Abstract
Widespread inflammatory lesions within the central nervous system grey and white matter are major hallmarks of multiple sclerosis. The development of full-blown demyelinating multiple sclerosis lesions might be preceded by preactive lesions which are characterized by focal microglia activation in close spatial relation to apoptotic oligodendrocytes. In this study, we investigated the expression of signaling molecules of oligodendrocytes that might be involved in initial microglia activation during preactive lesion formation. Sodium azide was used to trigger mitochondrial impairment and cellular stress in oligodendroglial cells in vitro. Among various chemokines and cytokines, IL6 was identified as a possible oligodendroglial cell-derived signaling molecule in response to cellular stress. Relevance of this finding for lesion development was further explored in the cuprizone model by applying short-term cuprizone feeding (2–4 days) on male C57BL/6 mice and subsequent analysis of gene expression, in situ hybridization and histology. Additionally, we analyzed the possible signaling of stressed oligodendroglial cells in vitro as well as in the cuprizone mouse model. In vitro, conditioned medium of stressed oligodendroglial cells triggered the activation of microglia cells. In cuprizone-fed animals, IL6 expression in oligodendrocytes was found in close vicinity of activated microglia cells. Taken together, our data support the view that stressed oligodendrocytes have the potential to activate microglia cells through a specific cocktail of chemokines and cytokines among IL6. Further studies will have to identify the temporal activation pattern of these signaling molecules, their cellular sources, and impact on neuroinflammation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Stressed oligodendrocytes in close proximity to foamy macrophages and clustered microglia expressing HLA-DR are found widespread within the normal appearing white matter of multiple sclerosis (MS) patients (De Groot et al. 2001; Zeis et al. 2009). These preactive lesions are thought to precede the development of full-blown demyelinating MS lesions (De Groot et al. 2001; Wuerfel et al. 2004; van der Valk and Amor 2009). Therefore, it seems reasonable to assume that the initial microglia activation and clustering as observed in preactive lesions might be triggered by oligodendrocyte-derived signaling molecules. It has been shown that oligodendrocytes are able to secrete a variety of signaling molecules such as chemokines and cytokines and other regulatory proteins which are known to be involved in the regulation of immunological processes (Cannella and Raine 2004; Balabanov et al. 2007; Kummer et al. 2007; Okamura et al. 2007; Tzartos et al. 2008; Merabova et al. 2012; Ramesh et al. 2012; Moyon et al. 2015). For example, IFNγ-treated primary rat oligodendrocytes significantly induced the expression of the chemokines CXCL10, CCL2, CCL3, and CCL5 (Balabanov et al. 2007). Furthermore, the cytokines IL6 and IL8 and the chemokine CCL2 were significantly induced in the human oligodendrocyte cell line MO3.13 when confronted with Borrelia burgdorferi. Exposure to the same bacteria caused the induction of IL8 and CCL2 in a dose-dependent manner in primary human oligodendrocytes (Ramesh et al. 2012).
So far, it is not completely understood which processes trigger oligodendrocyte stress and subsequent expression of signaling molecules. One factor leading to oligodendrocyte stress, impaired mitochondrial functions, and increased levels of reactive oxygen species (Wang et al. 2013) might be the accumulation of mutations within the mitochondrial genome. It is known that the mtDNA is prone to mutations with a mutation rate that is about tenfold higher than chromosomal DNA (Linnane et al. 1989). In comparison to other cell populations, oligodendrocytes have a reduced capacity to repair their mtDNA, possibly due to particularities in the expression of factors involved in DNA repairing mechanisms (Hollensworth et al. 2000). With respect to demyelination and oligodendrocyte pathology, both mutations in mitochondrial genes as well as oxidative stress play a role in lesion formation and disease progression in MS and MS-related animal models (Mahad et al. 2008; Mao and Reddy 2010; Su et al. 2013; Draheim et al. 2016).
As the sentinels and injury sensors of the central nervous system (CNS), microglia can be activated by many kinds of mechanical injury and pathological disturbances within the CNS (Perry et al. 1993; Gehrmann et al. 1995). In MS, the magnitude of myelin loss during demyelinating events positively correlates with the number of activated microglia cells (Clarner et al. 2012). Furthermore, microgliosis can be induced by leukocyte infiltration into the CNS (Scheld et al. 2016; Ruther et al. 2017). However, the listed factors are rather unlikely to contribute to the activation of microglia cells during the formation of preactive lesions, since the affected brain regions lack any signs of demyelination, leukocyte infiltration, astrogliosis or inciting agents such as viral or bacterial antigens (Gay et al. 1997; De Groot et al. 2001; Barnett and Prineas 2004; Marik et al. 2007; van der Valk and Amor 2009). A growing body of evidence suggests that stressed oligodendrocytes might be active contributors to MS lesion formation by initiating microglia reactivity. Barnett and Prineas have reported areas with an extensive oligodendrocyte apoptosis and concomitant microgliosis in the absence of infiltrating immune cells in the normal appearing white matter of MS patients, thus indicating that oligodendrocyte loss precedes inflammatory demyelination (Barnett and Prineas 2004).
In this study, we use the oligodendroglial cell line OLN93 to screen for oligodendrocyte-derived chemokines and cytokines that are potentially able to activate microglia cells. Subsequent culture experiments with the microglia cell line BV2 show that IL6, which is robustly expressed by stressed oligodendroglial cells, is a constituent of a cocktail of secreted proteins, which are responsible for microglia activation. Finally, IL6 expression was explored in the cuprizone model, which recapitulates distinct but important aspects of early MS lesion formation.
Materials and Methods
Cell Culture
Cells of the oligodendroglial linage cell line OLN93 were received from Dr. C. Richter-Landsberg (RRID: CVCL_5850; Oldenburg, Germany) and chosen as a model system because of strong similarities to primary oligodendrocytes regarding morphology and gene expression (Richter-Landsberg and Heinrich 1996). BV2 cell line was cultured according to Dr. E. Blasi (RRID: CVCL_0182; Modena, Italy) and chosen as a model system because of its suitability to study molecular mechanisms that control induction and expression of biological activities in microglia (Blasi et al. 1990). Cell lines were not authenticated prior to experiments. None of the cell lines used in these experiments is listed as a commonly misidentified cell line by the International Cell Line Authentication Committee. OLN93 oligodendroglial cell line and BV2 microglial cell lines were maintained in Dulbecco’s Modified Eagle Medium supplemented with 5% (OLN93) and 10% (BV2) heat-inactivated fetal bovine serum, penicillin G (10,000 units/mL), and streptomycin (10,000 μg/mL). For experiments, cells were seeded into 6-well dishes, 10 cm culture dishes or 75 cm2 flasks at densities of 3 × 105, 5 × 106, and 2.3 × 106 cells per well, respectively. Before treatment, cells were cultivated for 24 h in starving medium (OLN93: SATO with 1% penicillin G/streptomycin; BV2: DMEM supplemented with 0.5% fetal calf serum and 0.5% penicillin G/streptomycin). Cells were cultivated in a humidified atmosphere of 5% CO2 at 37 °C.
