conflicts of interest to report.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ning, W., Qiu, Z., Ji, X. et al. The Prognostic Value of EMT in Glioma and its Role in the Glioma Immune Microenvironment. J Mol Neurosci 70, 1501–1511 (2020). https://doi.org/10.1007/s12031-020-01583-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12031-020-01583-y