Oligodendroglial Cell-Conditioned Medium (OCM)
OLN93 cells were treated with 10 mM sodium azide (SA, Sigma Aldrich) or vehicle (UltraPure Distilled Water; Thermo Fisher Scientific) for 24 h. After washing twice with 1× phosphate buffered saline (PBS), cells were incubated with starving medium for a 24 h secretion period. The medium of each treatment group was pooled, centrifuged, and filtered through a 20 μm cell strainer. BV2 microglia were incubated with OCM from SA-treated cells (OCM-SA) for 6 h. Oligodendroglial starving medium and OCM from vehicle groups (OCM-vehicle) served as control. OCM was additionally used for ELISA analysis (see supplementary figure A for experimental overview).
Cell Viability and Metabolic Activity Assay
To investigate the toxic effects of the applied SA concentrations, CytoTox 96® Non-Radioactive Cytotoxicity Assay and CellTiter-Blue® Cell Viability Assay (Promega G1780, G8081) were performed according to the manufacturer’s instructions. Briefly, cells were seeded into an opaque-walled 96 tissue culture plate and treated with SA and vehicle for 24 h. Treatment of cells with a lysis solution served as a negative control as all cells are dead; medium without cells served as blank. Cells were incubated with CellTiter-Blue reagent until a change of color was observed; fluorescence was measured at 560/590 nm with the Tecan infinite M200 plate reader and processed with i-control 1.10 software. Medium of cells was incubated for 30 min with CytoTox96 reagent and the reaction was stopped with stop solution. Absorbance was measured at 490 nm with Tecan i-control software. Data are given in % of control fluorescence and absorbance values, respectively. No blinding was performed for the evaluation of these data. Experiments were performed with eight biological and two technical replicates.
Enzyme-linked Immunosorbent Assay
IL6 ELISA was conducted using cell culture supernatants of vehicle and SA-treated OLN93 cells (OCM) according to the manufacturer’s protocol (Quantikine ELISA, R&D Systems). Color development of substrate solution (stabilized hydrogen peroxide and stabilized chromogen (tetramethylbenzidine)) was monitored with a Tecan infinite M200 plate reader at 450 nm with a wavelength correction at 540 nm and processed with i-control 1.10 software. IL6 protein levels are displayed as absolute values in pg/ml. Experiments were performed with two biological and two technical replicates.
Animals and Cuprizone Intoxication
C57BL/6J male mice (19 ± 2 g) were obtained from Janvier and housed under standard laboratory conditions in the animal facility of the Uniklinik Aachen according to the Federation of European Laboratory Animal Science Association’s recommendations. Mice were maintained with food and water ad libitum in a 12 h light/dark cycle at controlled temperature and humidity (23 ± 2 °C; 55% ± 10% humidity). Experimental procedures, i.e., cuprizone feeding, were approved by the Review Board for the Care of Animal Subjects of the district government (Nordrhein-Westfalen, Germany). At noon, mice received a diet containing 0.25 % cuprizone (bis-cyclohexanone-oxaldihydrazone, Sigma Aldrich; choice of concentration via established protocols) for up to 2 days mixed into a ground standard rodent chow. The control group was fed with standard rodent chow. Animals were allocated to groups applying the following procedure: Animals were distributed across cages (three animals per cage; cage area 435 cm2) and each group consisted of mice with comparable weight. We used cards numbered from 1 to 2 for the respective experimental group (1 = control, 2 = 2 days cuprizone). The number on the card randomly assigned the cages to the respective group. Re-evaluation of cDNA samples of 1–4 days cuprizone-treated mice were performed using previously published work from our research group (Krauspe et al. 2015). Size of groups for each experiment is given in the appropriate figure legends and an experimental overview is shown in supplementary figure B.
Tissue Preparation
Mice were anaesthetized with ketamine/xylazine (100 mg/kg and 10 mg/kg; i.p. with 100 μl/10 g body weight) and transcardially perfused with either PBS or 3.7% formalin in PBS. Brains were removed to isolate RNA of the corpus callosum (CC) for gene expression analysis or whole brains were post-fixed in 3.7% formalin and subsequently embedded into paraffin for immunohistological analysis following established protocols (Clarner et al. 2015).
Immunohistological Staining and Fluorescence Labeling
For fluorescence and immunohistological analysis, 5 μm thick brain slides were cut with a microtome. Rabbit anti-IL6 (Abcam ab7737), mouse anti-OLIG2 (Millipore MABN50), donkey-anti-rabbit 488, and donkey-anti-mouse 594 were used for fluorescence labeling (Life Technologies A21206, A21203). Signal specificity was validated by incubating slices with the respective secondary antibody without pre-incubation with the first antibody (see supplementary figure C). Furthermore, cross reactivity of secondary antibodies with each other or the false primary antibody was additionally excluded (data not shown). For chromogen double labeling, anti-GFAP (Santa Cruz sc-6170), anti-IBA-1 (Millipore MABN92), and anti-APC (Millipore OP80) antibodies were visualized with a horseradish peroxidase enzyme (Vector Labs) and DAB substrate (Dako); anti-IL6 (Abcam ab6672) was visualized with an alkaline phosphatase (Zytomed Systems) and an AP Blue substrate that emits at 680 nm (Vector Laboratories).
In Situ Hybridization
Commercial fluorescence in situ hybridization kits (QuantiGene View RNA in situ hybridization tissue assay; Affymetrix-Panomics) were used for double labeling of formalin-fixed, paraffin-embedded tissue, following the manufacturer’s recommendations. Protease digestion time was adjusted to 20 min. Probes directed against Olig2 and Il6 were purchased from Affymetrix (Affymetrix-Panomics). Confocal images were captured using the LSM710 laser-scanning microscope station (Carl Zeiss).
Chemokine and Cytokine Array
RNA isolation was performed with RNeasy Micro Kit (Qiagen 74004). Cells were directly lysed with RLT buffer and homogenized with Precellys homogenizer. Further isolation procedures such as washing and DNA digestion were conducted in MinElute spin columns. RNA concentration was measured with NanoDrop 1000 spectrometer. Five hundred nanograms of RNA were reverse-transcribed with RT2 First Strand Kit (Qiagen 330404). Genomic DNA was eliminated and RNA reverse-transcribed according to the manufacturer’s instructions. Cytokine & Chemokine RT2 Profiler PCR Array (Qiagen PARN-150ZD) was performed with RT2 SYBR Green Mastermix and processed with Bio-Rad CFX connect cycler. PCR cycling program was composed of one 10 min 95 °C hot start cycle and 40 cycles of 95 °C 15 s and 60 °C 1 min to perform fluorescence data collection. Results were analyzed using the Data analysis center from Qiagen (https://www.qiagen.com/de/shop/genes-and-pathways/data-analysis-center-overview-page). β-Actin, β-2 microglobulin, hypoxanthine phosphoribosyltransferase 1, lactate dehydrogenase A, and ribosomal protein large P1 served as reference genes. Furthermore, the array contained one genomic DNA control, three replicate reverse-transcription controls to test reverse-transcription efficiency, and three replicate positive PCR controls to test PCR efficiency. Experiments were performed with one biological and one technical replicate.
Gene Expression Analysis
RNA for array validation and microglia gene expression analysis was isolated by using peqGold TriFast (Peqlab) and reverse-transcribed in a 20 μL reaction volume using a reverse-transcription kit (Thermo Fisher Scientific 28025-021). cDNA levels were then analyzed by qPCR using SensiMix SYBR® & Fluorescein Kit (Bioline QT615-05) and Bio-Rad CFX connect cycler. The expression levels were calculated relative to the reference genes coding for glyceraldehyde 3-phosphate dehydrogenase or cyclophilin A using the ΔΔCt method. Primer sequences are given in Table 1. No blinding was performed for the evaluation of these data. Experiments were performed with at least six biological and two technical replicates if not stated otherwise. Furthermore, a gene array (Affymetrix) from the corpus callosum (CC) of control animals and animals that were fed cuprizone for 2 days was re-evaluated with respect to those genes identified in the OLN93 gene expression study (Krauspe et al. 2015).
Statistical Analysis
Statistical analysis was performed using JMP10 and GraphPad Prism 5. Data are presented as arithmetic means ± SEM. To test for equal variances, Bartlett test was performed. Data transformations via Boxcox for homoscedasticity are indicated if necessary. Shapiro-Wilk test was used to test for normal distribution. Parametric data were analyzed with one-way ANOVA followed by Tukey’s post hoc test for multiple comparisons or with Student’s t test. Non-parametric data were analyzed with Kruskal-Wallis test followed by the Dunn’s multiple comparison or Mann-Whitney U test. p < 0.05 was considered statistically significant. The following symbols were used to indicate the level of significance: *p < 0.05, **p < 0.005, ***p < 0.001; ns indicates not significant. No outliers were excluded from the analyses. No sample size calculation was performed.
Results
Sodium Azide Induces Stress in Oligodendroglial Cells
SA is a potent inhibitor of the mitochondrial respiratory chain and inhibits the mitochondrial complex IV which is responsible for the transfer of cytochrome c to an oxygen molecule (Bennett et al. 1996; Teske et al. 2018). To induce a breakdown of the mitochondrial inner transmembrane potential (i.e., sub-lethal mitochondrial stress reaction), the oligodendroglial cell line OLN93 was stimulated with 10 mM sodium azide (SA) for 24 h as previously described by Teske et al. (2018). Afterwards, cells were kept in culture for additional 24 h to produce OCM (oligodendroglial cell-conditioned medium). To confirm that the used SA concentration was not inducing immediate cell death, lactate dehydrogenase (LDH) cytotoxicity assay and cell titer blue (CTB) viability assay were performed after 24 h of SA-treatment. Results are shown in Fig. 1a. LDH levels in the cell culture supernatants were significantly increased after SA treatment compared to vehicle, indicating cell death; LDH levels of lysis control were significantly increased compared to both treatment groups (left histogram in Fig. 1a). SA-induced LDH release was paralleled by a trend towards lower levels of metabolic activity in these cells (right histogram in Fig. 1a); however, this difference was not statistically significant. Additionally, 10 mM SA-treated OLN93 cells did not show major morphological changes or loss of cell numbers at the microscopic level (Fig. 1b). DDIT3 and ATF3 are members of the integrated stress response which is closely connected to oxidative stress. Gene expression analysis of Ddit3 (Rutkowski et al. 2006; Puthalakath et al. 2007) and Atf3 (Edagawa et al. 2014) was measured after the 24 h secretion phase via qPCR to confirm endoplasmic reticulum stress in SA-treated cells on the transcriptional level (Fig. 1c).
a The cytotoxicity (left) assay normalized to the lysis control and cell viability (right) assay normalized to the vehicle-treated cells of SA- and vehicle-treated OLN93 cells (eight culture wells, one experiment). b Microscopic pictures using the FLoidTM Cell Imaging station (Thermo Fisher Scientific) of vehicle and SA-treated oligodendroglial cells did not reveal obvious signs of cell loss or death (× 20 magnification). c Gene expression analysis of Ddit3 and Atf3 (BoxCox-Y transformed) in vehicle- and SA-treated oligodendroglial cells indicating metabolic stress (at least six culture wells, five independent experiments). ns, not significant; *p < 0.05; **p < 0.005; ***p < 0.001; vehicle, up H2O; scale bars 50 μm
Stressed Oligodendroglial Cells Secrete Various Cytokines and Chemokines
In a next step, we aimed to analyze the gene expression of secreted molecules that oligodendroglial cells produce upon SA-induced mitochondrial stress. To get a first hint about the possible identity of such signaling molecules, we performed PCR arrays that screened for mRNA expression levels of 84 cytokines and chemokines with one sample (n = 1). Out of the 84 investigated genes, 13 were induced by at least twofold when comparing the SA group with the vehicle group. Those 13 genes are displayed in Fig. 2a. IL6 and GDF15 were most robustly induced in stressed oligodendroglial cells compared to cells treated with vehicle. To confirm validity of the path finding gene array, we performed qPCR in independent samples from two additional experiments with each n = 6 for IL6, GDF15, and three randomly assigned genes (Fig. 2b). As indicated in Fig. 2b, IL6 gene expression and GDF15 gene expression were found to be induced > 12-fold in qPCR analysis in SA-treated OLN93 cells. Differences in between the gene expression measured in array analysis versus qPCR are likely due to different evaluation strategies, i.e., different reference genes and subsequent software analysis and the measuring of only one sample in the PCR array. In comparison, genes for SPP1, LIF, and CSF1 were induced 2.5- to 4-fold. In a next step, the relevance of the identified factors in vivo was investigated. Since short-term cuprizone intoxication mimics early lesion formation and induces stress in oligodendroglial cells, we re-evaluated gene array data from the CC of 2-day cuprizone-fed and control animals (Krauspe et al. 2015). Results of this evaluation are shown in Fig. 2c. Out of the 13 investigated genes, 7 were significantly induced. Although GDF15 and CCL7 were highest induced in the tissue samples from cuprizone-fed animals, oligodendroglial cells might be responsible—at least in part—for the increased IL6 expression in vivo. In a next step, we investigated whether oligodendroglial cells secrete IL6 on the protein level. Therefore, sandwich ELISA was performed using OCM from SA-treated and vehicle-treated oligodendroglial cells. Results are shown in Fig. 2e and demonstrate a fourfold increase in IL6 levels upon SA-treatment. Our data show that stressed oligodendroglial cells express various signaling molecules that potentially activate microglia cells. To further investigate this hypothesis, the activation state of microglia cells was evaluated in OCM-stimulated BV2 microglia. Morphologically, no differences were found between cells growing in control medium (DMEM 0.5 %) and those cells grown in OCM-SA. Representative pictures after 6 h incubation time are shown in Fig. 2d. Additionally, performed arborization analysis and counting of bipolar cells did not reveal morphological changes in response to OCM-SA (supplementary figure E).
a The fold difference of all chemokines and cytokines that were induced at least twofold in SA-treated OLN93 cells compared to control in the PCR array. b To validate the PCR array results, the gene expression was analyzed via qPCR on independent samples (at least six culture wells, two independent experiments). In c, the re-evaluation of a gene array from the CC of 2-day cuprizone-intoxicated mice is shown. Data are displayed as fold induction of control. Note that a selective induction of the identified factors in oligodendroglial cells might be obscured due to the fact that this array measured expression in the whole tissue and not exclusively in oligodendrocytes (three animals per group, one experiment, see Krauspe et al. 2015). d Representative pictures after 6 h incubation time of BV2 microglia cells with OCM-SA and DMEM 0.5% (control). e IL6 protein levels increased up to fourfold in stressed oligodendroglial cells versus unstressed oligodendroglial cells (BoxCox-Y transformed; six samples each group, two independent experiments). f The gene expression results of pro- and anti-inflammatory markers arginase 1 (BoxCox-Y transformed) and Nos2 of OCM-stimulated microglia cells (at least five culture wells, four independent experiments). g Induction of Nos2 expression by OCM was partly (not significant) counteracted by anti-IL6 antibody in the media. ns, not significant; *p < 0.05; **p < 0.005; ***p < 0.001; vehicle, up H2O; scale bars, 30 μm
Expression levels of typical pro- and anti-inflammatory microglia markers were measured by means of qPCR 6 h after beginning of treatment (see supplementary figure A for experimental overview). Results of these experiments are shown in Fig. 2f. Gene expression of the anti-inflammatory marker arginase 1 was induced when BV2 microglia cells were treated with OCM from SA-treated oligodendroglial cells compared to OCM-vehicle-treated BV2 microglia. One factor that is known to be induced in microglia cells upon pro-inflammatory stimulation is NOS2 (Kempuraj et al. 2016). OCM-SA-treated BV2 microglia displayed an induced expression of NOS2 compared to control. Blocking of IL6 with anti-IL6 antibody only partly counteracted OCM-SA effects (Fig. 2g), and none of the used concentrations of IL6 protein (10, 30, and 50 ng/ml) was sufficient to show BV2 microglia responses with respect to the gene expression of both Nos2 and Arg1, indicating that other secreted molecules contribute to microglia activation in this scenario (supplementary figure F).
Oligodendrocytes are a Source of IL6 In Vivo
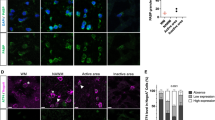
To investigate whether oligodendrocytes might be a relevant source of signaling molecules in vivo, we included short-term cuprizone-intoxicated mice (up to 4 days cuprizone) in the study. Since IL6 was highly induced in oligodendroglial cells in vitro, we focused on this molecule in this part of the study. Immunohistochemistry, qPCR, and in situ hybridization were performed on brain slices of cuprizone-intoxicated animals. After 2 days of cuprizone intoxication, IBA-1 immunohistochemistry revealed that microglia display an activated cell morphology in the CC (i.e., less ramified, swollen somata) compared to control mice (Fig. 3a). Similar morphological changes as well as an increase in microglia cell numbers in early cuprizone-induced lesions have been published previously by our group after a 2-day exposure to cuprizone (Clarner et al. 2015; Krauspe et al. 2015). Re-evaluation via qPCR revealed that IL6 gene expression was increased about fivefold in the CC after 2 days of cuprizone intoxication ((Krauspe et al. 2015), Fig. 3b, n ≥ 3). This induction further increased up to 17-fold after 4 days of cuprizone intoxication. In situ hybridization showed an increase of IL6 mRNA signals in the CC of cuprizone-intoxicated animals compared to controls (Fig. 3c, left panel). Double in situ hybridization (Fig. 3c, right panel) and validation by fluorescence double labeling of Olig2 and IL6 proteins (Fig. 3d) as well as double chromogen labeling of APC and IL6 proteins (Fig. 3e) revealed oligodendroglial cells as one source of IL6 at this early time point (see supplementary figure B for experimental overview). Although APC labels astrocytes and neurons in the brain as well, the double-positive cell in Fig. 3e resembles morphologically strongly an oligodendrocyte. Three cuprizone-fed animals were investigated and IL6/Olig2- and IL6/APC-positive cells were found in the CC in all three animals. Since confocal Z-stack analysis was necessary to undoubtedly identify double-positive cells, no quantification of the number of these cells could be performed. Note that oligodendrocytes are not the sole source of IL6 but also other glial cells such as astrocytes express IL6 after 2 days of cuprizone feeding (see supplementary figure D). Tezuka and colleagues also identified astrocytes as a source of IL6 in this model (Tezuka et al. 2013).
a IBA-1+ microglia cells display an activated morphology upon short-term cuprizone intoxication compared to control mice (scale bars 50 μm). b IL6 gene expression was increased in the CC after 2, 3, and 4 days of cuprizone intoxication compared to untreated animals (re-evaluation of cDNA of short-term cuprizone-fed mice; BoxCox-Y transformed). c At 2 days of 0.25% cuprizone intoxication, the signal for IL6 mRNA was increased compared to the control. In situ hybridization of OLIG2 and IL6 identified oligodendrocytes as a source of IL6 production after short-term cuprizone intoxication (scale bars for IL6 20 μm; for IL6/OLIG2 5 μm), which was further validated via d fluorescence double-labeling of OLIG2 and IL6 (scale bars for control 25 μm; for 2-day cup 10 μm). e Double labeling of IL6 and APC (scale bars 10 μm) (three animals per group, one experiment for figure a–e). ns, not significant; *p < 0.05; **p < 0.005; ***p < 0.001
Discussion
The contribution of stressed oligodendrocytes to the formation of inflammatory CNS lesions is only incompletely understood. Here, we induced oligodendrocyte stress by using the mitochondrial inhibitor SA in vitro and cuprizone intoxication in vivo. In vitro, this treatment caused a selective increase in the oligodendroglial expression of a variety of immune-modulatory factors such as IL6, GDF15, and a number of chemokines. The expression of IL6 in the in vivo situation was explored by qPCR analysis, ELISA, in situ hybridization, and immunohistological staining of tissue from short-term cuprizone-intoxicated mice. Fluorescence double-labeling and in situ hybridization of OLIG2/APC and IL6 identified oligodendrocytes as a possible source of this molecule in vivo (Ramesh et al. 2012). These results indicate that our in vitro data obtained from SA-treated oligodendroglial cells might be relevant for the in vivo situation. It has to be mentioned at this point, that oligodendrocytes are not the sole source of these molecules in vivo. With respect to IL6, astrocytes and microglia have been shown to be additional sources in cuprizone intoxication. We would like to point out that due to the high number of possibly involved molecules and the highly complex interplay of different glia cells in the formation of early inflammatory lesions, knock-out mice deficient for single molecules would be of limited use to further investigate the role of oligodendrocytes in this scenario.
Regarding the role of IL6 in CNS inflammation and regeneration in general, it has been shown that on one hand, it is neuroprotective via accelerating nerve regeneration following trauma or spinal cord injury (Hirota et al. 1996; Yang et al. 2012) and protects mice from demyelination in the cuprizone model by inducing a specific activation state in microglia (Petkovic et al. 2017). On the other hand, IL6 supports chronic inflammatory processes in disorders such as Alzheimer’s disease (Swardfager et al. 2010). In active demyelinating MS lesions, IL6 expression by astrocytes and macrophages is relevant for the preservation of oligodendrocytes (Schonrock et al. 2000). IL6 signaling and mitochondrial functions are closely linked, since IL6 protects cells against a loss of mitochondrial complex IV after bacterial infection in mice (Maiti et al. 2015). In the liver, IL6 is necessary for the repair of mitochondrial mutations caused by ethanol intoxication (Zhang et al. 2010). The physiological IL6 concentration in blood plasma samples from healthy humans is about 1–1.5 pg/ml, whereas the concentration in inflammatory conditions is highly increased and might reach up to 1,000 pg/ml in sever inflammation (Damas et al. 1992; Seino et al. 1994; Ridker et al. 2000). This is comparable to the concentrations we measured in the medium of stressed oligodendroglial cells (app. 800 pg/ml). Regarding the IL6 concentrations within the brain, both in healthy and inflamed tissue, only little is known. Therefore, further studies will have to show the local IL6 concentrations and precise source within brain tissue in distinct pathologies. With respect to microglia cells, it has been shown that a concentration of 100 ng/ml does activate Nos2 mRNA expression in BV2 microglia (Matsumoto et al. 2018). Furthermore, IL6 (50 ng/ml) increases the mitochondrial Ca2+ levels in a STAT3-dependent manner in CD4+ T cells (Yang et al. 2015). By using Luciferin-expressing mice, we recently demonstrated that cuprizone intoxication causes an early induction of Nrf2-ARE signaling within the brain (Draheim et al. 2016). Since Nrf2 has been shown to regulate IL6 expression (Wruck et al. 2011), the increase in oxidative stress and subsequent Nrf2-activation might be a possible mechanism by which IL6 expression is initially triggered in oligodendrocytes in this model. Further studies including Nrf2-deficient mice will have to show the link between oxidative stress and Nrf2 activity on one hand and the expression of IL6 in oligodendrocytes on the other hand. Given this data, we consider this early IL6 expression by oligodendrocytes during preactive lesion formation as a potential first “call for help” by metabolically dysfunctional or oxidatively challenged oligodendrocytes.
Another member of the IL6 cytokine family that was induced upon SA-stimulation in this study is LIF, which signals through the JAK/STAT pathway as well. It is required for the induction of inflammatory responses of microglia and astrocytes to brain damage (Holmberg and Patterson 2006). Furthermore, it has been shown that LIF plays a role in the initial infiltration of inflammatory cells into the CNS and in the neuronal response to brain injury (Sugiura et al. 2000). Another highly induced molecule in our study was GDF15, also known as macrophage inhibitory cytokine-1. Bonaterra et al. showed that GDF15 is functionally linked to IL6 signaling since it regulates inflammatory IL6-dependent processes in vascular injury (Bonaterra et al. 2012). A study on the tumorigenesis of prostate carcinoma indicated that the expression of GDF15 is upregulated by IL6 (Tsui et al. 2012). Despite these links to IL6 signaling, GDF15 plays a major role in regulating inflammatory pathways in injured tissues and is involved in pathological processes such as cancer, cardiovascular disorders, ischemia, and atherosclerosis (Schlittenhardt et al. 2005; Kempf et al. 2006; Jiang et al. 2016).
A number of chemokines were induced in SA-treated OLN93 cells that are known to be involved in microglia activation, including CXCL1 and CCL5 (Škuljec et al. 2011). The cytokine-like glycoprotein SPP1, also known as osteopontin, was induced about fourfold in stressed oligodendroglial cells. Osteopontin has been implicated in the pathogenesis of several autoimmune diseases such as rheumatoid arthritis, autoimmune hepatitis, and MS (Chabas et al. 2001; Yumoto et al. 2002; Mochida et al. 2004). Furthermore, osteopontin activity is found in MS lesions (Chen et al. 2009). It has been shown that the treatment of mixed cortical cultures with osteopontin leads to a stimulation of myelin basic protein expression and the formation of myelin sheaths indicating a putative role in remyelination and recovery (Selvaraju et al. 2004). CSF1, a macrophage colony-stimulating factor, was induced 2.5-fold in SA-stressed oligodendroglial cells and is involved in the proliferation, differentiation, and chemotactic activity of monocytes and macrophages. Interestingly, the survival of adult murine microglia cells seems to be fully dependent upon CSF1 receptor signaling, since all microglia cells can be eliminated from the CNS through CSF1R inhibitor administration (Elmore et al. 2014).
All of the abovementioned signaling molecules might account—either alone or in combination—for the effects, we observed in OCM-treated microglia cells. Our data indicate that not a sole oligodendroglial cell-derived molecule but rather a combination of different factors account for the activation of microglia cells.
In summary, our data supports the view that stressed oligodendrocytes have the potential to activate microglia cells through a specific cocktail of chemokines and cytokines such as IL6. Further studies will have to identify the temporal activation pattern of these signaling molecules, their cellular sources and their impact on neuroinflammation.
References
Balabanov R, Strand K, Goswami R, McMahon E, Begolka W, Miller SD, Popko B (2007) Interferon-gamma-oligodendrocyte interactions in the regulation of experimental autoimmune encephalomyelitis. J Neurosci 27(8):2013–2024
Barnett MH, Prineas JW (2004) Relapsing and remitting multiple sclerosis: pathology of the newly forming lesion. Ann Neurol 55(4):458–468
Bennett MC, Mlady GW, Kwon YH, Rose GM (1996) Chronic in vivo sodium azide infusion induces selective and stable inhibition of cytochrome c oxidase. J Neurochem 66(6):2606–2611
Blasi E, Barluzzi R, Bocchini V, Mazzolla R, Bistoni F (1990) Immortalization of murine microglial cells by a v-raf/v-myc carrying retrovirus. J Neuroimmunol 27(2-3):229–237
Bonaterra GA, Zugel S, Thogersen J, Walter SA, Haberkorn U, Strelau J, Kinscherf R (2012) Growth differentiation factor-15 deficiency inhibits atherosclerosis progression by regulating interleukin-6-dependent inflammatory response to vascular injury. J Am Heart Assoc 1(6):e002550
Cannella B, Raine CS (2004) Multiple sclerosis: cytokine receptors on oligodendrocytes predict innate regulation. Ann Neurol 55(1):46–57
Chabas D, Baranzini SE, Mitchell D, Bernard CC, Rittling SR, Denhardt DT, Sobel RA, Lock C, Karpuj M, Pedotti R, Heller R, Oksenberg JR, Steinman L (2001) The influence of the proinflammatory cytokine, osteopontin, on autoimmune demyelinating disease. Science 294(5547):1731–1735
Chen M, Chen G, Nie H, Zhang X, Niu X, Zang YC, Skinner SM, Zhang JZ, Killian JM, Hong J (2009) Regulatory effects of IFN-beta on production of osteopontin and IL-17 by CD4+ T Cells in MS. Eur J Immunol 39(9):2525–2536
Clarner T, Diederichs F, Berger K, Denecke B, Gan L, van der Valk P, Beyer C, Amor S, Kipp M (2012) Myelin debris regulates inflammatory responses in an experimental demyelination animal model and multiple sclerosis lesions. Glia 60(10):1468–1480
Clarner T, Janssen K, Nellessen L, Stangel M, Skripuletz T, Krauspe B, Hess FM, Denecke B, Beutner C, Linnartz-Gerlach B, Neumann H, Vallieres L, Amor S, Ohl K, Tenbrock K, Beyer C, Kipp M (2015) CXCL10 triggers early microglial activation in the cuprizone model. J Immunol 194(7):3400–3413
Damas P, Ledoux D, Nys M, Vrindts Y, De Groote D, Franchimont P, Lamy M (1992) Cytokine serum level during severe sepsis in human IL-6 as a marker of severity. Ann Surg 215(4):356–362
De Groot CJ, Bergers E, Kamphorst W, Ravid R, Polman CH, Barkhof F, van der Valk P (2001) Post-mortem MRI-guided sampling of multiple sclerosis brain lesions: increased yield of active demyelinating and (p)reactive lesions. Brain 124(Pt 8):1635–1645
Draheim T, Liessem A, Scheld M, Wilms F, Weissflog M, Denecke B, Kensler TW, Zendedel A, Beyer C, Kipp M, Wruck CJ, Fragoulis A, Clarner T (2016) Activation of the astrocytic Nrf2/ARE system ameliorates the formation of demyelinating lesions in a multiple sclerosis animal model. Glia 64(12):2219–2230
Edagawa M, Kawauchi J, Hirata M, Goshima H, Inoue M, Okamoto T, Murakami A, Maehara Y, Kitajima S (2014) Role of activating transcription factor 3 (ATF3) in endoplasmic reticulum (ER) stress-induced sensitization of p53-deficient human colon cancer cells to tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL)-mediated apoptosis through up-regulation of death receptor 5 (DR5) by zerumbone and celecoxib. J Biol Chem 289(31):21544–21561
Elmore MR, Najafi AR, Koike MA, Dagher NN, Spangenberg EE, Rice RA, Kitazawa M, Matusow B, Nguyen H, West BL, Green KN (2014) Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron 82(2):380–397
Gay FW, Drye TJ, Dick GW, Esiri MM (1997) The application of multifactorial cluster analysis in the staging of plaques in early multiple sclerosis. Identification and characterization of the primary demyelinating lesion. Brain 120(Pt 8):1461–1483
Gehrmann J, Matsumoto Y, Kreutzberg GW (1995) Microglia: intrinsic immuneffector cell of the brain. Brain Res Brain Res Rev 20(3):269–287
Hirota H, Kiyama H, Kishimoto T, Taga T (1996) Accelerated nerve regeneration in mice by upregulated expression of interleukin (IL) 6 and IL-6 receptor after trauma. J Exp Med 183(6):2627–2634
Hollensworth SB, Shen C, Sim JE, Spitz DR, Wilson GL, LeDoux SP (2000) Glial cell type-specific responses to menadione-induced oxidative stress. Free Radic Biol Med 28(8):1161–1174
Holmberg KH, Patterson PH (2006) Leukemia inhibitory factor is a key regulator of astrocytic, microglial and neuronal responses in a low-dose pilocarpine injury model. Brain Res 1075(1):26–35
Jiang J, Wen W, Sachdev PS (2016) Macrophage inhibitory cytokine-1/growth differentiation factor 15 as a marker of cognitive ageing and dementia. Curr Opin Psychiatry 29(2):181–186
Kempf T, Eden M, Strelau J, Naguib M, Willenbockel C, Tongers J, Heineke J, Kotlarz D, Xu J, Molkentin JD, Niessen HW, Drexler H, Wollert KC (2006) The transforming growth factor-beta superfamily member growth-differentiation factor-15 protects the heart from ischemia/reperfusion injury. Circ Res 98(3):351–360
Kempuraj D, Thangavel R, Natteru PA, Selvakumar GP, Saeed D, Zahoor H, Zaheer S, Iyer SS, Zaheer A (2016) Neuroinflammation induces neurodegeneration. J Neurol Neurosurg Spine 1(1)
Krauspe BM, Dreher W, Beyer C, Baumgartner W, Denecke B, Janssen K, Langhans CD, Clarner T, Kipp M (2015) Short-term cuprizone feeding verifies N-acetylaspartate quantification as a marker of neurodegeneration. J Mol Neurosci 55(3):733–748
Kummer JA, Broekhuizen R, Everett H, Agostini L, Kuijk L, Martinon F, van Bruggen R, Tschopp J (2007) Inflammasome components NALP 1 and 3 show distinct but separate expression profiles in human tissues suggesting a site-specific role in the inflammatory response. J Histochem Cytochem 55(5):443–452
Linnane AW, Marzuki S, Ozawa T, Tanaka M (1989) Mitochondrial DNA mutations as an important contributor to ageing and degenerative diseases. Lancet 1(8639):642–645
Mahad D, Lassmann H, Turnbull D (2008) Review: mitochondria and disease progression in multiple sclerosis. Neuropathol Appl Neurobiol 34(6):577–589
Maiti AK, Sharba S, Navabi N, Forsman H, Fernandez HR, Lindén SK (2015) IL-4 protects the mitochondria against TNFα and IFNγ induced insult during clearance of infection with citrobacter rodentium and Escherichia coli. Scientific Reports 5:15434
Mao P, Reddy PH (2010) Is multiple sclerosis a mitochondrial disease? Biochimica et biophysica acta 1802(1):66–79
Marik C, Felts PA, Bauer J, Lassmann H, Smith KJ (2007) Lesion genesis in a subset of patients with multiple sclerosis: a role for innate immunity? Brain 130(Pt 11):2800–2815
Matsumoto J, Dohgu S, Takata F, Machida T, Bolukbasi Hatip FF, Hatip-Al-Khatib I, Yamauchi A, Kataoka Y (2018) TNF-alpha-sensitive brain pericytes activate microglia by releasing IL-6 through cooperation between IkappaB-NFkappaB and JAK-STAT3 pathways. Brain Res 1692:34–44
Merabova N, Kaminski R, Krynska B, Amini S, Khalili K, Darbinyan A (2012) JCV agnoprotein-induced reduction in CXCL5/LIX secretion by oligodendrocytes is associated with activation of apoptotic signaling in neurons. J Cell Physiol 227(8):3119–3127
Mochida S, Yoshimoto T, Mimura S, Inao M, Matsui A, Ohno A, Koh H, Saitoh E, Nagoshi S, Fujiwara K (2004) Transgenic mice expressing osteopontin in hepatocytes as a model of autoimmune hepatitis. Biochem Biophys Res Commun 317(1):114–120
Moyon S, Dubessy AL, Aigrot MS, Trotter M, Huang JK, Dauphinot L, Potier MC, Kerninon C, Melik Parsadaniantz S, Franklin RJ, Lubetzki C (2015) Demyelination causes adult CNS progenitors to revert to an immature state and express immune cues that support their migration. J Neurosci 35(1):4–20
Okamura RM, Lebkowski J, Au M, Priest CA, Denham J, Majumdar AS (2007) Immunological properties of human embryonic stem cell-derived oligodendrocyte progenitor cells. J Neuroimmunol 192(1-2):134–144
Perry VH, Andersson PB, Gordon S (1993) Macrophages and inflammation in the central nervous system. Trends Neurosci 16(7):268–273
Petkovic F, Campbell IL, Gonzalez B, Castellano B (2017) Reduced cuprizone-induced cerebellar demyelination in mice with astrocyte-targeted production of IL-6 is associated with chronically activated, but less responsive microglia. J Neuroimmunol 310:97–102
Puthalakath H, O'Reilly LA, Gunn P, Lee L, Kelly PN, Huntington ND, Hughes PD, Michalak EM, McKimm-Breschkin J, Motoyama N, Gotoh T, Akira S, Bouillet P, Strasser A (2007) ER stress triggers apoptosis by activating BH3-only protein Bim. Cell 129(7):1337–1349
Ramesh G, Benge S, Pahar B, Philipp MT (2012) A possible role for inflammation in mediating apoptosis of oligodendrocytes as induced by the Lyme disease spirochete Borrelia burgdorferi. J Neuroinflammation 9:72
Richter-Landsberg C, Heinrich M (1996) OLN-93: a new permanent oligodendroglia cell line derived from primary rat brain glial cultures. J Neurosci Res 45(2):161–173
Ridker PM, Rifai N, Stampfer MJ, Hennekens CH (2000) Plasma concentration of interleukin-6 and the risk of future myocardial infarction among apparently healthy men. Circulation 101(15):1767–1772
Ruther BJ, Scheld M, Dreymueller D, Clarner T, Kress E, Brandenburg LO, Swartenbroekx T, Hoornaert C, Ponsaerts P, Fallier-Becker P, Beyer C, Rohr SO, Schmitz C, Chrzanowski U, Hochstrasser T, Nyamoya S, Kipp M (2017) Combination of cuprizone and experimental autoimmune encephalomyelitis to study inflammatory brain lesion formation and progression. Glia 65(12):1900–1913
Rutkowski DT, Arnold SM, Miller CN, Wu J, Li J, Gunnison KM, Mori K, Sadighi Akha AA, Raden D, Kaufman RJ (2006) Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol 4(11):e374
Scheld M, Ruther BJ, Grosse-Veldmann R, Ohl K, Tenbrock K, Dreymuller D, Fallier-Becker P, Zendedel A, Beyer C, Clarner T, Kipp M (2016) Neurodegeneration triggers peripheral immune cell recruitment into the forebrain. J Neurosci 36(4):1410–1415
Schlittenhardt D, Schmiedt W, Bonaterra GA, Metz J, Kinscherf R (2005) Colocalization of oxidized low-density lipoprotein, caspase-3, cyclooxygenase-2, and macrophage migration inhibitory factor in arteriosclerotic human carotid arteries. Cell Tissue Res 322(3):425–435
Schonrock LM, Gawlowski G, Bruck W (2000) Interleukin-6 expression in human multiple sclerosis lesions. Neurosci Lett 294(1):45–48
Seino Y, Ikeda U, Ikeda M, Yamamoto K, Misawa Y, Hasegawa T, Kano S, Shimada K (1994) Interleukin 6 gene transcripts are expressed in human atherosclerotic lesions. Cytokine 6(1):87–91
Selvaraju R, Bernasconi L, Losberger C, Graber P, Kadi L, Avellana-Adalid V, Picard-Riera N, Baron Van Evercooren A, Cirillo R, Kosco-Vilbois M, Feger G, Papoian R, Boschert U (2004) Osteopontin is upregulated during in vivo demyelination and remyelination and enhances myelin formation in vitro. Mol Cell Neurosci 25(4):707–721
Škuljec J, Sun H, Pul R, Bénardais K, Ragancokova D, Moharregh-Khiabani D, Kotsiari A, Trebst C, Stangel M (2011) CCL5 induces a pro-inflammatory profile in microglia in vitro. Cellular Immunology 270(2):164–171
Su K, Bourdette D, Forte M (2013) Mitochondrial dysfunction and neurodegeneration in multiple sclerosis. Front Physiol 4:169
Sugiura S, Lahav R, Han J, Kou SY, Banner LR, de Pablo F, Patterson PH (2000) Leukaemia inhibitory factor is required for normal inflammatory responses to injury in the peripheral and central nervous systems in vivo and is chemotactic for macrophages in vitro. Eur J Neurosci 12(2):457–466
Swardfager W, Lanctot K, Rothenburg L, Wong A, Cappell J, Herrmann N (2010) A meta-analysis of cytokines in Alzheimer's disease. Biol Psychiatry 68(10):930–941
Teske N, Liessem A, Fischbach F, Clarner T, Beyer C, Wruck C, Fragoulis A, Tauber SC, Victor M, Kipp M (2018) Chemical hypoxia-induced integrated stress response activation in oligodendrocytes is mediated by the transcription factor nuclear factor (erythroid-derived 2)-like 2 (NRF2). J Neurochem 144(3):285–301
Tezuka T, Tamura M, Kondo MA, Sakaue M, Okada K, Takemoto K, Fukunari A, Miwa K, Ohzeki H, Kano S, Yasumatsu H, Sawa A, Kajii Y (2013) Cuprizone short-term exposure: astrocytic IL-6 activation and behavioral changes relevant to psychosis. Neurobiol Dis 59:63–68
Tsui KH, Chang YL, Feng TH, Chung LC, Lee TY, Chang PL, Juang HH (2012) Growth differentiation factor-15 upregulates interleukin-6 to promote tumorigenesis of prostate carcinoma PC-3 cells. J Mol Endocrinol 49(2):153–163
Tzartos JS, Friese MA, Craner MJ, Palace J, Newcombe J, Esiri MM, Fugger L (2008) Interleukin-17 production in central nervous system-infiltrating T cells and glial cells is associated with active disease in multiple sclerosis. Am J Pathol 172(1):146–155
van der Valk P, Amor S (2009) Preactive lesions in multiple sclerosis. Curr Opin Neurol 22(3):207–213
Wang CH, Wu SB, Wu YT, Wei YH (2013) Oxidative stress response elicited by mitochondrial dysfunction: implication in the pathophysiology of aging. Exp Biol Med (Maywood) 238(5):450–460
Wruck CJ, Streetz K, Pavic G, Gotz ME, Tohidnezhad M, Brandenburg LO, Varoga D, Eickelberg O, Herdegen T, Trautwein C, Cha K, Kan YW, Pufe T (2011) Nrf2 induces interleukin-6 (IL-6) expression via an antioxidant response element within the IL-6 promoter. J Biol Chem 286(6):4493–4499
Wuerfel J, Bellmann-Strobl J, Brunecker P, Aktas O, McFarland H, Villringer A, Zipp F (2004) Changes in cerebral perfusion precede plaque formation in multiple sclerosis: a longitudinal perfusion MRI study. Brain 127(Pt 1):111–119
Yang P, Wen H, Ou S, Cui J, Fan D (2012) IL-6 promotes regeneration and functional recovery after cortical spinal tract injury by reactivating intrinsic growth program of neurons and enhancing synapse formation. Exp Neurol 236(1):19–27
Yang R, Lirussi D, Thornton TM, Jelley-Gibbs DM, Diehl SA, Case LK, Madesh M, Taatjes DJ, Teuscher C, Haynes L, Rincon M (2015) Mitochondrial Ca(2)(+) and membrane potential, an alternative pathway for Interleukin 6 to regulate CD4 cell effector function. Elife 4
Yumoto K, Ishijima M, Rittling SR, Tsuji K, Tsuchiya Y, Kon S, Nifuji A, Uede T, Denhardt DT, Noda M (2002) Osteopontin deficiency protects joints against destruction in anti-type II collagen antibody-induced arthritis in mice. Proc Natl Acad Sci U S A 99(7):4556–4561
Zeis T, Probst A, Steck AJ, Stadelmann C, Bruck W, Schaeren-Wiemers N (2009) Molecular changes in white matter adjacent to an active demyelinating lesion in early multiple sclerosis. Brain Pathol 19(3):459–466
Zhang X, Tachibana S, Wang H, Hisada M, Williams GM, Gao B, Sun Z (2010) Interleukin-6 is an important mediator for mitochondrial DNA repair after alcoholic liver injury in mice. Hepatology 52(6):2137–2147
Acknowledgements
The excellent support by Helga Helten, Petra Ibold and Uta Zahn is appreciated. We thank Prof. Dr. Reinhard Windoffer and Dr. Volker Buck for their assistance with confocal microscopy and PD Dr. Claudia Krusche for technical support.
Funding
Grant sponsor: This study was funded by the START program of the medical faculty of the RWTH Aachen University (M.S.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(A) Schematic illustration of the in vitro experimental setup for oligodendroglial cell stimulation and subsequent microglia stimulation with OCM. Oligodendroglial cells (OLN93) were stressed with 10 mM SA for 24 h, medium changed and incubated for 24 h with or without SA to produce OCM-vehicle and OCM-SA, respectively. OCM was then used to stimulate microglia cells for 6 h. (B) Time-line diagram of the 2-d cuprizone feeding in vivo experiments. (C) Signal specificity was validated by incubating slices with the respective secondary antibody without pre-incubation with the first antibody. Results for IL6 and OLIG2 negative controls are shown. Note that no signal was found within the examined tissue regions for any secondary antibody (scale bars 50 μm). Cross reactivity of secondary antibodies with each other or the false primary antibody was additionally excluded (data not shown). (D) Double labeling of IL6 with GFAP and IBA-1 (scale bars 10 μm) shows that also other glial cells than oligodendrocytes are possible sources of IL6 after 2 d cuprizone intoxication. No signal was found in the wildtype negative control (lower row) (scale bars 20 μm). (E) No morphological changes such as an increase in the number of bipolar cells or an increased arborization were found in OCM-SA-stimulated compared to OCM-Veh-stimulated BV2 microglia cells. (F) Shows the gene expression results of arginase 1 and Nos2 (BoxCox-Y transformed) of IL6-stimulated microglia cells (four culture wells, one experiment). LPS stimulation served as a positive control for Nos2 induction. *p < 0.05, vehicle, up H2O (PDF 1494 kb)
Rights and permissions
About this article
Cite this article
Scheld, M., Fragoulis, A., Nyamoya, S. et al. Mitochondrial Impairment in Oligodendroglial Cells Induces Cytokine Expression and Signaling. J Mol Neurosci 67, 265–275 (2019). https://doi.org/10.1007/s12031-018-1236-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12031-018-1236-6